Abstract
Protein–protein interactions (PPIs) are fundamental to many critical biological processes and are crucial in mediating essential cellular functions across diverse organisms, including bacteria, parasites, and viruses. A notable example is the interaction between the SARS-CoV-2 spike (S) protein and the human angiotensin-converting enzyme 2 (hACE2), which initiates a series of events leading to viral replication. Interrupting this interaction offers a promising strategy for blocking or significantly reducing infection, highlighting its potential as a target for anti-SARS-CoV-2 therapies. This review focuses on the hACE2 and SARS-CoV-2 spike protein interaction, exemplifying the latest advancements in peptide-based strategies for developing PPI inhibitors. We discuss various approaches for creating peptide-based inhibitors that target this critical interaction, aiming to provide potential treatments for COVID-19.
1. Introduction
For many decades, peptides were underrated as therapeutic agents due to inherent challenges such as high molecular weight, poor solubility, membrane impermeability, and biological instability [1]. However, the development of peptide-based drugs has recently become a prominent topic in the pharmaceutical industry and in scientific research [2]. As essential functional molecules in nature, peptides are well suited to interact with biological targets, leading to their resurgence as valuable building blocks in medicinal chemistry and promising candidates for drug discovery. Peptides possess unique features that are often unattainable by small molecules, such as a high specificity, strong efficacy, low toxicity, and the ability to target larger surface areas, including protein receptors and protein–protein interaction (PPI) interfaces [3,4,5,6,7].
PPIs are involved in many critical biological processes in living organisms and play a crucial role in mediating many important cellular functions, including vital biosynthetic pathways in bacteria, parasites, and viruses [8]. A subset of pathogen PPIs, which are essential for pathogenesis, engage in competitive binding with host proteins—a phenomenon known as cross-species interactions [9]. By mimicking the interface between host proteins, pathogens can hijack critical cellular functions for their own benefit, such as blocking the host’s immune response or facilitating viral entry into cells [10].
Paradoxically, one well-studied system is the interaction between the SARS-CoV-2 spike protein (S) and its human protein receptor, angiotensin-converting enzyme 2 (hACE2), which was only recently discovered and characterized during the outbreak of coronavirus disease (COVID-19) [11]. The interaction between the S protein and the hACE2 receptor triggers a cascade of events that ultimately lead to viral replication [12]. Disrupting the recognition process of the hACE2 receptor by the S protein could block or significantly reduce infection by the novel pathogen, making it a promising target for potential anti-SARS-CoV-2 therapies. Thus, many studies have focused on identifying small molecules that target the S protein or the hACE2 receptor to disrupt their complex formation and potentially prevent SARS-CoV-2 infection [13,14,15]. An approach that uses peptides to inhibit this crucial recognition process, which is essential for viral cell penetration, may offer an innovative treatment option to prevent the spread of the SARS-CoV-2 virus.
We chose the interaction between hACE2 and the SARS-CoV-2 spike protein as an exemplary case of current, state-of-the-art advancements in peptide-based strategies for designing and developing protein–protein interaction inhibitors. In this review, we also summarize the various approaches to developing peptide-based inhibitors, targeting the hACE2 and S interaction as potential treatments for COVID-19. To examine previously reported therapeutic peptides as inhibitors of S-hACE2 interaction, we searched PubMed using the keywords “peptides as inhibitors of SARS-CoV-2”. From the results, we focused exclusively on studies that experimentally demonstrated the ability of these peptides to block the interaction or to interact with one of the protein partners involved in the complex formation. The studies we found were divided into two groups: the first group contained native peptides, while the second group consisted of optimized peptides, primarily modified by natural amino acid residues.
2. Complex Structure of hACE and Spike Proteins
The SARS-CoV-2 contains envelope (E), membrane (M), nucleocapsid (N), and spike (S) structural proteins [16,17]. The latter, which is crucial for the virus’s entry into host cells, is composed of two subunits, S1 and S2. The S1 subunit includes the receptor-binding domain (RBD) that specifically attaches to the hACE2 receptor on host cells, primarily epithelial cells in the respiratory tract. The S2 subunit facilitates the fusion of the viral membrane with the host–cell membrane, a critical step for viral entry [18,19]. The S protein by RBD recognizes the extracellular peptidase domain (PD) of the hACE2 receptor of host epithelial cells, which initiates infection by virus attachment and its entry into the cells [11,20,21].
