Specific Multiomic Profiling in Aortic Stenosis in Bicuspid Aortic Valve Disease
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. Aortic Tissue Extraction
2.3. Tissue Homogenization
2.4. RNA-Seq
2.5. Gene Enrichment Analysis
2.6. Lipid Profile and Low-Molecular-Weight Metabolite Quantification by 1H-NMR
2.7. Statistical Analysis and Multiomics Integrative Analysis
2.8. Ethics
3. Results
3.1. Clinical Characteristics of the Cohort
3.2. RNA Pattern Depends on Aortic Valve Morphology in AS
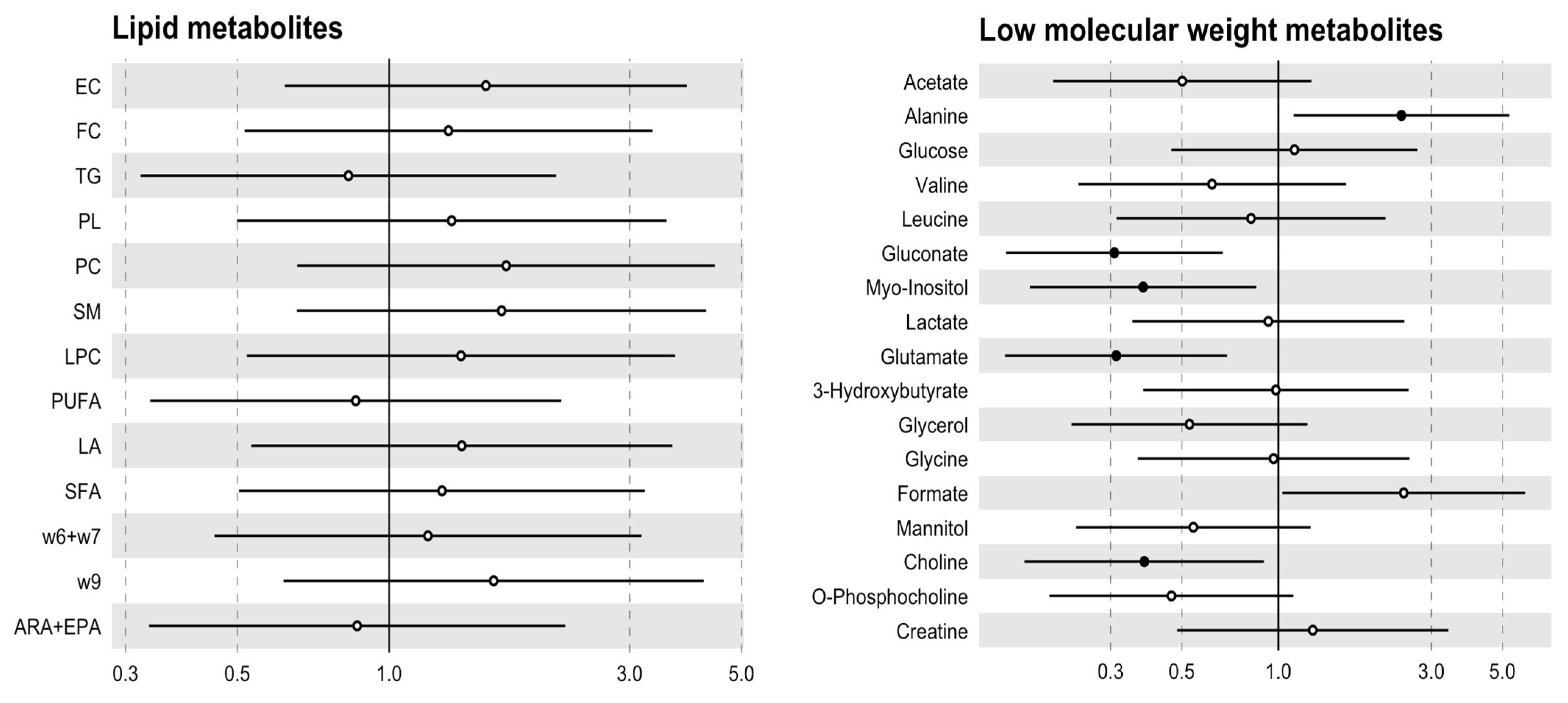
3.3. The Metabolic Profile Seems to Be Specified by the Aortic Valve Morphology
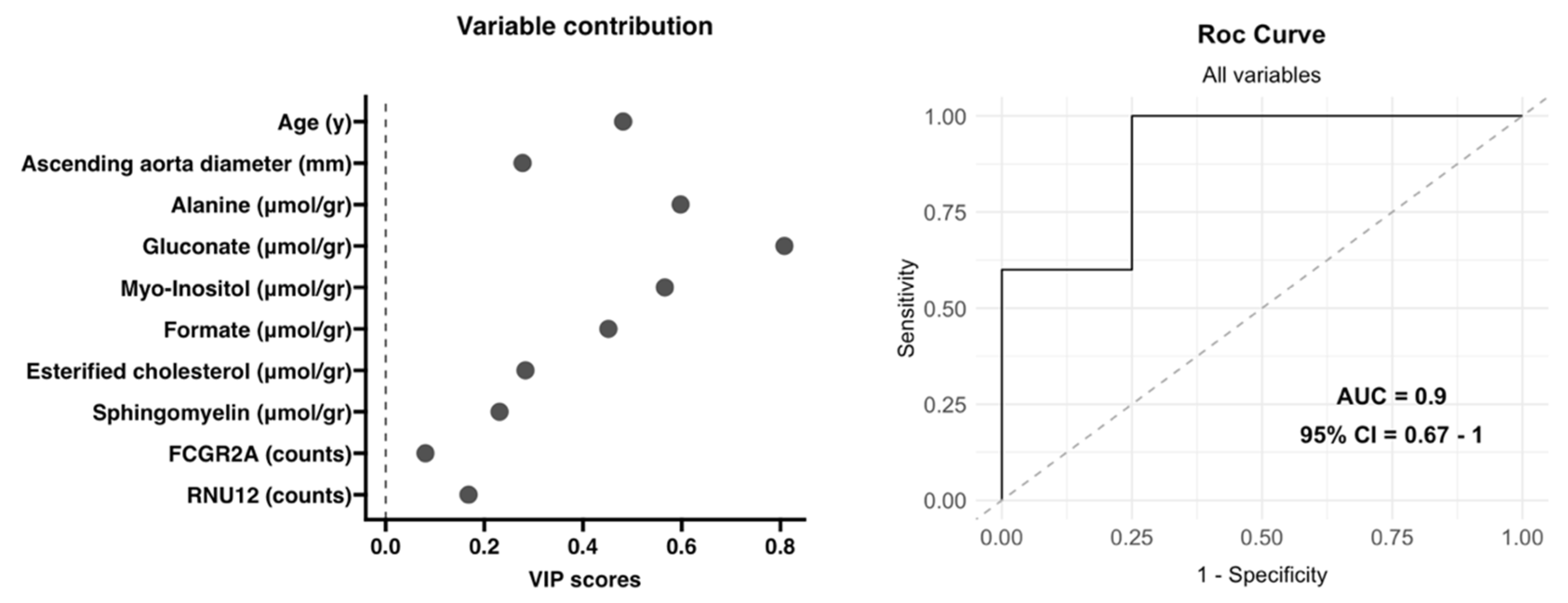
3.4. Integrative Analysis of Clinical, 1H-NMR and RNA-Seq Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AS | aortic stenosis |
| BAV | bicuspid aortic valve |
| mTAVG | mean transaortic valvular gradient |
| RNA-seq | RNA sequencing |
| TAV | tricuspid aortic valve |
| 1H-NMR | proton nuclear magnetic resonance spectroscopy |
References
- Joseph, J.; Naqvi, S.Y.; Giri, J.; Goldberg, S. Aortic Stenosis: Pathophysiology, Diagnosis, and Therapy. Am. J. Med. 2017, 130, 253–263. [Google Scholar] [CrossRef]
- Boskovski, M.T.; Gleason, T.G. Current Therapeutic Options in Aortic Stenosis. Circ. Res. 2021, 128, 1398–1417. [Google Scholar] [CrossRef] [PubMed]
- Vincent, F.; Ternacle, J.; Denimal, T.; Shen, M.; Redfors, B.; Delhaye, C.; Simonato, M.; Debry, N.; Verdier, B.; Shahim, B.; et al. Transcatheter Aortic Valve Replacement in Bicuspid Aortic Valve Stenosis. Circulation 2021, 143, 1043–1061. [Google Scholar] [CrossRef] [PubMed]
- Ward, C. Clinical significance of the bicuspid aortic valve. Heart 2000, 83, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Osler, S.W. The Bicuspid Condition of the Aortic Valves; Wiliam J. Dornan: Philadelphia, PA, USA, 1886. [Google Scholar]
- Hoffman, J.I.E.; Kaplan, S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002, 39, 1890–1900. [Google Scholar] [CrossRef] [PubMed]
