The Perspectives of Platelet Proteomics in Health and Disease
Abstract
1. Introduction
2. The Principal Role of Platelet: Hemostasis and Thrombosis
3. Activation of Platelets
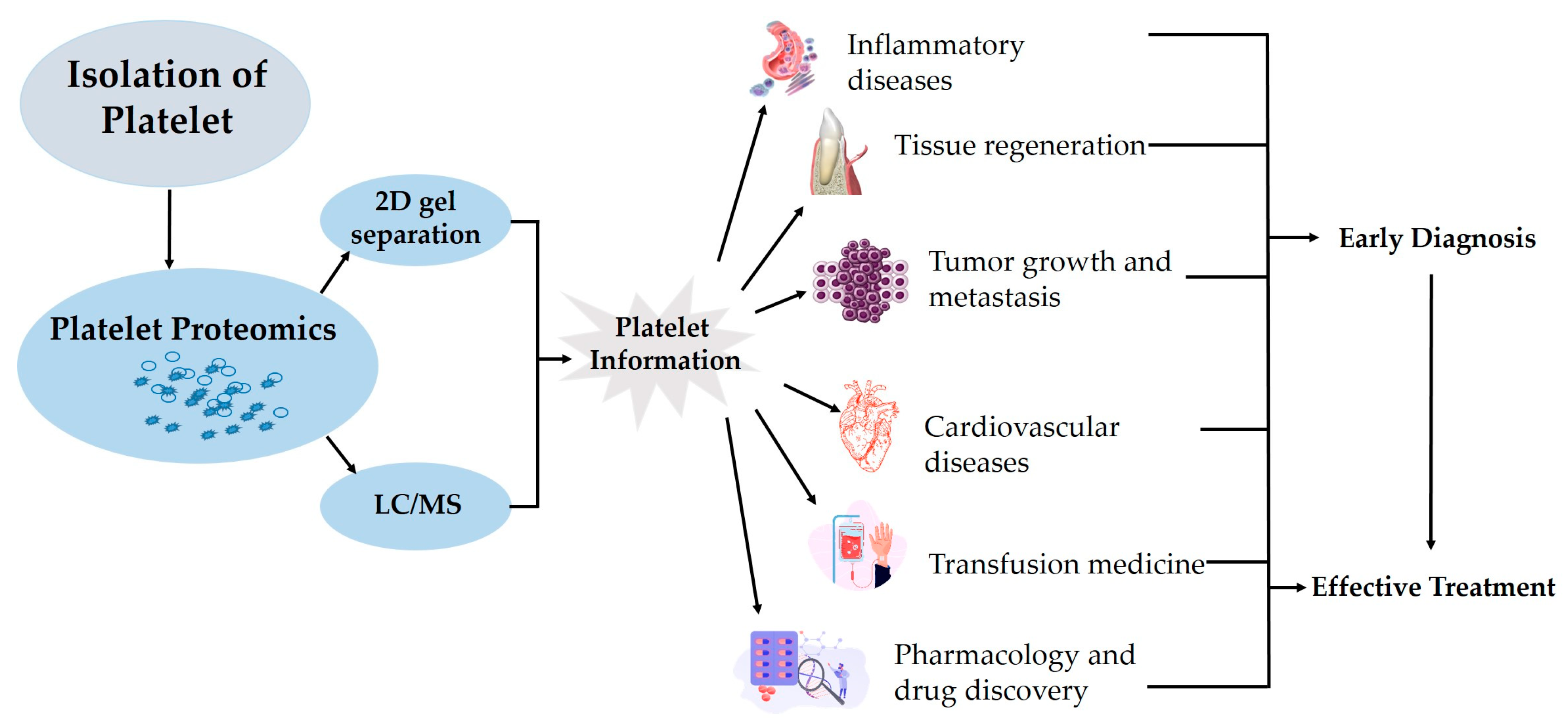
4. Platelet Proteomics
5. Methods of Platelet Proteomic Analysis
6. Platelet Proteome in Health and Diseases
7. Platelet Proteomics in Transfusion Medicine
8. Hurdles in Accurate Platelet Proteome Research
9. Is Clinical Translation between Mouse Platelet Proteome to Human Clinical Studies Possible?
10. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Chaudhary, P.K.; Kim, S.; Kim, S. An insight into recent advances on platelet function in health and disease. Int. J. Mol. Sci. 2022, 23, 6022. [Google Scholar] [CrossRef]
- Villa-Fajardo, M.; Palma, M.C.Y.; Acebes-Huerta, A.; Martínez-Botía, P.; Meinders, M.; Nolte, M.A.; Cuesta, C.B.; Eble, J.A.; Del Castillo, J.G.; Martín-Sánchez, F.J. Platelet number and function alterations in preclinical models of sterile inflammation and sepsis patients: Implications in the pathophysiology and treatment of inflammation. Transfus. Apher. Sci. 2022, 61, 103413. [Google Scholar] [CrossRef] [PubMed]
- Gianazza, E.; Brioschi, M.; Baetta, R.; Mallia, A.; Banfi, C.; Tremoli, E. Platelets in healthy and disease states: From biomarkers discovery to drug targets identification by proteomics. Int. J. Mol. Sci. 2020, 21, 4541. [Google Scholar] [CrossRef]
- Antunes-Ferreira, M.; Koppers-Lalic, D.; Würdinger, T. Circulating platelets as liquid biopsy sources for cancer detection. Mol. Oncol. 2021, 15, 1727–1743. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, J. Origin of blood platelets. Nature 1955, 176, 38. [Google Scholar] [CrossRef] [PubMed]
- Pagel, O.; Walter, E.; Jurk, K.; Zahedi, R.P. Taking the stock of granule cargo: Platelet releasate proteomics. Platelets 2017, 28, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, H.; Van Der Sluijs, P. Platelet secretory behaviour: As diverse as the granules… or not? J. Thromb. 2015, 13, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Thon, J.N.; Peters, C.G.; Machlus, K.R.; Aslam, R.; Rowley, J.; Macleod, H.; Devine, M.T.; Fuchs, T.A.; Weyrich, A.S.; Semple, J.W. T granules in human platelets function in TLR9 organization and signaling. J. Cell Biol. 2012, 198, 561–574. [Google Scholar] [CrossRef]
- Michelson, A.D.; Cattaneo, M.; Frelinger, A.; Newman, P. Platelets; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Ghoshal, K.; Bhattacharyya, M. Overview of platelet physiology: Its hemostatic and nonhemostatic role in disease pathogenesis. Sci. World 2014, 20, 14. [Google Scholar] [CrossRef]
- Nuyttens, B.P.; Thijs, T.; Deckmyn, H.; Broos, K. Platelet adhesion to collagen. Thromb. J. 2011, 127, S26–S29. [Google Scholar] [CrossRef]
- Schmaier, A.A.; Zou, Z.; Kazlauskas, A.; Emert-Sedlak, L.; Fong, K.P.; Neeves, K.B.; Maloney, S.F.; Diamond, S.L.; Kunapuli, S.P.; Ware, J. Molecular priming of Lyn by GPVI enables an immune receptor to adopt a hemostatic role. Proc. Natl. Acad. Sci. USA 2009, 106, 21167–21172. [Google Scholar] [CrossRef]
- Heemskerk, J.W.; Harper, M.T.; Cosemans, J.M.; Poole, A.W. Unravelling the different functions of protein kinase C isoforms in platelets. FEBS Lett. 2011, 585, 1711–1716. [Google Scholar] [CrossRef]
- Crittenden, J.R.; Bergmeier, W.; Zhang, Y.; Piffath, C.L.; Liang, Y.; Wagner, D.D.; Housman, D.E.; Graybiel, A.M. CalDAG-GEFI integrates signaling for platelet aggregation and thrombus formation. Nat. Med. 2004, 10, 982–986. [Google Scholar] [CrossRef]
- Durrant, T.N.; van den Bosch, M.T.; Hers, I. Integrin αIIbβ3 outside-in signaling. Blood Am. Soc. Hematol. 2017, 130, 1607–1619. [Google Scholar] [CrossRef]
- Sang, Y.; Roest, M.; de Laat, B.; de Groot, P.G.; Huskens, D. Interplay between platelets and coagulation. Blood Rev. 2021, 46, 100733. [Google Scholar] [CrossRef] [PubMed]
- Koupenova, M.; Clancy, L.; Corkrey, H.A.; Freedman, J.E. Circulating platelets as mediators of immunity, inflammation, and thrombosis. Circ. Res. 2018, 122, 337–351. [Google Scholar] [CrossRef]
- Huang, J.; Swieringa, F.; Solari, F.A.; Provenzale, I.; Grassi, L.; De Simone, I.; Baaten, C.C.; Cavill, R.; Sickmann, A.; Frontini, M. Assessment of a complete and classified platelet proteome from genome-wide transcripts of human platelets and megakaryocytes covering platelet functions. Sci. Rep. 2021, 11, 12358. [Google Scholar] [CrossRef]
- Dowal, L.; Yang, W.; Freeman, M.R.; Steen, H.; Flaumenhaft, R. Proteomic analysis of palmitoylated platelet proteins. Blood Am. J. Hematol. 2011, 118, e62–e73. [Google Scholar] [CrossRef]
- Lee, H.; Chae, S.; Park, J.; Bae, J.; Go, E.-B.; Kim, S.-J.; Kim, H.; Hwang, D.; Lee, S.-W.; Lee, S.-Y. Comprehensive proteome profiling of platelet identified a protein profile predictive of responses to an antiplatelet agent sarpogrelate. Mol. Cell Proteom. 2016, 15, 3461–3472. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, J.M.; Vaudel, M.; Gambaryan, S.; Radau, S.; Walter, U.; Martens, L.; Geiger, J.; Sickmann, A.; Zahedi, R.P. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood Am. Soc. Hematol. 2012, 120, e73–e82. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, J.M.; Gambaryan, S.; Watson, S.P.; Jurk, K.; Walter, U.; Sickmann, A.; Heemskerk, J.W.; Zahedi, R.P. What can proteomics tell us about platelets? Circ. Res. 2014, 114, 1204–1219. [Google Scholar] [CrossRef]
- Thon, J.N.; Schubert, P.; Devine, D.V. Platelet storage lesion: A new understanding from a proteomic perspective. Transfus. Med. Rev. 2008, 22, 268–279. [Google Scholar] [CrossRef]
- Handtke, S.; Steil, L.; Palankar, R.; Conrad, J.; Cauhan, S.; Kraus, L.; Ferrara, M.; Dhople, V.; Wesche, J.; Völker, U. Role of platelet size revisited—Function and protein composition of large and small platelets. Thromb. Haemost. 2019, 119, 407–420. [Google Scholar] [CrossRef]
- Kim, S.; Chaudhary, P.K.; Kim, S. Role of Prednisolone in Platelet Activation by Inhibiting TxA(2) Generation through the Regulation of cPLA(2) Phosphorylation. Animals 2023, 13, 1299. [Google Scholar] [CrossRef]
- Patrono, C.; Rocca, B. Aspirin, 110 years later. J. Thromb. Haemost. 2009, 7, 258–261. [Google Scholar] [CrossRef]
- Egidi, M.G.; D’Alessandro, A.; Mandarello, G.; Zolla, L. Troubleshooting in platelet storage temperature and new perspectives through proteomics. Blood Transfus. 2010, 8, 73–81. [Google Scholar]
- Canas, B.; Pineiro, C.; Calvo, E.; Lopez-Ferrer, D.; Gallardo, J.M. Trends in sample preparation for classical and second generation proteomics. J. Chromatogr. A 2007, 1153, 235–258. [Google Scholar] [CrossRef]
- Barber, A.J.; Pepper, D.S.; Jamieson, G.A. A comparison of methods for platelet lysis and the isolation of platelet membranes. Thromb. Diath. Haemorrh. 1971, 26, 38–57. [Google Scholar] [CrossRef] [PubMed]