The structure of hACE2 and the spike of the SARS-CoV-2 (RBD) proteins complex were first proposed by homology modeling [22]; these have since been supported by experimental techniques such as cryo-electron microscopy [23] and X-ray diffraction crystallography [11,21,23,24]. The SARS-CoV-2 spike RBD has a significant interaction interface with the N-terminal α1 helix (residues S19-T52) of hACE2. This fragment engages with the outer surface of the viral receptor-binding domain (RBD) and features a substantial buried surface area of 1688 Å2. This area is composed of 19 residues from hACE2 (825 Å2) and 17 residues from the viral protein (863 Å2) [11] (Table 1/Figure 1). The interaction is predominantly polar, involving 11 hydrogen bonds and one salt bridge between the K417 of the RBD and the D38 of hACE2 (Table 1/Figure 1). These interactions stabilize the complex, resulting in an equilibrium dissociation constant (KD) of approximately 14.7 nM [25].

Table 1.
Interaction interface data.
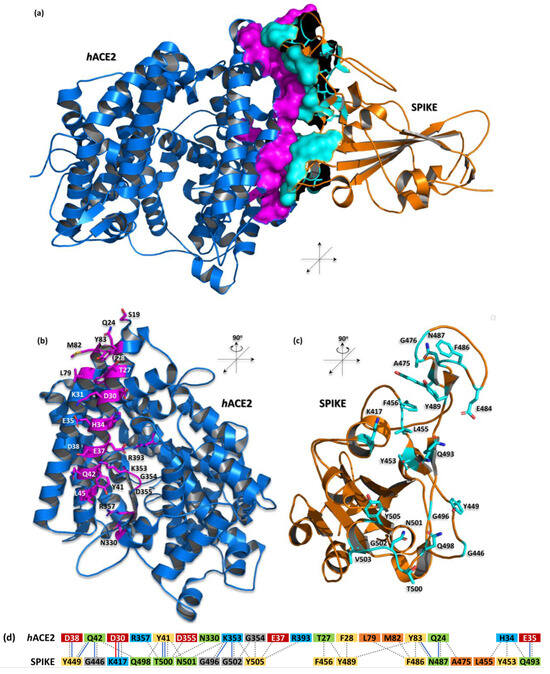
Figure 1.
The complex structure of hACE2 and SARS-CoV-2 spike proteins (PDB id: 6m0j)— computer-generated, cartoon representation. (a) The interaction interface between hACE2 (blue) and the spike (orange) is shown as the solvent accessible surface area and highlighted by magenta and cyan colors, respectively; (b,c) depicted amino acid residues forming the interface for a particular protein are shown as sticks in this representation; (d) schematic diagram of interactions between proteins. Residues are colored according to the type: positive (H, K, R); negative (D, E); S, T, N, Q = neutral; A, V, L, I, M = aliphatic; F, Y, W = aromatic; G = Gly. Type of contacts: hydrogen bonds (blue line); salt bridges (red line); nonbonded contacts (gray dash line). Protein visualization was prepared by the PyMOL Molecular Graphics System, Version 3.0.0 Schrödinger, LLC.
3. Native Peptides
Several studies have demonstrated that peptides derived directly from the native sequence of one of the interacting partners serve as valuable starting points for developing effective PPI inhibitors [26,27,28,29]. Since the structure of the hACE2 and SARS-CoV-2 spike protein complex has been known since the early days of the pandemic [23], efforts to find peptides that effectively block this interaction began immediately after the first COVID-19 cases, employing various approaches including small molecules screening, peptide design, the production of monoclonal antibodies, and chimeric proteins.
However, it is important to note that the concept of blocking the interaction between the hACE2 receptor and viral protein was not new, even within the field of coronaviruses. The initial studies demonstrating this antiviral strategy were published by Ho and colleagues nearly two decades before first COVID-19 case was detected in humans [30]. Their research showed that small peptides derived from the viral S protein of SARS-CoV could effectively block the interaction between hACE2 and the S protein of the closely related virus to SARS-CoV-2. Out of the 14 peptides tested, 4 were identified as active and successfully inhibited the hACE2-S interactions, with a half maximal inhibitory concentration (IC50) in the low nanomolar range [30]. This foundational work provided a basis for similar approaches during the COVID-19 pandemic.