- Atkins, S.K.; Sucosky, P. Etiology of bicuspid aortic valve disease: Focus on hemodynamics. World J. Cardiol. 2014, 6, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- Grewal, N.; Girdauskas, E.; DeRuiter, M.; Goumans, M.-J.; Poelmann, R.E.; Klautz, R.J.M.; Groot, A.C.G.-D. The role of hemodynamics in bicuspid aortopathy: A histopathologic study. Cardiovasc. Pathol. 2019, 41, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Çiçek, M.S. Altered hemodynamics in bicuspid aortic valve disease: Leaning more toward nurture! J. Thorac. Cardiovasc. Surg. 2017, 153, S63–S64. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dutta, P.; James, J.F.; Kazik, H.; Lincoln, J. Genetic and Developmental Contributors to Aortic Stenosis. Circ. Res. 2021, 128, 1330–1343. [Google Scholar] [CrossRef]
- Bhatia, N.; Basra, S.S.; Skolnick, A.H.; Wenger, N.K. Aortic valve disease in the older adult. J. Geriatr. Cardiol. 2016, 13, 941–944. [Google Scholar] [CrossRef]
- Huntley, G.D.; Thaden, J.J.; Alsidawi, S.; Michelena, H.I.; Maleszewski, J.J.; Edwards, W.D.; Scott, C.G.; Pislaru, S.V.; Pellikka, P.A.; Greason, K.L.; et al. Comparative study of bicuspid vs. tricuspid aortic valve stenosis. Eur. Heart J. Cardiovasc. Imaging 2018, 19, 3–8. [Google Scholar] [CrossRef]
- Yotti, R.; Bermejo, J.; Gutiérrez-Ibañes, E.; del Villar, C.P.; Mombiela, T.; Elízaga, J.; Benito, Y.; González-Mansilla, A.; Barrio, A.; Rodríguez-Pérez, D.; et al. Systemic Vascular Load in Calcific Degenerative Aortic Valve Stenosis: Insight from Percutaneous Valve Replacement. J. Am. Coll. Cardiol. 2015, 65, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Kaden, J.J.; Dempfle, C.-E.; Grobholz, R.; Tran, H.-T.; Kiliç, R.; Sarikoç, A.; Brueckmann, M.; Vahl, C.; Hagl, S.; Haase, K.K.; et al. Interleukin-1 beta promotes matrix metalloproteinase expression and cell proliferation in calcific aortic valve stenosis. Atherosclerosis 2003, 170, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Zemskov, E.A.; Lu, Q.; Ornatowski, W.; Klinger, C.N.; Desai, A.A.; Maltepe, E.; Yuan, J.X.-J.; Wang, T.; Fineman, J.R.; Black, S.M. Biomechanical Forces and Oxidative Stress: Implications for Pulmonary Vascular Disease. Antioxid. Redox Signal. 2019, 31, 819–842. [Google Scholar] [CrossRef]
- Akahori, H.; Tsujino, T.; Masuyama, T.; Ishihara, M. Mechanisms of aortic stenosis. J. Cardiol. 2018, 71, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Pedriali, G.; Morciano, G.; Patergnani, S.; Cimaglia, P.; Morelli, C.; Mikus, E.; Ferrari, R.; Gasbarro, V.; Giorgi, C.; Wieckowski, M.R.; et al. Aortic Valve Stenosis and Mitochondrial Dysfunctions: Clinical and Molecular Perspectives. Int. J. Mol. Sci. 2020, 21, 4899. [Google Scholar] [CrossRef] [PubMed]