- Zufferey, A.; Fontana, P.; Reny, J.L.; Nolli, S.; Sanchez, J.C. Platelet proteomics. Mass. Spectrom. Rev. 2012, 31, 331–351. [Google Scholar] [CrossRef] [PubMed]
- Gorg, A.; Obermaier, C.; Boguth, G.; Harder, A.; Scheibe, B.; Wildgruber, R.; Weiss, W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 2000, 21, 1037–1053. [Google Scholar] [CrossRef]
- Aebersold, R.; Goodlett, D.R. Mass spectrometry in proteomics. Nature 2001, 101, 269–295. [Google Scholar] [CrossRef]
- Burkhart, J.M.; Schumbrutzki, C.; Wortelkamp, S.; Sickmann, A.; Zahedi, R.P. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J. Proteom. 2012, 75, 1454–1462. [Google Scholar] [CrossRef]
- Shevchuk, O.; Begonja, A.J.; Gambaryan, S.; Totzeck, M.; Rassaf, T.; Huber, T.B.; Greinacher, A.; Renne, T.; Sickmann, A. Proteomics: A Tool to Study Platelet Function. Int. J. Mol. Sci. 2021, 22, 4776. [Google Scholar] [CrossRef]
- Hunt, D.F.; Yates, J.R.; Shabanowitz, J.; Winston, S.; Hauer, C.R. Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 1986, 83, 6233–6237. [Google Scholar] [CrossRef] [PubMed]
- Pratt, J.M.; Robertson, D.H.; Gaskell, S.J.; Riba-Garcia, I.; Hubbard, S.J.; Sidhu, K.; Oliver, S.G.; Butler, P.; Hayes, A.; Petty, J.; et al. Stable isotope labelling in vivo as an aid to protein identification in peptide mass fingerprinting. Proteomics 2002, 2, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Mnatsakanyan, R.; Shema, G.; Basik, M.; Batist, G.; Borchers, C.H.; Sickmann, A.; Zahedi, R.P. Detecting post-translational modification signatures as potential biomarkers in clinical mass spectrometry. Expert. Rev. Proteom. 2018, 15, 515–535. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hasan, M.A.; Chen, J.Y. Pathway and network analysis in proteomics. J. Theor. Biol. 2014, 362, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, M.; Birschmann, I.; Mietner, S.; Sickmann, A.; Walter, U.; Dandekar, T. Platelet protein interactions: Map, signaling components, and phosphorylation groundstate. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1326–1331. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, M.; Moser, M.; Mann, M. Copy number analysis of the murine platelet proteome spanning the complete abundance range. Mol. Cell Proteom. 2014, 13, 3435–3445. [Google Scholar] [CrossRef]
- Winkler, W.; Zellner, M.; Diestinger, M.; Babeluk, R.; Marchetti, M.; Goll, A.; Zehetmayer, S.; Bauer, P.; Rappold, E.; Miller, I. Biological variation of the platelet proteome in the elderly population and its implication for biomarker research. Mol. Cell Proteom. 2008, 7, 193–203. [Google Scholar] [CrossRef]
- Maguire, P.B.; Parsons, M.E.; Szklanna, P.B.; Zdanyte, M.; Münzer, P.; Chatterjee, M.; Wynne, K.; Rath, D.; Comer, S.P.; Hayden, M. Comparative platelet releasate proteomic profiling of acute coronary syndrome versus stable coronary artery disease. Front. Cardiovasc. Med. 2020, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, C.; Joshi, A.; Mayr, M. Platelet “-omics” in health and cardiovascular disease. Atherosclerosis 2020, 307, 87–96. [Google Scholar] [CrossRef]
- Fernandez Parguina, A.; Grigorian-Shamajian, L.; Agra, R.M.; Teijeira-Fernández, E.; Rosa, I.; Alonso, J.; Viñuela-Roldán, J.E.; Seoane, A.; González-Juanatey, J.R.; Garcia, A. Proteins involved in platelet signaling are differentially regulated in acute coronary syndrome: A proteomic study. PLoS ONE 2010, 5, e13404. [Google Scholar] [CrossRef] [PubMed]
- López-Farré, A.J.; Zamorano-Leon, J.J.; Azcona, L.; Modrego, J.; Mateos-Cáceres, P.J.; González-Armengol, J.; Villarroel, P.; Moreno-Herrero, R.; Rodríguez-Sierra, P.; Segura, A. Proteomic changes related to “bewildered” circulating platelets in the acute coronary syndrome. Proteomics 2011, 11, 3335–3348. [Google Scholar] [CrossRef] [PubMed]
- Vélez, P.; Ocaranza-Sánchez, R.; López-Otero, D.; Grigorian-Shamagian, L.; Rosa, I.; Bravo, S.B.; González-Juanatey, J.R.; García, Á. 2D-DIGE-based proteomic analysis of intracoronary versus peripheral arterial blood platelets from acute myocardial infarction patients: Upregulation of platelet activation biomarkers at the culprit site. Proteom. Clin. Appl. 2016, 10, 851–858. [Google Scholar] [CrossRef]
- Vélez, P.; Izquierdo, I.; Rosa, I.; García, Á. A 2D-DIGE-based proteomic analysis reveals differences in the platelet releasate composition when comparing thrombin and collagen stimulations. Sci. Rep. 2015, 5, 8198. [Google Scholar] [CrossRef]
- Hell, L.; Lurger, K.; Mauracher, L.-M.; Grilz, E.; Reumiller, C.M.; Schmidt, G.J.; Ercan, H.; Koder, S.; Assinger, A.; Basilio, J. Altered platelet proteome in lupus anticoagulant (LA)-positive patients—Protein disulfide isomerase and NETosis as new players in LA-related thrombosis. Exp. Mol. Med. 2020, 52, 66–78. [Google Scholar] [CrossRef]
- Loroch, S.; Trabold, K.; Gambaryan, S.; Reiß, C.; Schwierczek, K.; Fleming, I.; Sickmann, A.; Behnisch, W.; Zieger, B.; Zahedi, R.P. Alterations of the platelet proteome in type I Glanzmann thrombasthenia caused by different homozygous delG frameshift mutations in ITGA2B. Thromb. Haemost. 2017, 117, 556–569. [Google Scholar] [CrossRef]
- van Kruchten, R.; Mattheij, N.J.; Saunders, C.; Feijge, M.A.; Swieringa, F.; Wolfs, J.L.; Collins, P.W.; Heemskerk, J.W.; Bevers, E.M. Both TMEM16F-dependent and TMEM16F-independent pathways contribute to phosphatidylserine exposure in platelet apoptosis and platelet activation. Blood Am. Soc. Hematol. 2013, 121, 1850–1857. [Google Scholar] [CrossRef]
- Bergemalm, D.; Ramström, S.; Kardeby, C.; Hultenby, K.; Eremo, A.G.; Sihlbom, C.; Bergström, J.; Palmblad, J.; Åström, M. Platelet proteome and function in X− linked thrombocytopenia with thalassemia and in silico comparisons with gray platelet syndrome. Haematologica 2021, 106, 2947. [Google Scholar] [CrossRef]
- Sims, M.C.; Mayer, L.; Collins, J.H.; Bariana, T.K.; Megy, K.; Lavenu-Bombled, C.; Seyres, D.; Kollipara, L.; Burden, F.S.; Greene, D. Novel manifestations of immune dysregulation and granule defects in gray platelet syndrome. Blood Am. Soc. Hematol. 2020, 136, 1956–1967. [Google Scholar] [CrossRef]
- Sabrkhany, S.; Kuijpers, M.J.; Knol, J.C.; Damink, S.W.O.; Dingemans, A.-M.C.; Verheul, H.M.; Piersma, S.R.; Pham, T.V.; Griffioen, A.W.; Oude Egbrink, M.G. Exploration of the platelet proteome in patients with early-stage cancer. J. Proteom. 2018, 177, 65–74. [Google Scholar] [CrossRef]
- Jalal, D.I.; Chonchol, M.; Targher, G. In Disorders of hemostasis associated with chronic kidney disease. Semin. Thromb. Hemost. 2010, 2010, 34–40. [Google Scholar] [CrossRef]