Following the identification of SARS-CoV-2 [31,32], the first research papers demonstrating the efficacy of native fragment peptides emerged a few months later [33]. This initial research paved the way for a prolific output of studies on peptides and peptide-derived molecules, which have shown considerable therapeutic potential, as documented extensively in the scientific literature.
The knowledge about the structure of complex SARS-CoV-2 and hACE2 proteins, along with the experience gained with SARS-CoV [34], initiated studies which resulted in a number of publications that demonstrated promising outcomes on the use of peptide inhibitors; these were designed by combining the critical residues in the protein interface to prevent the entrance of SARS-CoV-2.
One of the first studies that experimentally validated peptides that may block the interaction between hACE2 and the S protein was conducted by Larue and colleagues [33]. Their study involved a rational analysis of the interacting interfaces of both proteins, focusing primarily on the 27-TFLDKFNHEAEDLFYQ-42 fragment of the N-terminal α1 helix of the hACE2 receptor. They proposed four native sequence fragments from α1 for peptide synthesis: SAP1, SAP2, SAP5, and SAP6, containing 16, 9, 13, and 6 residues, respectively (Table 2). Additionally, they investigated two peripheral regions of the hACE2 interface, resulting in peptides SAP3 and SAP4, which contained 7 residues from α3 and 8 residues from α11, respectively. The synthetic peptides SAP1 (16-mer) and SAP6 (6-mer) have similar binding affinities to the viral protein, with KD values of 0.53 ± 0.01 mM and 1.36 ± 0.14 mM, respectively.

Table 2.
Sequences of native peptides based on hACE2.
Based on the luciferase assay system results, they revealed that the inhibitory effects of the hACE2-derived peptides on pseudovirus infection were comparable, with IC50 values of 2.39 mM for SAP1 and 1.90 mM for SAP6. In contrast, SAP3, SAP4, and SAP5 were identified as poor or non-inhibitors of spike-mediated virus infection. According to their findings, the minimum number of residues required for the effective inhibition of SARS-CoV-2 was found in the 6-mer peptide SAP6 (37-EDLFYQ-42) [33].
Another research group, as presented by Pei et al., investigated short native sequence peptides derived from the α1-helix of hACE2 [35]. They designed the octapeptide SI4α and the heptapeptide SI5α. Notably, these peptides did not include the crucial binding sequence to the spike protein identified by Larue [33]. Specifically, SI4α consists of residues Q24-K31, while SI5α is a shortened version ending before K31. Consequently, both peptides partially overlap with the sequence of SAP5, which was identified as a poor inhibitor by Larue [33].
Despite this, Pei et al. found that SI5α was one of the shortest native peptides with significant antiviral activity, demonstrating a half maximal effective concentration (EC50) value of 1.59 μM against the model coronavirus GX_P2V. Additionally, SI5α inhibited the interaction between the RBD and hACE2 by approximately 75% and 81% at concentrations of 50 µM and 100 µM, respectively [35].
Unlike the studies by Larue and Pei, Abouhajar et al. focused on synthesizing and testing slightly longer hACE2-derived peptides aimed at disrupting the ACE2/RBD complex [36]. They designed 23-mer and 13-mer peptides based on analyses of ACE2/SARS-CoV-2 S-protein interaction residues from prior ACE2/RBD crystal structure studies. Their objective was to evaluate how variations in peptide length affect the blocking efficiency of the SARS-CoV-2 spike protein. The researchers created two potential peptide inhibitors: Peptide [22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44] and Peptide [30,31,32,33,34,35,36,37,38,39,40,41,42], designed to bind to the SARS-CoV-2 S protein and disrupt its interaction with the human ACE2 receptor. The binding of these peptides to the spike protein was assessed using a surface plasmon resonance (SPR) competitive assay. The results showed that increasing the concentration of both peptides reduced the binding signal of the hACE2/RBD complex. Interestingly, the 13-mer peptide (D30–Q42), a truncated version of the 23-mer peptide (E22–S44), exhibited more effective inhibition, with an IC50 value of 0.65 μg/mL, compared to the 23-mer peptide, which had an IC50 of 2.00 μg/mL. This suggests that longer peptides do not always enhance binding affinity toward the S protein [36].