- Milin, A.C.; Vorobiof, G.; Aksoy, O.; Ardehali, R. Insights Into Aortic Sclerosis and Its Relationship With Coronary Artery Disease. J. Am. Heart Assoc. 2014, 3, e001111. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zhang, X.; An, P.; Luo, J.; Luo, Y. Mitochondrial Homeostasis in VSMCs as a Central Hub in Vascular Remodeling. Int. J. Mol. Sci. 2023, 24, 3483. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.C.; Kraler, S.; Lüscher, T.F.; Aikawa, E. Multi-Omics Approaches to Define Calcific Aortic Valve Disease Pathogenesis. Circ. Res. 2021, 128, 1371–1397. [Google Scholar] [CrossRef]
- Aboumsallem, J.P.; Shi, C.; Wit, S.D.; Markousis-Mavrogenis, G.; Bracun, V.; Eijgenraam, T.R.; Hoes, M.F.; Meijers, W.C.; Screever, E.M.; Schouten, M.E.; et al. Multi-omics analyses identify molecular signatures with prognostic values in different heart failure aetiologies. J. Mol. Cell. Cardiol. 2023, 175, 13–28. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Löfgren, L.; Ståhlman, M.; Forsberg, G.-B.; Saarinen, S.; Nilsson, R.; Hansson, G.I. The BUME method: A novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res. 2012, 53, 1690–1700. [Google Scholar] [CrossRef] [PubMed]
- Barrilero, R.; Gil, M.; Amigó, N.; Dias, C.B.; Wood, L.G.; Garg, M.L.; Ribalta, J.; Heras, M.; Vinaixa, M.; Correig, X. LipSpin: A New Bioinformatics Tool for Quantitative 1H NMR Lipid Profiling. Anal. Chem. 2018, 90, 2031–2040. [Google Scholar] [CrossRef] [PubMed]
- Vinaixa, M.; Rodríguez, M.A.; Rull, A.; Beltrán, R.; Bladé, C.; Brezmes, J.; Cañellas, N.; Joven, J.; Correig, X. Metabolomic assessment of the effect of dietary cholesterol in the progressive development of fatty liver disease. J. Proteome Res. 2010, 9, 2527–2538. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.-Z.; Jiménez, J.M.; Ou, K.; McCormick, M.E.; Zhang, L.-D.; Davies, P.F. Hemodynamic Disturbed Flow Induces Differential DNA Methylation of Endothelial Kruppel-Like Factor 4 (KLF4) Promoter in vitro and in vivo. Circ. Res. 2014, 115, 32–43. [Google Scholar] [CrossRef]