- Lutz, J.; Menke, J.; Sollinger, D.; Schinzel, H.; Thürmel, K. Haemostasis in chronic kidney disease. Nephrol. Dial. Transpl. 2014, 29, 29–40. [Google Scholar] [CrossRef]
- Ravid, J.D.; Chitalia, V.C. Molecular mechanisms underlying the cardiovascular toxicity of specific uremic solutes. Cells 2020, 9, 2024. [Google Scholar] [CrossRef] [PubMed]
- Bijak, M.; Olejnik, A.; Rokita, B.; Morel, A.; Dziedzic, A.; Miller, E.; Saluk-Bijak, J. Increased level of fibrinogen chains in the proteome of blood platelets in secondary progressive multiple sclerosis patients. J. Cell. Mol. Med. 2019, 23, 3476–3482. [Google Scholar] [CrossRef] [PubMed]
- Trugilho, M.R.d.O.; Hottz, E.D.; Brunoro, G.V.F.; Teixeira-Ferreira, A.; Carvalho, P.C.; Salazar, G.A.; Zimmerman, G.A.; Bozza, F.A.; Bozza, P.T.; Perales, J. Platelet proteome reveals novel pathways of platelet activation and platelet-mediated immunoregulation in dengue. PLoS Pathog. 2017, 13, e1006385. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Liu, Y.; He, B.; He, T.; Chen, C.; He, J.; Yang, X.; Wang, J.Z. Platelet biomarkers for a descending cognitive function: A proteomic approach. Aging Cell 2021, 20, e13358. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; Weinrauch, Y.; Brinkmann, V.; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007, 176, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Labib, D.A.; Ashmawy, I.; Elmazny, A.; Helmy, H.; Ismail, R.S. Toll-like receptors 2 and 4 expression on peripheral blood lymphocytes and neutrophils of Egyptian multiple sclerosis patients. Int. J. Neurosci. 2022, 132, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Semeraro, F.; Ammollo, C.T.; Morrissey, J.H.; Dale, G.L.; Friese, P.; Esmon, N.L.; Esmon, C.T. Extracellular histones promote thrombin generation through platelet-dependent mechanisms: Involvement of platelet TLR2 and TLR4. Blood 2011, 118, 1952–1961. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.A.; Brill, A.; Duerschmied, D.; Schatzberg, D.; Monestier, M.; Myers, D.D., Jr.; Wrobleski, S.K.; Wakefield, T.W.; Hartwig, J.H.; Wagner, D.D. Extracellular DNA traps promote thrombosis. Proc. Natl. Acad. Sci. USA 2010, 107, 15880–15885. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, B.; Massberg, S. Thrombosis as an intravascular effector of innate immunity. Nat. Rev. Immunol. 2013, 13, 34–45. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, J.; Haslbauer, J.D.; Stalder, A.K.; Romanens, A.; Mertz, K.D.; Studt, J.D.; Siegemund, M.; Buser, A.; Holbro, A.; Tzankov, A. Von Willebrand factor and the thrombophilia of severe COVID-19: In situ evidence from autopsies. Res. Pract. Thromb. Haemost. 2023, 7, 100182. [Google Scholar] [CrossRef] [PubMed]
- Kaltenmeier, C.; Simmons, R.L.; Tohme, S.; Yazdani, H.O. Neutrophil Extracellular Traps (NETs) in Cancer Metastasis. Cancers 2021, 13, 6131. [Google Scholar] [CrossRef] [PubMed]
- Etulain, J.; Martinod, K.; Wong, S.L.; Cifuni, S.M.; Schattner, M.; Wagner, D.D. P-selectin promotes neutrophil extracellular trap formation in mice. Blood 2015, 126, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Demers, M.; Krause, D.S.; Schatzberg, D.; Martinod, K.; Voorhees, J.R.; Fuchs, T.A.; Scadden, D.T.; Wagner, D.D. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc. Natl. Acad. Sci. USA 2012, 109, 13076–13081. [Google Scholar] [CrossRef]
- Guglietta, S.; Chiavelli, A.; Zagato, E.; Krieg, C.; Gandini, S.; Ravenda, P.S.; Bazolli, B.; Lu, B.; Penna, G.; Rescigno, M. Coagulation induced by C3aR-dependent NETosis drives protumorigenic neutrophils during small intestinal tumorigenesis. Nat. Commun. 2016, 7, 11037. [Google Scholar] [CrossRef]
- Yalavarthi, S.; Gould, T.J.; Rao, A.N.; Mazza, L.F.; Morris, A.E.; Nunez-Alvarez, C.; Hernandez-Ramirez, D.; Bockenstedt, P.L.; Liaw, P.C.; Cabral, A.R.; et al. Release of neutrophil extracellular traps by neutrophils stimulated with antiphospholipid antibodies: A newly identified mechanism of thrombosis in the antiphospholipid syndrome. Arthritis Rheumatol. 2015, 67, 2990–3003. [Google Scholar] [CrossRef]
- Popp, S.K.; Vecchio, F.; Brown, D.J.; Fukuda, R.; Suzuki, Y.; Takeda, Y.; Wakamatsu, R.; Sarma, M.A.; Garrett, J.; Giovenzana, A.; et al. Circulating platelet-neutrophil aggregates characterize the development of type 1 diabetes in humans and NOD mice. JCI Insight 2022, 7, e153993. [Google Scholar] [CrossRef]
- Ren, J.; He, J.; Zhang, H.; Xia, Y.; Hu, Z.; Loughran, P.; Billiar, T.; Huang, H.; Tsung, A. Platelet TLR4-ERK5 Axis Facilitates NET-Mediated Capturing of Circulating Tumor Cells and Distant Metastasis after Surgical Stress. Cancer Res. 2021, 81, 2373–2385. [Google Scholar] [CrossRef]
- Pang, A.; Cui, Y.; Chen, Y.; Cheng, N.; Delaney, M.K.; Gu, M.; Stojanovic-Terpo, A.; Zhu, C.; Du, X. Shear-induced integrin signaling in platelet phosphatidylserine exposure, microvesicle release, and coagulation. Blood Am. Soc. Hematol. 2018, 132, 533–543. [Google Scholar] [CrossRef]
- Zimmerman, G.A.; Weyrich, A.S. Signal-dependent protein synthesis by activated platelets: New pathways to altered phenotype and function. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 17–24. [Google Scholar] [CrossRef]
- Ng, M.S.Y.; Tung, J.-P.; Fraser, J.F. Platelet storage lesions: What more do we know now? Transfus. Med. Rev. 2018, 32, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Prudova, A.; Serrano, K.; Eckhard, U.; Fortelny, N.; Devine, D.V.; Overall, C.M. TAILS N-terminomics of human platelets reveals pervasive metalloproteinase-dependent proteolytic processing in storage. Blood Am. Soc. Hematol. 2014, 124, e49–e60. [Google Scholar] [CrossRef] [PubMed]
- Thiele, T.; Braune, J.; Dhople, V.; Hammer, E.; Scharf, C.; Greinacher, A.; Völker, U.; Steil, L. Proteomic profile of platelets during reconstitution of platelet counts after apheresis. Proteom. Clin. Appl. 2016, 10, 831–838. [Google Scholar] [CrossRef]
- Rijkers, M.; van den Eshof, B.L.; van der Meer, P.F.; van Alphen, F.P.; de Korte, D.; Leebaeek, F.W.; Meijer, A.B.; Voorberg, J.; Jansen, A.G. Monitoring storage induced changes in the platelet proteome employing lable free quantative mass spectrometry. Sci. Rep. 2017, 7, 11045. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jiang, T.; Fan, Y.; Zhao, S. A proteomic approach reveals the variation in human platelet protein composition after storage at different temperatures. Platelets 2019, 30, 403–412. [Google Scholar] [CrossRef]
- Wood, B.; Padula, M.P.; Marks, D.C.; Johnson, L. Refrigerated storage of platelets initiates changes in platelet surface marker expression and localization of intracellular proteins. Transfusion 2016, 56, 2548–2559. [Google Scholar] [CrossRef]