Another study also focused on the hACE2 α1 helix and identified one of the longest peptides, Pep 1, containing 27 native residues (S19–L45) from a human receptor, which was further used for the optimization process (see next chapter) [37].
4. Mutated Peptides
The studies presented above demonstrate that carefully selected and extracted fragments of protein–protein interaction interfaces can play a crucial role in developing peptides that effectively block unwanted complex formation. The thorough analysis of the 3D structure of these complexes, including the types of amino acid residues and the complementarity between interacting surfaces based on their physicochemical, geometrical, and distance-related properties, can provide valuable insights for designing such peptides. However, peptide fragments extracted from the protein context often exhibit increased flexibility and may face challenges in becoming effective drug candidates. Therefore, these peptides typically require further optimization. This optimization is usually a multistep, iterative process that results in various derivatives. These derivatives may or may not possess improved properties, which can enhance their efficacy as inhibitors of protein–protein interactions and increase their therapeutic potential.
The peptides optimization process may be oriented on different aspects of such molecules like (1) blocking their degradation/improving their stability in the organism, (2) increasing their solubility, (3) ameliorating cell membrane penetration, or (4) improving some ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) properties, just to name a few. In this paper, we would like to highlight the methods of optimization which solely modify their sequences, leading to new optimized molecules which chemically remain pure protein amino acid peptides (with a few exceptions that are standard procedure in peptide synthesis chemistry like capping N- and C- terminals). More sophisticated chemical modifications, such as adding functional groups or stapling peptide structures, which involve complex alterations, are beyond the scope of this review.
One of the first optimized peptides targeting the recognition of the hACE2 receptor by the SARS-CoV-2 RBD spike protein was proposed by Karoyan et al. [38]. They started with the N-terminal α1 helix of hACE2 (19S-L45) as the base sequence for further optimization (Table 3). Their approach aimed to achieve two main objectives through peptide modifications. First, the optimized peptides needed to maintain comparable binding affinity to the spike RBD as the native hACE2, while also exhibiting low antigenicity to avoid triggering a neutralizing immune response. This was accomplished by preserving crucial residues from the native hACE2 that are responsible for binding, while substituting non-essential residues with Ala and/or Leu, which have higher helical folding propensities. This strategy aimed to reduce the entropy penalty upon peptide binding. The second goal was to minimize antigenicity. This was achieved using a semi-empirical method [39] to identify potential antigenic determinants, followed by the iterative testing of each new sequence. Among the ten different 27-mer and 29-mer peptides tested, three peptides—P8, P9, and P10—were found to inhibit SARS-CoV-2 effectively, with inhibitory concentrations in the nanomolar range and high helical folding propensities ranging from 53% to 70%.

Table 3.
Sequences of mutated peptides based on hACE2.
Among the peptides designed to target ACE2, P10 emerged as one of the most promising. It was demonstrated to block SARS-CoV-2 infection in human pulmonary cells, with a binding affinity of ~0.03 nM as measured by biolayer interferometry and an IC50 value of ~42 nM as assessed by ELISA assays. Notably, the peptides P8, P9, and P10 exhibited no cytotoxicity in Vero-E5 and Calu-3 cell cultures [38].