- Tzima, E.; Irani-Tehrani, M.; Kiosses, W.B.; Dejana, E.; Schultz, D.A.; Engelhardt, B.; Cao, G.; DeLisser, H.; Schwartz, M.A. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature 2005, 437, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.B.; Ettenson, D.S.; Jonas, M.; Nugent, M.A.; Iozzo, R.V.; Edelman, E.R. Endothelial Cells Provide Feedback Control for Vascular Remodeling Through a Mechanosensitive Autocrine TGF-β Signaling Pathway. Circ. Res. 2008, 103, 289–297. [Google Scholar] [CrossRef]
- Niu, N.; Xu, S.; Xu, Y.; Little, P.J.; Jin, Z. Targeting mechanosensitive transcription factors in atherosclerosis. Trends Pharmacol. Sci. 2019, 40, 253–266. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, C.K.; Yi, M.; Lui, K.O.; Huang, Y. Targeting endothelial dysfunction and inflammation. J. Mol. Cell. Cardiol. 2022, 168, 58–67. [Google Scholar] [CrossRef]
- Katayama, S.; Umetani, N.; Hisada, T.; Sugiura, S. Bicuspid aortic valves undergo excessive strain during opening: A simulation study. J. Thorac. Cardiovasc. Surg. 2013, 145, 1570–1576. [Google Scholar] [CrossRef][Green Version]
- Yamamoto, K.; Nogimori, Y.; Imamura, H.; Ando, J. Shear stress activates mitochondrial oxidative phosphorylation by reducing plasma membrane cholesterol in vascular endothelial cells. Proc. Natl. Acad. Sci. USA 2020, 117, 33660–33667. [Google Scholar] [CrossRef] [PubMed]
- Antequera-González, B.; Martínez-Micaelo, N.; Alegret, J.M. Bicuspid Aortic Valve and Endothelial Dysfunction: Current Evidence and Potential Therapeutic Targets. Front. Physiol. 2020, 11, 1015. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-H.; Chang, H.-C.; Ting, P.-C.; Wang, D.L. Laminar shear stress promotes mitochondrial homeostasis in endothelial cells. J. Cell. Physiol. 2018, 233, 5058–5069. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Shkurat, T.P.; Melnichenko, A.A.; Grechko, A.V.; Orekhov, A.N. The role of mitochondrial dysfunction in cardiovascular disease: A brief review. Ann. Med. 2018, 50, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial Reactive Oxygen Species (ROS) and ROS-Induced ROS Release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.W.; Nargund, A.M.; Haynes, C.M. Signaling the mitochondrial unfolded protein response. Biochim. Biophys. Acta 2013, 1833, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Vásquez-Trincado, C.; García-Carvajal, I.; Pennanen, C.; Parra, V.; Hill, J.A.; Rothermel, B.A.; Lavandero, S. Mitochondrial dynamics, mitophagy and cardiovascular disease. J. Physiol. 