- Schubert, P.; Culibrk, B.; Karwal, S.; Goodrich, R.P.; Devine, D.V. Protein translation occurs in platelet concentrates despite riboflavin/UV light pathogen inactivation treatment. Proteom. Clin. Appl. 2016, 10, 839–850. [Google Scholar] [CrossRef]
- Sonego, G.; Abonnenc, M.; Crettaz, D.; Lion, N.; Tissot, J.-D.; Prudent, M. Irreversible oxidations of platelet proteins after riboflavin-UVB pathogen inactivation. Transfus. Clin. Biol. 2020, 27, 36–42. [Google Scholar] [CrossRef]
- Hermida-Nogueira, L.; Barrachina, M.N.; Izquierdo, I.; García-Vence, M.; Lacerenza, S.; Bravo, S.; Castrillo, A.; García, Á. Proteomic analysis of extracellular vesicles derived from platelet concentrates treated with Mirasol® identifies biomarkers of platelet storage lesion. J. Proteom. 2020, 210, 103529. [Google Scholar] [CrossRef]
- Greening, D.W.; Sparrow, R.L.; Simpson, R.J. Preparation of platelet concentrates. Plasma Proteom. Methods Protoc. 2011, 728, 267–278. [Google Scholar]
- Wrzyszcz, A.; Urbaniak, J.; Sapa, A.; Woźniak, M. An efficient method for isolation of representative and contamination-free population of blood platelets for proteomic studies. Platelets 2017, 28, 43–53. [Google Scholar] [CrossRef]
- Kim, T.; Chen, I.R.; Parker, B.L.; Humphrey, S.J.; Crossett, B.; Cordwell, S.J.; Yang, P.; Yang, J.Y.H. QCMAP: An Interactive Web-Tool for Performance Diagnosis and Prediction of LC-MS Systems. Proteomics 2019, 19, 1900068. [Google Scholar] [CrossRef] [PubMed]
- Van Houtven, J.; Agten, A.; Boonen, K.; Baggerman, G.; Hooyberghs, J.; Laukens, K.; Valkenborg, D. Qcquan: A web tool for the automated assessment of protein expression and data quality of labeled mass spectrometry experiments. J. Proteome Res. 2019, 18, 2221–2227. [Google Scholar] [CrossRef] [PubMed]
- Chiva, C.; Olivella, R.; Borras, E.; Espadas, G.; Pastor, O.; Sole, A.; Sabido, E. QCloud: A cloud-based quality control system for mass spectrometry-based proteomics laboratories. PLoS ONE 2018, 13, e0189209. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Hou, J.; Tanner, J.J.; Cheng, J. Bioinformatics methods for mass spectrometry-based proteomics data analysis. Int. J. Mol. Sci. 2020, 21, 2873. [Google Scholar] [CrossRef] [PubMed]
- Solari, F.A.; Dell’Aica, M.; Sickmann, A.; Zahedi, R.P. Why phosphoproteomics is still a challenge. Mol. Biosyst. 2015, 11, 1487–1493. [Google Scholar] [CrossRef] [PubMed]
- Swieringa, F.; Solari, F.A.; Pagel, O.; Beck, F.; Huang, J.; Feijge, M.A.; Jurk, K.; Körver-Keularts, I.M.; Mattheij, N.J.; Faber, J. Impaired iloprost-induced platelet inhibition and phosphoproteome changes in patients with confirmed pseudohypoparathyroidism type Ia, linked to genetic mutations in GNAS. Sci. Rep. 2020, 10, 11389. [Google Scholar] [CrossRef] [PubMed]
- Looße, C.; Swieringa, F.; Heemskerk, J.W.; Sickmann, A.; Lorenz, C. Platelet proteomics: From discovery to diagnosis. Expert. Rev. Proteom. 2018, 15, 467–476. [Google Scholar] [CrossRef]
- Chen, L.; Kashina, A. Post-translational Modifications of the Protein Termini. Front. Cell. Dev. Biol. 2021, 9, 719590. [Google Scholar] [CrossRef]
- Martínez-Botía, P.; Villar, P.; Carbajo-Argüelles, G.; Jaiteh, Z.; Acebes-Huerta, A.; Gutiérrez, L. Proteomics-wise, how similar are mouse and human platelets? Platelets 2023, 34, 2220415. [Google Scholar] [CrossRef]
- Monaco, G.; van Dam, S.; Casal Novo Ribeiro, J.L.; Larbi, A.; de Magalhães, J.P. A comparison of human and mouse gene co-expression networks reveals conservation and divergence at the tissue, pathway and disease levels. BMC Evol. Biol. 2015, 15, 259. [Google Scholar] [CrossRef]
- Schmitt, A.; Guichard, J.; Massé, J.-M.; Debili, N.; Cramer, E.M. Of mice and men: Comparison of the ultrastructure of megakaryocytes and platelets. Exp. Hematol. 2001, 29, 1295–1302. [Google Scholar] [CrossRef]
- Thijs, T.; Deckmyn, H.; Broos, K. Model systems of genetically modified platelets. Blood Am. Soc. Hematol. 2012, 119, 1634–1642. [Google Scholar] [CrossRef]
- Jirouskova, M.; Shet, A.; Johnson, G.J. A guide to murine platelet structure, function, assays, and genetic alterations. Thromb. Haemost. 2007, 5, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, A.; Miyata, K.; Fujita, N. Platelet-activating factor podoplanin: From discovery to drug development. Cancer Metastasis Rev. 2017, 36, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Herzog, B.H.; Fu, J.; Wilson, S.J.; Hess, P.R.; Sen, A.; McDaniel, J.M.; Pan, Y.; Sheng, M.; Yago, T.; Silasi-Mansat, R.; et al. Podoplanin maintains high endothelial venule integrity by interacting with platelet CLEC-2. Nature 2013, 502, 105–109. [Google Scholar] [CrossRef]
- Mobarrez, F.; He, S.; Broijersen, A.; Wiklund, B.; Antovic, A.; Antovic, J.; Egberg, N.; Jorneskog, G.; Wallen, H. Atorvastatin reduces thrombin generation and expression of tissue factor, P-selectin and GPIIIa on platelet-derived microparticles in patients with peripheral arterial occlusive disease. Thromb. Haemost. 2011, 106, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Kidokoro, M.; Tanaka, M.; Inoue, S.; Tsuji, T.; Akatuska, H.; Okada, C.; Iida, Y.; Okada, Y.; Suzuki, Y.; et al. Podoplanin is indispensable for cell motility and platelet-induced epithelial-to-mesenchymal transition-related gene expression in esophagus squamous carcinoma TE11A cells. Cancer Cell Int. 2020, 20, 263. [Google Scholar] [CrossRef]
- Kato, Y.; Kaneko, M.K.; Kunita, A.; Ito, H.; Kameyama, A.; Ogasawara, S.; Matsuura, N.; Hasegawa, Y.; Suzuki-Inoue, K.; Inoue, O.; et al. Molecular analysis of the pathophysiological binding of the platelet aggregation-inducing factor podoplanin to the C-type lectin-like receptor CLEC-2. Cancer Sci. 2008, 99, 54–61. [Google Scholar] [CrossRef]
- Suzuki-Inoue, K.; Kato, Y.; Inoue, O.; Kaneko, M.K.; Mishima, K.; Yatomi, Y.; Yamazaki, Y.; Narimatsu, H.; Ozaki, Y. Involvement of the snake toxin receptor CLEC-2, in podoplanin-mediated platelet activation, by cancer cells. J. Biol. Chem. 2007, 282, 25993–26001. [Google Scholar] [CrossRef]
- Mach, F.; Schonbeck, U.; Libby, P. CD40 signaling in vascular cells: A key role in atherosclerosis? Atherosclerosis 1998, 137, 89–95. [Google Scholar] [CrossRef]
- de Lemos, J.A.; Zirlik, A.; Schonbeck, U.; Varo, N.; Murphy, S.A.; Khera, A.; McGuire, D.K.; Stanek, G.; Lo, H.S.; Nuzzo, R.; et al. Associations between soluble CD40 ligand, atherosclerosis risk factors, and subclinical atherosclerosis: Results from the Dallas Heart Study. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 2192–2196. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lacy, M.; Burger, C.; Shami, A.; Ahmadsei, M.; Winkels, H.; Nitz, K.; van Tiel, C.M.; Seijkens, T.T.P.; Kusters, P.J.H.; Karshovka, E.; et al. Cell-specific and divergent roles of the CD40L-CD40 axis in atherosclerotic vascular disease. Nat. Commun. 2021, 12, 3754. [Google Scholar] [CrossRef]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef] [PubMed]