Chitsike et al. focused on the same fragment of the hACE2 N-terminal α1 helix (S19-L45) as Karoyan et al., proposing various modified peptides based on this native sequence [37]. They selected this fragment based on computational alanine-scanning mutagenesis (using Rosetta and Bude), which identified S19, Q24, T27, F28, E30, K31, H34, E35, E37, D38, Y41, and L45 as key residues with a significant impact on binding energy. Pep 1, as described above, was directly adopted from the native sequence, whereas Pep 2 was developed by combining the part of the Pep 1 sequence (24–45) with an additional fragment of the hACE2 interface, residues 351-LGKGDFR-360, linked via the glycine residue to the C-terminal of Pep1. For the next three peptide variants, the original sequence was truncated at the N-terminal, and new mutations were introduced to enhance binding affinity and/or improve structural stability in the solution. Thus, Pep 3 consisted of hACE2 residues Q24-L45 with four mutations (A25V, T27Y, L39R, F40D) which were found to enhance binding affinity by other studies [40]. Pep 4 and Pep 5 were designed based on the T27-A46 fragment, incorporating several mutations to enhance helical folding by introducing an amphipathic α-helical pattern, with both peptides also having their N-terminal phenylalanine residues replaced with proline to reduce the likelihood of proteolytic degradation. Additionally, the Pep 5 includes a covalent side-chain cyclization designed to stabilize the α-helical structure. Pep 6, on the other hand, was derived from the S1 RBD fragment (483V–505Y), which is critical for host receptor recognition during viral attachment. An AlphaScreen™ assay was used to evaluate the ability of these peptides, both with and without modifications, to block ACE2-RBD binding. Notably, the SARS-CoV-2 RBD-derived 23-mer Pep 6 exhibited the highest activity, with an IC50 of 27 μM, outperforming most of ACE2-derived peptides, which had IC50 values ranging from 42 μM to 363 μM [37].
Chopra and colleagues initially identified a 24-mer fragment of hACE2 (S19-Q42) that encompasses most of the known contact residues. They discovered that shifting this sequence of nine residues toward the N-terminal direction of hACE2 protein increased the binding affinity to the SARS-CoV-2 RBD [41]. Following this, they systematically permuted the shifted hACE2 fragment (S10–N33) to identify peptides with higher binding affinities to the viral protein. Ultimately, they identified 22 peptides with a three-fold greater binding affinity compared to the initially proposed shifted peptide. Among these, only one peptide, BP19, effectively inhibited the interaction between the viral protein and the human receptor, with an IC50 of 2.08 ± 0.38 μM, as measured by a competitive ELISA assay for hACE2 protein interaction. BP19 competed with approximately 13.48 μM of hACE2. Notably, all mutations except one (N33I) were introduced in the shifted 9-residue N-terminal fragment, below the serine at position 19.
Rajpoot et al. initiated their peptide development using a slightly shorter 13-mer fragment of the hACE2 α1 helix, starting from residue F28. They performed mutation studies to enhance binding affinity. Their research highlighted the importance of substituting specific amino acid residues (E8N, A9F, and E10K) to increase binding affinity without disrupting the RBD binding site or adversely affecting the peptide’s physicochemical properties. Among the peptides tested, the 13AApi peptide demonstrated the ability to inhibit the interaction between the SARS-CoV-2 spike protein and hACE2, achieving a minimal inhibition of 40% at a 100 µM concentration, as assessed by ELISA [42].
One of the shorter peptides were proposed by Odolczyk et al. [27]. Based on their findings that the interaction between hACE2 and RBD is primarily mediated by the D30-Q42 fragment [27], they designed three short peptides: pep1c, pep1d, and pep1e, each incorporating a single mutation compared to the native hACE2 sequence. The rationale behind these mutations was to prevent the aggregation of the peptide fragments with the hACE2 protein during its folding process, which could be an undesired effect for potential future drug molecules. Among the peptides developed, the 9-mer pep1c and the 6-mer pep1d both bound to the spike protein with similar affinities (KD ~280 nM and ~210 nM, respectively). However, only pep1c was tested for its ability to inhibit the RBD-hACE2 complex formation, achieving an IC50 of approximately 3.3 mM.
In the continuation of their studies, Odolczyk et al. applied in silico modeling to optimize the sequence of pep1d, aiming to enhance peptide–protein interactions and increase binding affinity to the spike protein [43]. The in silico analysis, complemented by microscale thermophoresis (MST) and ELISA assays, led to the development of an hACE2-based peptide named J3. The J3 peptide, with the sequence DYGNHE, showed a significantly improved binding affinity (KD ~50 nM) compared to the previous peptides. This enhancement was achieved by substituting the positively charged Lys at the second position of the 6-mer peptide with the neutral hydrophobic Tyr [43]. The J3 peptide effectively diminished the hACE2–spike interaction, with the measured KD changing from ~151 nM in the absence of the peptide to ~720 nM in the presence of 1 mM of peptide. The experimental data of peptides activity verification were summarized in Table 4.

Table 4.
The activity characteristics of the hACE2-derived peptides against hACE2-RBD interaction.