2016, 594, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C. Mitochondrial Dynamics and Its Involvement in Disease. Annu. Rev. Pathol. 2020, 15, 235–259. [Google Scholar] [CrossRef]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef]
- Wollenman, L.C.; Ploeg, M.R.V.; Miller, M.L.; Zhang, Y.; Bazil, J.N. The effect of respiration buffer composition on mitochondrial metabolism and function. PLoS ONE 2017, 12, e0187523. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-C.; Zhang, X.; Wang, G.; Zhang, W.; Cai, Z.; Pan, B.-S.; Gu, H.; Xu, C.; Jin, G.; Xu, X.; et al. Inositol serves as a natural inhibitor of mitochondrial fission by directly targeting AMPK. Mol. Cell 2021, 81, 3803–3819.e7. [Google Scholar] [CrossRef]
- Shetewy, A.; Shimada-Takaura, K.; Warner, D.; Jong, C.J.; Mehdi, A.-B.A.; Alexeyev, M.; Takahashi, K.; Schaffer, S.W. Mitochondrial defects associated with β-alanine toxicity: Relevance to hyper-beta-alaninemia. Mol. Cell. Biochem. 2016, 416, 11–22. [Google Scholar] [CrossRef]
- Hofmann, S.; Lichtner, P.; Schuffenhauer, S.; Gerbitz, K.D.; Meitinger, T. Assignment of the human genes coding for cytochrome c oxidase subunits Va (COX5A), VIc (COX6C) and VIIc (COX7C) to chromosome bands 15q25, 8q22→q23 and 5q14 and of three pseudogenes (COX5AP1, COX6CP1, COX7CP1) to 14q22, 16p12 and 13q14→q21 by FISH and radiation hybrid mapping. Cytogenet. Cell Genet. 1998, 83, 226–227. [Google Scholar] [CrossRef] [PubMed]
- Seelan, R.S.; Grossman, L.I. Structural organization and promoter analysis of the bovine cytochrome c oxidase subunit VIIc gene. A functional role for YY1. J. Biol. Chem. 1997, 272, 10175–10181. [Google Scholar] [CrossRef]
- Frey, J.; Janes, M.; Engelhardt, W.; Afting, E.G.; Geerds, C.; Möller, B. Fc gamma-receptor-mediated changes in the plasma membrane potential induce prostaglandin release from human fibroblasts. Eur. J. Biochem. 1986, 158, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.-F.; Wu, J.; Wu, Y.; Huang, W.; Liu, M.; Dong, Z.-R.; Xu, B.-Y.; Jin, Y.; Wang, F.; Zhang, X.-M. The lncRNA SCARNA2 mediates colorectal cancer chemoresistance through a conserved microRNA-342-3p target sequence. J. Cell. Physiol. 2019, 234, 10157–10165. [Google Scholar] [CrossRef]