- Schmahl, J.; Raymond, C.S.; Soriano, P. PDGF signaling specificity is mediated through multiple immediate early genes. Nat. Genet. 2007, 39, 52–60. [Google Scholar] [CrossRef]
- Zeller, J.A.; Tschoepe, D.; Kessler, C. Circulating platelets show increased activation in patients with acute cerebral ischemia. Thromb. Haemost. 1999, 81, 373–377. [Google Scholar] [PubMed]
- O’Connor, C.M.; Gurbel, P.A.; Serebruany, V.L. Usefulness of soluble and surface-bound P-selectin in detecting heightened platelet activity in patients with congestive heart failure. Am. J. Cardiol. 1999, 83, 1345–1349. [Google Scholar] [CrossRef]
- Tschoepe, D.; Schultheiss, H.P.; Kolarov, P.; Schwippert, B.; Dannehl, K.; Nieuwenhuis, H.K.; Kehrel, B.; Strauer, B.; Gries, F.A. Platelet membrane activation markers are predictive for increased risk of acute ischemic events after PTCA. Circulation 1993, 88, 37–42. [Google Scholar] [CrossRef]
- Blann, A.D.; Nadar, S.K.; Lip, G.Y. The adhesion molecule P-selectin and cardiovascular disease. Eur. Heart J. 2003, 24, 2166–2179. [Google Scholar] [CrossRef]
- Bielinski, S.J.; Berardi, C.; Decker, P.A.; Kirsch, P.S.; Larson, N.B.; Pankow, J.S.; Sale, M.; de Andrade, M.; Sicotte, H.; Tang, W.; et al. P-selectin and subclinical and clinical atherosclerosis: The Multi-Ethnic Study of Atherosclerosis (MESA). Atherosclerosis 2015, 240, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Fullard, J.F. The role of the platelet glycoprotein IIb/IIIa in thrombosis and haemostasis. Curr. Pharm. Des. 2004, 10, 1567–1576. [Google Scholar] [CrossRef]
- Lippi, G.; Montagnana, M.; Danese, E.; Favaloro, E.J.; Franchini, M. Glycoprotein IIb/IIIa inhibitors: An update on the mechanism of action and use of functional testing methods to assess antiplatelet efficacy. Biomark. Med. 2011, 5, 63–70. [Google Scholar] [CrossRef]
- Schneider, D.J. Anti-platelet therapy: Glycoprotein IIb-IIIa antagonists. Br. J. Clin. Pharmacol. 2011, 72, 672–682. [Google Scholar] [CrossRef]
- Pellitero, S.; Reverter, J.L.; Tassies, D.; Pizarro, E.; Monteagudo, J.; Salinas, I.; Aguilera, E.; Sanmarti, A.; Reverter, J.C. Polymorphisms in platelet glycoproteins Ia and IIIa are associated with arterial thrombosis and carotid atherosclerosis in type 2 diabetes. Thromb. Haemost. 2010, 103, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Befekadu, R.; Christiansen, K.; Larsson, A.; Grenegard, M. Increased plasma cathepsin S and trombospondin-1 in patients with acute ST-segment elevation myocardial infarction. Cardiol. J. 2019, 26, 385–393. [Google Scholar] [CrossRef]
- van Almen, G.C.; Verhesen, W.; van Leeuwen, R.E.; van de Vrie, M.; Eurlings, C.; Schellings, M.W.; Swinnen, M.; Cleutjens, J.P.; van Zandvoort, M.A.; Heymans, S.; et al. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell 2011, 10, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Palma Dos Reis, R.; Rodrigues, R.; Sousa, A.C.; Gomes, S.; Borges, S.; Ornelas, I.; Freitas, A.I.; Guerra, G.; Henriques, E.; et al. Association of ADAMTS7 gene polymorphism with cardiovascular survival in coronary artery disease. Physiol. Genom. 2016, 48, 810–815. [Google Scholar] [CrossRef][Green Version]
- Bauer, R.C.; Tohyama, J.; Cui, J.; Cheng, L.; Yang, J.; Zhang, X.; Ou, K.; Paschos, G.K.; Zheng, X.L.; Parmacek, M.S.; et al. Knockout of Adamts7, a novel coronary artery disease locus in humans, reduces atherosclerosis in mice. Circulation 2015, 131, 1202–1213. [Google Scholar] [CrossRef]
- Hofmann, B.; Adam, A.C.; Jacobs, K.; Riemer, M.; Erbs, C.; Bushnaq, H.; Simm, A.; Silber, R.E.; Santos, A.N. Advanced glycation end product associated skin autofluorescence: A mirror of vascular function? Exp. Gerontol. 2013, 48, 38–44. [Google Scholar] [CrossRef]
- de Vos, L.C.; Lefrandt, J.D.; Dullaart, R.P.; Zeebregts, C.J.; Smit, A.J. Advanced glycation end products: An emerging biomarker for adverse outcome in patients with peripheral artery disease. Atherosclerosis 2016, 254, 291–299. [Google Scholar] [CrossRef]
- Singh, V.P.; Bali, A.; Singh, N.; Jaggi, A.S. Advanced glycation end products and diabetic complications. Korean J. Physiol. Pharmacol. 2014, 18, 1–14. [Google Scholar] [CrossRef]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D.; Executive Group on behalf of the Joint European Society of Cardiology/American College of Cardiology/American Heart Association/World Heart Federation Task Force for the Universal Definition of Myocardial Infarction. Fourth Universal Definition of Myocardial Infarction (2018). J. Am. Coll. Cardiol. 2018, 72, 2231–2264. [Google Scholar] [CrossRef]
- Samman Tahhan, A.; Sandesara, P.; Hayek, S.S.; Hammadah, M.; Alkhoder, A.; Kelli, H.M.; Topel, M.; O’Neal, W.T.; Ghasemzadeh, N.; Ko, Y.A.; et al. High-Sensitivity Troponin I Levels and Coronary Artery Disease Severity, Progression, and Long-Term Outcomes. J. Am. Heart Assoc. 2018, 7, e007914. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.W.; Dobbin, S.J.H.; Pettit, S.J.; Di Angelantonio, E.; Willeit, P. High-Sensitivity Cardiac Troponin and New-Onset Heart Failure: A Systematic Review and Meta-Analysis of 67,063 Patients With 4,165 Incident Heart Failure Events. JACC Heart Fail. 2018, 6, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Filion, K.B.; Agarwal, S.K.; Ballantyne, C.M.; Eberg, M.; Hoogeveen, R.C.; Huxley, R.R.; Loehr, L.R.; Nambi, V.; Soliman, E.Z.; Alonso, A. High-sensitivity cardiac troponin T and the risk of incident atrial fibrillation: The Atherosclerosis Risk in Communities (ARIC) study. Am. Heart J. 2015, 169, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Zethelius, B.; Berglund, L.; Sundstrom, J.; Ingelsson, E.; Basu, S.; Larsson, A.; Venge, P.; Arnlov, J. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N. Engl. J. Med. 2008, 358, 2107–2116. [Google Scholar] [CrossRef]
- Shin, D.S.; Kim, H.N.; Shin, K.D.; Yoon, Y.J.; Kim, S.J.; Han, D.C.; Kwon, B.M. Cryptotanshinone inhibits constitutive signal transducer and activator of transcription 3 function through blocking the dimerization in DU145 prostate cancer cells. Cancer Res. 2009, 69, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Oi, T.; Asanuma, K.; Matsumine, A.; Matsubara, T.; Nakamura, T.; Iino, T.; Asanuma, Y.; Goto, M.; Okuno, K.; Kakimoto, T.; et al. STAT3 inhibitor, cucurbitacin I, is a novel therapeutic agent for osteosarcoma. Int. J. Oncol. 2016, 49, 2275–2284. [Google Scholar] [CrossRef][Green Version]