5. Discussion
SARS-CoV-2 is responsible for the acute respiratory infection that has caused large numbers of cases and deaths worldwide. For a long time, there was no specific therapy for COVID-19 disease; this created an urgent need to develop a new drug against the virus [44,45]. However, the traditional drug discovery process takes a long time, which is not suitable for the fast reaction that is needed for new emerging threats caused by rapidly spreading pathogens.
Research has consistently shown that peptides derived directly from the native sequences of interacting partners within protein–protein interaction (PPI) interfaces provide an effective foundation for developing protein–protein interaction inhibitors [26,27,28,29]. These peptides are particularly valuable because they mimic the natural interaction sites and may offer a targeted approach to modulate biological pathways involved in diseases.
Therapeutic peptides are new and promising alternatives to treat numerous diseases, including cancer, metabolic disorders, infectious, and cardiovascular and neurodegenerative diseases [2,46], including COVID-19 [47,48,49,50,51,52]. The features that make peptides suitable for therapeutic candidates include their accessibility, structural and functional diversity, high specificity and affinity in low concentrations, high level of biological activity, low production costs, low immunogenicity, and ease of synthesis and storage [3,4,5,6,7]. They are small in size and do not accumulate in specific organs, which helps to reduce their toxic side effects.
Another advantage of antiviral peptides is that in many cases they are designed to act extracellularly and do not need to pass the cell membrane, reducing the possibility of virus–host cell interactions [2]. Because of their simplicity of synthesis, peptide sequences can also be easily modified via chemical or molecular biology approaches, for example, to react in response to emerging new drug-resistant strains of a virus.
However, peptides also encounter several challenges related to their direct applications as drug molecules. Thus, it is crucial to address to main limitations associated with their use in living organisms and to explore potential strategies to overcome these obstacles. One of the main challenges with peptide drug molecules is their poor stability, as they are highly susceptible to degradation by proteolytic enzymes. This results in a short half-life and reduced efficacy, limiting their therapeutic potential.
To address this issue, peptide stapling has emerged as a promising approach. This technique involves incorporating lactam-based i and i + 4 linkages between amino acids to stabilize the peptide’s structure, thereby enhancing its resistance to proteases. For example, the stapled peptide hACE2_21-55 (A36K-F40E) has demonstrated an effective inhibition of the SARS-CoV-2 spike protein binding to hACE2, with promising in vitro results (IC50 = 3.6 μM, KD = 2.1 μM) [53]. Introducing such constraints may also lock the peptide molecule in a bioactive conformation, resulting in more efficient interactions with its target. However, it is worth mentioning that this approach may also reduce binding affinity when the target recognizes the unstructured peptide. In such cases, stapled peptides may have fewer opportunities to bind, which could negatively impact their binding affinity [54].
Proteolysis often initiates at the N- or C-termini due to the action of various proteases and peptidases. Different amino acid residues at the termini exhibit varying degrees of resistance to degradation, with some being more susceptible to proteolysis [55]. For instance, N-terminal residues like Met, Ser, Ala, and Gly tend to enhance stability, while sequences rich in Pro, Glu, or Thr are more prone to degradation. By modifying the terminal sequences without compromising the peptide’s targeting or binding properties, degradation can be minimized, leading to enhanced bioavailability. Common modifications include C-terminal amidation and N-terminal acetylation, which shield the peptide from enzymatic attack [3,56].
Another significant challenge is the poor bioavailability of peptides, particularly when administered orally. Their hydrophilic nature makes it difficult for peptides to cross biological membranes, and they are often degraded in the gastrointestinal tract. To address this limitation, alternative delivery methods, such as non-invasive approaches like nasal administration, are being explored [57]. For example, in a study by de Vries et al., researchers developed an intranasal lipopeptide designed to inhibit viral fusion [58]. Administered via the nasal route, this peptide demonstrated substantial efficacy in preventing the direct-contact transmission of SARS-CoV-2 in ferret models. This approach underscores the potential of nasal delivery systems for peptides, providing a practical solution to enhance their therapeutic potential.
Peptide drug molecules are typically weak immunogens; however, several strategies can be applied during peptide design to reduce their immunogenic risks. Reducing the size of the peptide or mutating non-essential hydrophobic residues, which are more easily recognized by the host immune system, can help to mitigate these risks and improve their therapeutic effectiveness.