- Elsaid, M.F.; Chalhoub, N.; Ben-Omran, T.; Kumar, P.; Kamel, H.; Ibrahim, K.; Mohamoud, Y.; Al-Dous, E.; Al-Azwani, I.; Malek, J.A.; et al. Mutation in noncoding RNA RNU12 causes early onset cerebellar ataxia. Ann. Neurol. 2017, 81, 68–78. [Google Scholar] [CrossRef]
- Bouchareb, R.; Guauque-Olarte, S.; Snider, J.; Zaminski, D.; Anyanwu, A.; Stelzer, P.; Lebeche, D. Proteomic Architecture of Valvular Extracellular Matrix: FNDC1 and MXRA5 Are New Biomarkers of Aortic Stenosis. JACC Basic Transl. Sci. 2021, 6, 25–39. [Google Scholar] [CrossRef]
- Surendran, A.; Edel, A.; Chandran, M.; Bogaert, P.; Hassan-Tash, P.; Asokan, A.K.; Hiebert, B.; Solati, Z.; Sandhawalia, S.; Raabe, M.; et al. Metabolomic Signature of Human Aortic Valve Stenosis. JACC Basic Transl. Sci. 2020, 5, 1163–1177. [Google Scholar] [CrossRef]
- Martínez-Micaelo, N.; Ligero, C.; Antequera-González, B.; Junza, A.; Yanes, O.; Alegret, J.M. Plasma Metabolomic Profiling Associates Bicuspid Aortic Valve Disease and Ascending Aortic Dilation with a Decrease in Antioxidant Capacity. J. Clin. Med. 2020, 9, 2215. [Google Scholar] [CrossRef] [PubMed]
- Poggio, P.; Songia, P.; Moschetta, D.; Valerio, V.; Myasoedova, V.; Perrucci, G.L.; Pompilio, G. MiRNA profiling revealed enhanced susceptibility to oxidative stress of endothelial cells from bicuspid aortic valve. J. Mol. Cell. Cardiol. 2019, 131, 146–154. [Google Scholar] [CrossRef] [PubMed]
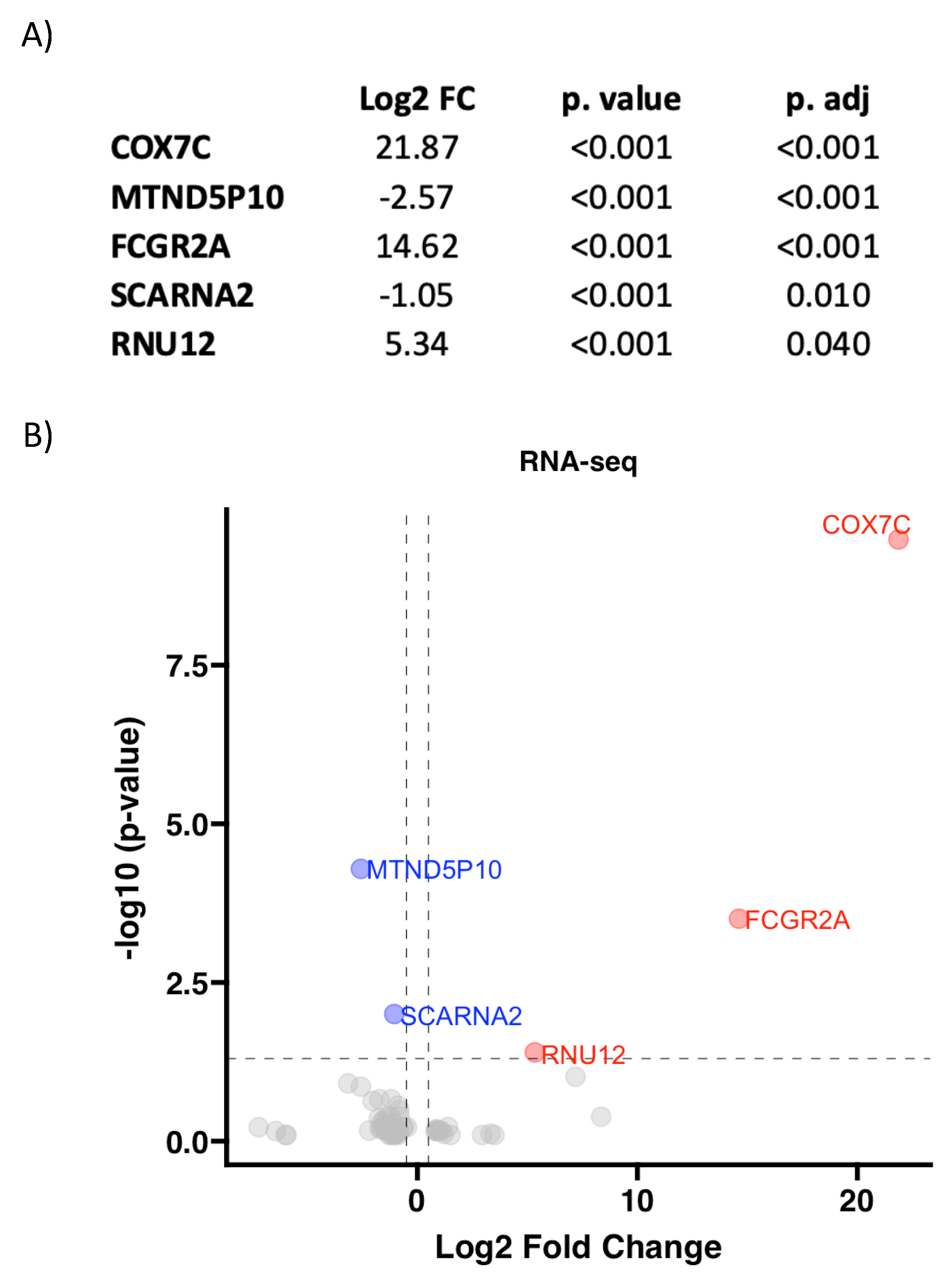


| TAV (n = 8) | BAV (n = 10) | p Value | |
|---|---|---|---|
| Sex | 1.000 | ||
| Female | 2 (25.0%) | 3 (30.0%) | |
| Age (year) | 77.5 [76.8;78.5] | 65.5 [61.2;73.2] | 0.008 |
| Weight (kg) | 78.0 [66.2;106] | 75.5 [66.5;82.8] | 0.699 |
| Height (cm) | 168 [162;172] | 168 [156;176] | 0.948 |
| BMI (kg/m2) | 27.5 [26.4;36.1] | 26.9 [25.5;28.1] | 0.439 |
| Hypertension | 7 (87.5%) | 4 (40.0%) | 0.066 |
| Diabetes mellitus | 6 (75.0%) | 2 (20.0%) | 0.054 |
| Dyslipidemia | 6 (75.0%) | 3 (30.0%) | 0.153 |
| Smoking | 0.588 | ||
| Ex | 2 (25.0%) | 1 (10.0%) | |
| No | 6 (75.0%) | 7 (70.0%) | |
| Yes | 0 (0.00%) | 2 (20.0%) | |