- Pidgeon, G.P.; Barr, M.P.; Harmey, J.H.; Foley, D.A.; Bouchier-Hayes, D.J. Vascular endothelial growth factor (VEGF) upregulates BCL-2 and inhibits apoptosis in human and murine mammary adenocarcinoma cells. Br. J. Cancer 2001, 85, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Bachelder, R.E.; Lipscomb, E.A.; Shaw, L.M.; Mercurio, A.M. Integrin (alpha 6 beta 4) regulation of eIF-4E activity and VEGF translation: A survival mechanism for carcinoma cells. J. Cell Biol. 2002, 158, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) and Its Receptor (VEGFR) Signaling in Angiogenesis: A Crucial Target for Anti- and Pro-Angiogenic Therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, T.; Wincup, C.; Buchholz, I.; Pericleous, C.; Giles, I.; Ripoll, V.; Cohen, H.; Delcea, M.; Rahman, A. The role of beta-2-glycoprotein I in health and disease associating structure with function: More than just APS. Blood Rev. 2020, 39, 100610. [Google Scholar] [CrossRef] [PubMed]
- Maiolino, G.; Rossitto, G.; Caielli, P.; Bisogni, V.; Rossi, G.P.; Calo, L.A. The role of oxidized low-density lipoproteins in atherosclerosis: The myths and the facts. Mediat. Inflamm. 2013, 2013, 714653. [Google Scholar] [CrossRef] [PubMed]
- Pitari, G.M.; Cotzia, P.; Ali, M.; Birbe, R.; Rizzo, W.; Bombonati, A.; Palazzo, J.; Solomides, C.; Shuber, A.P.; Sinicrope, F.A.; et al. Vasodilator-Stimulated Phosphoprotein Biomarkers Are Associated with Invasion and Metastasis in Colorectal Cancer. Biomark. Cancer 2018, 10, 1179299X18774551. [Google Scholar] [CrossRef]
- Aratani, Y. Myeloperoxidase: Its role for host defense, inflammation, and neutrophil function. Arch. Biochem. Biophys. 2018, 640, 47–52. [Google Scholar] [CrossRef] [PubMed]
- van der Veen, B.S.; de Winther, M.P.; Heeringa, P. Myeloperoxidase: Molecular mechanisms of action and their relevance to human health and disease. Antioxid. Redox Signal 2009, 11, 2899–2937. [Google Scholar] [CrossRef]
- Arnhold, J. The Dual Role of Myeloperoxidase in Immune Response. Int. J. Mol. Sci. 2020, 21, 8057. [Google Scholar] [CrossRef]
- Lipkova, J.; Parenica, J.; Duris, K.; Helanova, K.; Tomandl, J.; Kubkova, L.; Vasku, A.; Goldbergova Pavkova, M. Association of circulating levels of RANTES and -403G/A promoter polymorphism to acute heart failure after STEMI and to cardiogenic shock. Clin. Exp. Med. 2015, 15, 405–414. [Google Scholar] [CrossRef]
- Appay, V.; Rowland-Jones, S.L. RANTES: A versatile and controversial chemokine. Trends Immunol. 2001, 22, 83–87. [Google Scholar] [CrossRef]
- Ueba, T.; Nomura, S.; Inami, N.; Yokoi, T.; Inoue, T. Elevated RANTES level is associated with metabolic syndrome and correlated with activated platelets associated markers in healthy younger men. Clin. Appl. Thromb. Hemost. 2014, 20, 813–818. [Google Scholar] [CrossRef]
- Amabile, N.; Rautou, P.E.; Tedgui, A.; Boulanger, C.M. Microparticles: Key protagonists in cardiovascular disorders. Semin. Thromb. Hemost. 2010, 36, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Margolis, J.; Kenrick, K.G. 2-dimensional resolution of plasma proteins by combination of polyacrylamide disc and gradient gel electrophoresis. Nature 1969, 221, 1056–1057. [Google Scholar] [CrossRef]
- Tan, K.T.; Tayebjee, M.H.; Lynd, C.; Blann, A.D.; Lip, G.Y. Platelet microparticles and soluble P selectin in peripheral artery disease: Relationship to extent of disease and platelet activation markers. Ann. Med. 2005, 37, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Namba, M.; Tanaka, A.; Shimada, K.; Ozeki, Y.; Uehata, S.; Sakamoto, T.; Nishida, Y.; Nomura, S.; Yoshikawa, J. Circulating platelet-derived microparticles are associated with atherothrombotic events: A marker for vulnerable blood. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 255–256. [Google Scholar] [CrossRef] [PubMed]
- Badimon, L.; Suades, R.; Fuentes, E.; Palomo, I.; Padro, T. Role of Platelet-Derived Microvesicles as Crosstalk Mediators in Atherothrombosis and Future Pharmacology Targets: A Link between Inflammation, Atherosclerosis, and Thrombosis. Front. Pharmacol. 2016, 7, 293. [Google Scholar] [CrossRef]
- Chiva-Blanch, G.; Suades, R.; Padro, T.; Vilahur, G.; Pena, E.; Ybarra, J.; Pou, J.M.; Badimon, L. Microparticle Shedding by Erythrocytes, Monocytes and Vascular Smooth Muscular Cells Is Reduced by Aspirin in Diabetic Patients. Rev. Esp. Cardiol. Engl. Ed. 2016, 69, 672–680. [Google Scholar] [CrossRef]
- Getts, D.R.; Terry, R.L.; Getts, M.T.; Deffrasnes, C.; Muller, M.; van Vreden, C.; Ashhurst, T.M.; Chami, B.; McCarthy, D.; Wu, H.; et al. Therapeutic inflammatory monocyte modulation using immune-modifying microparticles. Sci. Transl. Med. 2014, 6, 219. [Google Scholar] [CrossRef]
- Chiva-Blanch, G.; Crespo, J.; Suades, R.; Arderiu, G.; Padro, T.; Vilahur, G.; Cubedo, J.; Corella, D.; Salas-Salvado, J.; Aros, F.; et al. CD142+/CD61+, CD146+ and CD45+ microparticles predict cardiovascular events in high risk patients following a Mediterranean diet supplemented with nuts. Thromb. Haemost. 2016, 116, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Almog, N.; Henke, V.; Flores, L.; Hlatky, L.; Kung, A.L.; Wright, R.D.; Berger, R.; Hutchinson, L.; Naumov, G.N.; Bender, E.; et al. Prolonged dormancy of human liposarcoma is associated with impaired tumor angiogenesis. FASEB J. 2006, 20, 947–949. [Google Scholar] [CrossRef] [PubMed]
- Naumov, G.N.; Bender, E.; Zurakowski, D.; Kang, S.Y.; Sampson, D.; Flynn, E.; Watnick, R.S.; Straume, O.; Akslen, L.A.; Folkman, J.; et al. A model of human tumor dormancy: An angiogenic switch from the nonangiogenic phenotype. J. Natl. Cancer Inst. 2006, 98, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Cervi, D.; Yip, T.T.; Bhattacharya, N.; Podust, V.N.; Peterson, J.; Abou-Slaybi, A.; Naumov, G.N.; Bender, E.; Almog, N.; Italiano, J.E., Jr.; et al. Platelet-associated PF-4 as a biomarker of early tumor growth. Blood 2008, 111, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Hansson, G.K. Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 2005, 352, 1685–1695. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, K.; Muller, M.A.; Varlamov, O.; Tavori, H.; Packwood, W.; Mueller, P.A.; Xie, A.; Ruggeri, Z.; Chung, D.; Lopez, J.A.; et al. Proteolysis of Von Willebrand Factor Influences Inflammatory Endothelial Activation and Vascular Compliance in Atherosclerosis. JACC Basic Transl. Sci. 2020, 5, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
- Takaya, H.; Namisaki, T.; Kitade, M.; Kaji, K.; Nakanishi, K.; Tsuji, Y.; Shimozato, N.; Moriya, K.; Seki, K.; Sawada, Y.; et al. VWF/ADAMTS13 ratio as a potential biomarker for early detection of hepatocellular carcinoma. BMC Gastroenterol. 2019, 19, 167. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, S.F.; Stoll, G.; Wagner, D.D.; Kleinschnitz, C. von Willebrand factor: An emerging target in stroke therapy. Stroke 2012, 43, 599–606. [Google Scholar] [CrossRef]