In conclusion, while peptide inhibitors offer considerable therapeutic promise, overcoming their limitations through chemical modifications, innovative delivery systems, and advancements in peptide engineering will be essential for enhancing their clinical viability and effectiveness. Beyond peptides, other classes of inhibitors, such as small-molecule inhibitors and monoclonal antibodies, have also been explored for therapeutic purposes.
Despite the challenges associated with peptides, the case study of the hACE2 protein and SARS-CoV-2 spike protein highlights the significant potential of the structure-guided development of peptidic inhibitors targeting protein–protein interactions. This potential has been supported not only by in vitro studies but also by in vivo experiments, as recently presented by Oliveira et al. [59]. The authors identified a wild-type 25-mer mimetic peptide (E25-A46) of hACE2 as a promising drug candidate. In their study, they treated mice, already infected with SARS-CoV-2, via the nasal instillation of the peptide. Administering the peptide 24 h post-infection led to noticeable improvements in clinical symptoms associated with experimental COVID-19, as evidenced by the body weight and clinical scores of the mice. These factors were monitored daily over a five-day period, showing significantly better outcomes in the treated group compared to untreated controls [59].
Overall, both in vitro and in vivo studies support the use of peptides as viable candidates for preventing SARS-CoV-2 infection and mitigating COVID-19-related symptoms.
Peptides are worth considering as a promising alternative to small molecules, such as the recently developed cepharanthine, which may be associated with different side effects [60,61].
Using hACE2 and spike protein interaction as an example, this review demonstrated that PPIs with peptide-based approaches can develop new inhibitors and should be considered as new potential drug targets against many human pathogens, particularly in the era of increasing microbial antibiotic resistance, which poses a high risk of new pandemic episodes [62].
Author Contributions
Writing—original draft preparation, A.R.; writing—review and editing, N.O.; visualization, N.O.; supervision, P.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the Polish Ministry of Science and Higher Education, under the projects (79/E35/SPUB/SP/2019) and POL-OPENSCREEN (DIR/Wk/2018/06).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant data are presented within the paper.
Acknowledgments
During the preparation of this work, the authors used ChatGPT (Model GPT-4o) and Grammarly services for language corrections. After using these tools/services, the authors reviewed and edited the content as needed and therefore take full responsibility for the content of the publication.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Henninot, A.; Collins, J.C.; Nuss, J.M. The Current State of Peptide Drug Discovery: Back to the Future? J. Med. Chem. 2018, 61, 1382–1414. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic Peptides: Current Applications and Future Directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.L.; Dunn, M.K. Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions. Bioorganic Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Cabri, W.; Cantelmi, P.; Corbisiero, D.; Fantoni, T.; Ferrazzano, L.; Martelli, G.; Mattellone, A.; Tolomelli, A. Therapeutic Peptides Targeting PPI in Clinical Development: Overview, Mechanism of Action and Perspectives. Front. Mol. Biosci. 2021, 8, 697586. [Google Scholar] [CrossRef]
- Apostolopoulos, V.; Bojarska, J.; Chai, T.-T.; Elnagdy, S.; Kaczmarek, K.; Matsoukas, J.; New, R.; Parang, K.; Lopez, O.P.; Parhiz, H.; et al. A Global Review on Short Peptides: Frontiers and Perspectives. Molecules 2021, 26, 430. [Google Scholar] [CrossRef]