| Statins treatment | 5 (62.5%) | 3 (30.0%) | 0.342 |
| Renal insufficiency | 1 (12.5%) | 1 (10.0%) | 1.000 |
| Ischemic heart disease | 5 (62.5%) | 2 (20.0%) | 0.145 |
| mTAVG (mmHg) | 45.0 [42.5;50.0] | 55.5 [49.0;62.8] | 0.062 |
| AR (≤II) | 1 (12.5%) | 4 (40.0%) | 0.348 |
| LVDD (mm) | 48.0 [45.0;52.0] | 46.0 [40.8;48.0] | 0.447 |
| LVSD (mm) | 33.0 [29.5;37.5] | 30.0 [24.0;35.0] | 0.306 |
| LVEF (%) | 60.0 [46.8;64.6] | 60.5 [56.2;69.5] | 0.475 |
| Ascending aorta (mm) | 36.5 [33.5;38.8] | 41.5 [37.8;43.0] | 0.068 |
| Aortic root (mm) | 36.0 [31.0;37.0] | 36.5 [34.0;42.8] | 0.349 |
| Metabolites | TAV (n = 8) | BAV (n = 10) | p Value |
|---|---|---|---|
| Acetate (μmol/gr) | 0.79 [0.18;1.43] | 0.50 [0.21;0.67] | 0.183 |
| Alanine (μmol/gr) | 0.08 [0.06;0.13] | 0.15 [0.11;0.19] | 0.046 |
| Glucose (μmol/gr) | 1.05 [0.83;1.83] | 1.48 [0.74;1.73] | 0.753 |
| Valine (μmol/gr) | 0.07 [0.07;0.08] | 0.07 [0.06;0.08] | 0.549 |
| Isoleucine (μmol/gr) | 0.03 [0.03;0.04] | 0.04 [0.03;0.06] | 0.201 |
| Leucine (μmol/gr) | 0.06 [0.04;0.07] | 0.05 [0.04;0.07] | 0.655 |
| Gluconate (μmol/gr) | 0.65 [0.45;1.19] | 0.27 [0.09;0.44] | 0.045 |
| Myo-Inositol (μmol/gr) | 0.38 [0.36;0.42] | 0.23 [0.19;0.31] | 0.188 |
| Lactate (μmol/gr) | 2.93 [2.08;3.76] | 2.96 [2.29;4.17] | 0.929 |
| Glutamate (μmol/gr) | 0.38 [0.30;0.45] | 0.25 [0.13;0.31] | 0.126 |
| Pyruvate (μmol/gr) | 0.02 [0.01;0.02] | 0.02 [0.01;0.02] | 0.462 |
| Glutamine (μmol/gr) | 0.28 [0.19;0.29] | 0.23 [0.18;0.31] | 0.655 |
| 3-Hydroxybutyrate (μmol/gr) | 0.04 [0.03;0.18] | 0.08 [0.07;0.10] | 0.380 |
| Glycerol (μmol/gr) | 0.04 [0.03;0.07] | 0.04 [0.02;0.05] | 0.286 |
| Glycine (μmol/gr) | 0.11 [0.06;0.14] | 0.14 [0.09;0.17] | 0.657 |
| Formate (μmol/gr) | 0.16 [0.14;0.19] | 0.22 [0.18;0.27] | 0.050 |
| Mannitol (μmol/gr) | 0.92 [0.56;1.43] | 0.34 [0.26;0.75] | 0.167 |
| Choline (μmol/gr) | 0.08 [0.06;0.09] | 0.05 [0.04;0.06] | 0.041 |
| O-Phosphocholine (μmol/gr) | 0.16 [0.09;0.16] | 0.10 [0.08;0.12] | 0.149 |
| Creatine (μmol/gr) | 0.04 [0.04;0.05] | 0.04 [0.03;0.07] | 0.886 |
| Histidine (μmol/gr) | 0.08 [0.08;0.08] | 6.47 [4.02;8.91] | 0.221 |
| Lipids | TAV (n = 8) | BAV (n = 10) | p Value |
|---|---|---|---|
| Esterified cholesterol (μmol/gr) | 61.3 [53.5;73.5] | 76.1 [68.6;93.0] | 0.214 |