- Qin, F.; Impeduglia, T.; Schaffer, P.; Dardik, H. Overexpression of von Willebrand factor is an independent risk factor for pathogenesis of intimal hyperplasia: Preliminary studies. J. Vasc. Surg. 2003, 37, 433–439. [Google Scholar] [CrossRef]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef]
- Perneczky, R.; Guo, L.H.; Kagerbauer, S.M.; Werle, L.; Kurz, A.; Martin, J.; Alexopoulos, P. Soluble amyloid precursor protein beta as blood-based biomarker of Alzheimer’s disease. Transl. Psychiatry 2013, 3, 227. [Google Scholar] [CrossRef]
- O’Brien, R.J.; Wong, P.C. Amyloid precursor protein processing and Alzheimer’s disease. Annu. Rev. Neurosci. 2011, 34, 185–204. [Google Scholar] [CrossRef]
- Mai, W.; Liao, Y. Targeting IL-1beta in the Treatment of Atherosclerosis. Front. Immunol. 2020, 11, 589654. [Google Scholar] [CrossRef] [PubMed]
- Libby, P. Interleukin-1 Beta as a Target for Atherosclerosis Therapy: Biological Basis of CANTOS and Beyond. J. Am. Coll. Cardiol. 2017, 70, 2278–2289. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.; Rivera, K.; Andia, M.E.; Martinez Rodriguez, G. The IL-1 Family and Its Role in Atherosclerosis. Int. J. Mol. Sci. 2022, 24, 17. [Google Scholar] [CrossRef] [PubMed]
- Peiro, C.; Lorenzo, O.; Carraro, R.; Sanchez-Ferrer, C.F. IL-1beta Inhibition in Cardiovascular Complications Associated to Diabetes Mellitus. Front. Pharmacol. 2017, 8, 363. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Nazy, I.; Smith, J.W.; Kelton, J.G.; Arnold, D.M. Platelet autoantibodies in the bone marrow of patients with immune thrombocytopenia. Blood Adv. 2020, 4, 2962–2966. [Google Scholar] [CrossRef] [PubMed]
- Zito, F.; Drummond, F.; Bujac, S.R.; Esnouf, M.P.; Morrissey, J.H.; Humphries, S.E.; Miller, G.J. Epidemiological and genetic associations of activated factor XII concentration with factor VII activity, fibrinopeptide A concentration, and risk of coronary heart disease in men. Circulation 2000, 102, 2058–2062. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miller, G.J.; Esnouf, M.P.; Burgess, A.I.; Cooper, J.A.; Mitchell, J.P. Risk of coronary heart disease and activation of factor XII in middle-aged men. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 2103–2106. [Google Scholar] [CrossRef]
- Ishii, K.; Oguchi, S.; Murata, M.; Mitsuyoshi, Y.; Takeshita, E.; Ito, D.; Tanahashi, N.; Fukuuchi, Y.; Oosumi, K.; Matsumoto, K.; et al. Activated factor XII levels are dependent on factor XII 46C/T genotypes and factor XII zymogen levels, and are associated with vascular risk factors in patients and healthy subjects. Blood Coagul. Fibrinolysis 2000, 11, 277–284. [Google Scholar]
- Johansson, K.; Jansson, J.H.; Johansson, L.; Bylesjo, I.; Nilsson, T.K.; Eliasson, M.; Soderberg, S.; Lind, M. Factor XII as a Risk Marker for Hemorrhagic Stroke: A Prospective Cohort Study. Cerebrovasc. Dis. Extra 2017, 7, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Bonnez, Q.; Sakai, K.; Vanhoorelbeke, K. ADAMTS13 and Non-ADAMTS13 Biomarkers in Immune-Mediated Thrombotic Thrombocytopenic Purpura. J. Clin. Med. 2023, 12, 6169. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Joshi, M.B.; Philippova, M.; Erne, P.; Hasler, P.; Hahn, S.; Resink, T.J. Activated endothelial cells induce neutrophil extracellular traps and are susceptible to NETosis-mediated cell death. FEBS Lett. 2010, 584, 3193–3197. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, E.; Yalavarthi, S.; Berthier, C.C.; Hodgin, J.B.; Khandpur, R.; Lin, A.M.; Rubin, C.J.; Zhao, W.; Olsen, S.H.; Klinker, M.; et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J. Immunol. 2011, 187, 538–552. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, M.; Petretto, A.; Santucci, L.; Vaglio, A.; Pratesi, F.; Migliorini, P.; Bertelli, R.; Lavarello, C.; Bartolucci, M.; Candiano, G.; et al. Neutrophil Extracellular Traps protein composition is specific for patients with Lupus nephritis and includes methyl-oxidized alphaenolase (methionine sulfoxide 93). Sci. Rep. 2019, 9, 7934. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.J.; Radic, M. Neutrophil extracellular traps: Double-edged swords of innate immunity. J. Immunol. 2012, 189, 2689–2695. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.S.; Chen, C.H.; Cheng, C.C.; Wang, C.C.; Lin, H.C.; Luo, T.Y.; Lien, G.S.; Chang, J. Neutrophil elastase as a diagnostic marker and therapeutic target in colorectal cancers. Oncotarget 2014, 5, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Luo, Q.; Wu, J.; Shi, Y.; Guan, Q. Neutrophil elastase as a potential biomarker related to the prognosis of gastric cancer and immune cell infiltration in the tumor immune microenvironment. Sci. Rep. 2023, 13, 13447. [Google Scholar] [CrossRef]
- Taylor, S.; Dirir, O.; Zamanian, R.T.; Rabinovitch, M.; Thompson, A.A.R. The Role of Neutrophils and Neutrophil Elastase in Pulmonary Arterial Hypertension. Front. Med. 2018, 5, 217. [Google Scholar] [CrossRef]
- Thalin, C.; Aguilera, K.; Hall, N.W.; Marunde, M.R.; Burg, J.M.; Rosell, A.; Daleskog, M.; Mansson, M.; Hisada, Y.; Meiners, M.J.; et al. Quantification of citrullinated histones: Development of an improved assay to reliably quantify nucleosomal H3Cit in human plasma. J. Thromb. Haemost. 2020, 18, 2732–2743. [Google Scholar] [CrossRef]
- Mauracher, L.M.; Posch, F.; Martinod, K.; Grilz, E.; Daullary, T.; Hell, L.; Brostjan, C.; Zielinski, C.; Ay, C.; Wagner, D.D.; et al. Citrullinated histone H3, a biomarker of neutrophil extracellular trap formation, predicts the risk of venous thromboembolism in cancer patients. J. Thromb. Haemost. 2018, 16, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Heo, K.S.; Chang, E.; Le, N.T.; Cushman, H.; Yeh, E.T.; Fujiwara, K.; Abe, J. De-SUMOylation enzyme of sentrin/SUMO-specific protease 2 regulates disturbed flow-induced SUMOylation of ERK5 and p53 that leads to endothelial dysfunction and atherosclerosis. Circ. Res. 2013, 112, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmsen, K.; Xu, F.; Farrar, K.; Tran, A.; Khakpour, S.; Sundar, S.; Prakash, A.; Wang, J.; Gray, N.S.; Hellman, J. Extracellular signal-regulated kinase 5 promotes acute cellular and systemic inflammation. Sci. Signal 2015, 8, 86. [Google Scholar] [CrossRef]
- Kim, J.; Lee, Y.R.; Lee, C.H.; Choi, W.H.; Lee, C.K.; Kim, J.; Bae, Y.M.; Cho, S.; Kim, B. Mitogen-activated protein kinase contributes to elevated basal tone in aortic smooth muscle from hypertensive rats. Eur. J. Pharmacol. 2005, 514, 209–215. [Google Scholar] [CrossRef]
- McLimans, K.E.; Willette, A.A. Alzheimer’s Disease Neuroimaging Initiative: Autotaxin is Related to Metabolic Dysfunction and Predicts Alzheimer’s Disease Outcomes. J. Alzheimers Dis. 2017, 56, 403–413. [Google Scholar] [CrossRef]
- Araki, T.; Okumura, T.; Hiraiwa, H.; Mizutani, T.; Kimura, Y.; Kazama, S.; Shibata, N.; Oishi, H.; Kuwayama, T.; Kondo, T.; et al. Serum autotaxin as a novel prognostic marker in patients with non-ischaemic dilated cardiomyopathy. ESC Heart Fail. 2022, 9, 1304–1313. [Google Scholar] [CrossRef]
- Linton, M.F.; Fazio, S. Cyclooxygenase products and atherosclerosis. Drug Discov. Today Ther. Strateg. 2008, 5, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Schonbeck, U.; Sukhova, G.K.; Graber, P.; Coulter, S.; Libby, P. Augmented expression of cyclooxygenase-2 in human atherosclerotic lesions. Am. J. Pathol. 1999, 155, 1281–1291. [Google Scholar] [CrossRef] [PubMed]

| Granule | Type | Contents | Role |
|---|---|---|---|
| α-granules | Adhesive proteins | P-selectin, Fibrinogen, von Willebrand factor, Fibronectin, Thrombospondin-1/2, Laminin-8, Vitronectin |
|
| Growth factors | Epidermal-growth factor, Insulin-like-growth factor, Hepatocyte-growth factor, Platelet-derived-growth factor |
| |
| Angiogenic factors | Growth factor from vascular endothelium, Platelet-derived growth factor, Fibroblast |
| |
| Chemokines | CXCL8/7/1/5/2/6/12 |
| |
| CCL5/3/2/7 |
| ||
| IL1β |
| ||
| Clotting factors | Factor V |
| |
| Protein S |
| ||
| Factor XI |
| ||
| Factor XIII |
| ||
| Kininogens |
| ||
| Plasminogen |
| ||
| Integral membrane proteins | Integrin αIIbβ3, GPIba-IX-V, GPVI, TLT-1, P-selectin |
| |
| Immune mediators | Complement C3/C4 precursor Factor D/H, C1 inhibitor, Immunoglobulins |
| |
| Protease inhibitors | α2-antiplasmin, PAI-1, α2-antitrypsin, α2-macroglobulin, TFPI, C1-inhibitor |
| |
| Proteoglycans | MMP2/9 |
| |
| Dense granules | Amines | Serotonin, Histamine |
|
| Bivalent cations | Ca2+, Mg2+ | ||
| Nucleotides | ATP, ADP, GTP, GDP | ||
| Lysosome granules | Acid proteases | Cathepsin D and E, Carboxypeptidases (A, B), Prolinecarboxypeptidase, Collagenase, Acid phosphatase, Arylsulphatase | |
| Glycohydrolases | Heparinase, β-N-acetyl-glucosaminidase | ||
| Gel-Based (2D) | Gel-Free (LCMS/MS) | |
|---|---|---|
| Advantages |
|
|
| Disadvantages |
|
|
| Biomarker | Disease Conditions | References |
|---|---|---|
| Podoplanin | Tumor-induced platelet activation and tumor metastasis and invasion. | [99,100,101,102,103,104] |
| CD40 ligand | Acute coronary syndromes, coronary revascularization procedures, atherosclerosis, and inflammatory processes. | [105,106,107] |
| Platelet-derived growth factors (PDGFs) | Gliomas, sarcomas, leukemias, and epithelial cancers. | [108,109] |
| P-selectin | Coronary heart disease, hypertension, arterial fibrillation, congestive heart failure, stroke, atherosclerosis. | [110,111,112,113,114] |
| Glycoprotein IIb/IIIa | Platelet aggregation, thrombosis, hemostasis, carotid atherosclerosis, and diabetes. | [115,116,117,118] |
| Thrombospondin | Myocardial infarction, heart failure, coronary artery disease, coronary heart disease, abdominal aortic aneurysms. | [119,120,121,122] |
| Advanced glycation end products | Peripheral artery disease increases thrombotic effect in diabetes and coronary heart diseases. | [123,124,125] |
| Troponin | Myocardial infarction, heart failure, arterial fibrillation, Takotsubo cardiomyopathy, stroke, atherosclerosis. | [126,127,128,129,130], |
| Signal transducer and activator of transcription | Chronic inflammation, osteosarcoma, and prostate cancer. | [131,132] |
| Vascular endothelial growth factor | Breast cancer progression, invasion, and migration, angiogenesis. | [133,134,135] |
| β2-Glycoprotein I | Autoimmune condition antiphospholipid syndrome, thrombosis. | [136] |
| Oxidized LDL receptors | Atherosclerosis. | [137] |
| Vasodilator stimulated phosphoprotein | Metastasis in colorectal cancer. | [138] |
| Myeloperoxidase | Atherosclerosis, coronary artery disease, myocardial infarction, heart failure, inflammation, colon cancer, breast cancer. | [139,140,141] |
| RANTES | Acute coronary syndrome, atherosclerosis, inflammation. Development and progression of atherosclerosis, inflammation, thrombosis, diabetes, myocardial infarction, and atherothrombosis. | [142,143,144,145,146,147,148,149,150,151,152] |
| Platelet factor 4 | Liposarcoma, mammary adenocarcinoma, and osteosarcoma, inflammation, atherosclerosis, myocardial infraction. | [153,154,155,156] |
| VWF | Atherosclerosis, hepatic carcinoma, hemostasis and thrombus formation. | [157,158,159,160] |
| Beta amyloid precursor protein II | Alzheimer’s disease. | [161,162,163] |
| IL-1B | Atherosclerosis, inflammation, diabetes, coronary heart disease, stroke, peripheral vascular disease. | [164,165,166,167] |
| Autoantibody against platelet protein | Immune thrombocytopenia. | [168] |
| Facotor XII | Coronary heart disease, atherosclerosis, ischemic and hemorrhagic stroke, myocardial infraction. | [169,170,171,172] |
| ADAMTS13 | Thrombotic microangiopathies, Thrombotic thrombocytopenic purpura, hepatocellular carcinoma, peripheral arterial disease, coronary heart disease, stroke, heart failure, myocardial infarction, liver cirrhosis. | [158,173] |
| Neutrophil extracellular traps (NETs) interacting protein | Autoimmune and inflammatory disorders, atherosclerosis, thrombosis. | [174,175,176,177] |
| Neutrophil elastase | Colorectal cancer, gastric cancer, pulmonary arterial hypertension. | [178,179,180] |
| Citrullinated histones | Thrombosis, inflammation, thromboembolism. | [181,182] |
| ERK5 | Inflammation, atherosclerosis, hypertension. | [183,184,185] |
| Autotaxin | Alzheimer’s disease, ischemic dilated cardiomyopathy, calcified aortic valve stenosis. | [186,187] |
| Cyclooxygenase | Atherosclerosis, aneurysm | [188,189] |
| Platelet-derived microvesicle | Development and progression of atherosclerosis, inflammation, thrombosis, diabetes, myocardial infarction, and atherothrombosis. | [145,146,147,148,149,150,151,152] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaudhary, P.K.; Upadhayaya, S.; Kim, S.; Kim, S. The Perspectives of Platelet Proteomics in Health and Disease. Biomedicines 2024, 12, 585. https://doi.org/10.3390/biomedicines12030585
Chaudhary PK, Upadhayaya S, Kim S, Kim S. The Perspectives of Platelet Proteomics in Health and Disease. Biomedicines. 2024; 12(3):585. https://doi.org/10.3390/biomedicines12030585
Chicago/Turabian StyleChaudhary, Preeti Kumari, Sachin Upadhayaya, Sanggu Kim, and Soochong Kim. 2024. "The Perspectives of Platelet Proteomics in Health and Disease" Biomedicines 12, no. 3: 585. https://doi.org/10.3390/biomedicines12030585
APA StyleChaudhary, P. K., Upadhayaya, S., Kim, S., & Kim, S. (2024). The Perspectives of Platelet Proteomics in Health and Disease. Biomedicines, 12(3), 585. https://doi.org/10.3390/biomedicines12030585