| Free cholesterol (μmol/gr) | 43.5 [36.9;47.8] | 51.0 [35.4;56.7] | 0.477 |
| Triglycerides (μmol/gr) | 12.0 [9.29;14.2] | 15.3 [9.77;19.4] | 0.594 |
| Glycerophospholipids (μmol/gr) | 10.8 [10.3;11.1] | 11.5 [10.8;12.0] | 0.286 |
| Phosphatidylcholine (μmol/gr) | 8.23 [7.48;9.45] | 9.57 [7.83;10.3] | 0.248 |
| Sphingomyelin (μmol/gr) | 13.7 [11.6;15.1] | 17.7 [12.2;22.2] | 0.214 |
| Lysophosphatidylcholine (μmol/gr) | 1.76 [1.60;1.92] | 2.12 [1.49;2.20] | 0.328 |
| Polyunsaturated fatty acids (μmol/gr) | 129 [115;158] | 128 [110;143] | 0.790 |
| Linoleic acid (μmol/gr) | 53.6 [47.9;56.9] | 59.1 [48.9;68.7] | 0.286 |
| Saturated fatty acids (μmol/gr) | 77.2 [71.6;81.2] | 81.1 [67.8;91.8] | 0.424 |
| Omega-6 and fatty acids (μmol/gr) | 16.9 [13.6;18.2] | 19.6 [14.8;21.6] | 0.374 |
| Omega-9 fatty acids (μmol/gr) | 49.4 [46.3;72.8] | 69.9 [57.7;84.5] | 0.248 |
| ARA + EPA (μmol/gr) | 16.1 [15.2;22.1] | 16.3 [15.4;19.6] | 0.790 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antequera-González, B.; Martínez-Micaelo, N.; Sureda-Barbosa, C.; Galian-Gay, L.; Siliato-Robles, M.S.; Ligero, C.; Evangelista, A.; Alegret, J.M. Specific Multiomic Profiling in Aortic Stenosis in Bicuspid Aortic Valve Disease. Biomedicines 2024, 12, 380. https://doi.org/10.3390/biomedicines12020380
Antequera-González B, Martínez-Micaelo N, Sureda-Barbosa C, Galian-Gay L, Siliato-Robles MS, Ligero C, Evangelista A, Alegret JM. Specific Multiomic Profiling in Aortic Stenosis in Bicuspid Aortic Valve Disease. Biomedicines. 2024; 12(2):380. https://doi.org/10.3390/biomedicines12020380
Chicago/Turabian StyleAntequera-González, Borja, Neus Martínez-Micaelo, Carlos Sureda-Barbosa, Laura Galian-Gay, M. Sol Siliato-Robles, Carmen Ligero, Artur Evangelista, and Josep M. Alegret. 2024. "Specific Multiomic Profiling in Aortic Stenosis in Bicuspid Aortic Valve Disease" Biomedicines 12, no. 2: 380. https://doi.org/10.3390/biomedicines12020380
APA StyleAntequera-González, B., Martínez-Micaelo, N., Sureda-Barbosa, C., Galian-Gay, L., Siliato-Robles, M. S., Ligero, C., Evangelista, A., & Alegret, J. M. (2024). Specific Multiomic Profiling in Aortic Stenosis in Bicuspid Aortic Valve Disease. Biomedicines, 12(2), 380. https://doi.org/10.3390/biomedicines12020380