- Guarracino, D.A.; Iannaccone, J.; Cabrera, A.; Kancharla, S. Harnessing the Therapeutic Potential and Biological Activity of Antiviral Peptides. ChemBioChem 2022, 23, e202200415. [Google Scholar] [CrossRef]
- Kahan, R.; Worm, D.J.; de Castro, G.V.; Ng, S.; Barnard, A. Modulators of Protein–Protein Interactions as Antimicrobial Agents. RSC Chem. Biol. 2021, 2, 387–409. [Google Scholar] [CrossRef]
- Poluri, K.M.; Gulati, K.; Tripathi, D.K.; Nagar, N. Protein-Protein Interactions in Host–Pathogen Interactions. In Protein-Protein Interactions: Pathophysiological and Therapeutic Aspects: Volume II; Poluri, K.M., Gulati, K., Tripathi, D.K., Nagar, N., Eds.; Springer Nature: Singapore, 2023; pp. 207–264. ISBN 978-981-9924-23-3. [Google Scholar]
- de Groot, N.S.; Burgas, M.T. Bacteria Use Structural Imperfect Mimicry to Hijack the Host Interactome. PLoS Comput. Biol. 2020, 16, e1008395. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, Y.; Yao, S.; Ge, H.; Zhu, Y.; Chen, K.; Chen, W.; Zhang, Y.; Zhu, W.; Wang, H.; et al. Discovery of Potential Small Molecular SARS-CoV-2 Entry Blockers Targeting the Spike Protein. Acta Pharmacol. Sin. 2022, 43, 788–796. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, Y.; Cardoze, S.M.; Obineche, O.W.; Melo, C.; Persaud, A.; Fernández Romero, J.A. Small Molecules Targeting SARS-CoV-2 Spike Glycoprotein Receptor-Binding Domain. ACS Omega 2022, 7, 28779–28789. [Google Scholar] [CrossRef]
- Lau, E.Y.; Negrete, O.A.; Bennett, W.F.D.; Bennion, B.J.; Borucki, M.; Bourguet, F.; Epstein, A.; Franco, M.; Harmon, B.; He, S.; et al. Discovery of Small-Molecule Inhibitors of SARS-CoV-2 Proteins Using a Computational and Experimental Pipeline. Front. Mol. Biosci. 2021, 8, 678701. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. In Coronaviruses: Methods and Protocols; Maier, H.J., Bickerton, E., Britton, P., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; pp. 1–23. ISBN 978-1-4939-2438-7. [Google Scholar]
- Li, G.; Fan, Y.; Lai, Y.; Han, T.; Li, Z.; Zhou, P.; Pan, P.; Wang, W.; Hu, D.; Liu, X.; et al. Coronavirus Infections and Immune Responses. J. Med. Virol. 2020, 92, 424–432. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Satarker, S.; Nampoothiri, M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch. Med. Res. 2020, 51, 482–491. [Google Scholar] [CrossRef]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular Interaction and Inhibition of SARS-CoV-2 Binding to the ACE2 Receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the Novel Coronavirus from the Ongoing Wuhan Outbreak and Modeling of Its Spike Protein for Risk of Human Transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Bachiller, M.I.; Brzozowska, I.; Odolczyk, N.; Zielenkiewicz, U.; Zielenkiewicz, P.; Rademann, J. Mapping Protein–Protein Interactions of the Resistance-Related Bacterial Zeta Toxin–Epsilon Antitoxin Complex (Ε2ζ2) with High Affinity Peptide Ligands Using Fluorescence Polarization. Toxins 2016, 8, 222. [Google Scholar] [CrossRef]
- Odolczyk, N.; Marzec, E.; Winiewska-Szajewska, M.; Poznański, J.; Zielenkiewicz, P. Native Structure-Based Peptides as Potential Protein-Protein Interaction Inhibitors of SARS-CoV-2 Spike Protein and Human ACE2 Receptor. Molecules 2021, 26, 2157. [Google Scholar] [CrossRef]
- Pelay-Gimeno, M.; Glas, A.; Koch, O.; Grossmann, T.N. Structure-Based Design of Inhibitors of Protein–Protein Interactions: Mimicking Peptide Binding Epitopes. Angew. Chem. Int. Ed. 2015, 54, 8896–8927. [Google Scholar] [CrossRef]
- Rezaei Araghi, R.; Keating, A.E. Designing Helical Peptide Inhibitors of Protein–Protein Interactions. Curr. Opin. Struct. Biol. 2016, 39, 27–38. [Google Scholar] [CrossRef]
- Ho, T.-Y.; Wu, S.-L.; Chen, J.-C.; Wei, Y.-C.; Cheng, S.-E.; Chang, Y.-H.; Liu, H.-J.; Hsiang, C.-Y. Design and Biological Activities of Novel Inhibitory Peptides for SARS-CoV Spike Protein and Angiotensin-Converting Enzyme 2 Interaction. Antivir. Res. 2006, 69, 70–76. [Google Scholar] [CrossRef]
- Han, Y.; Král, P. Computational Design of ACE2-Based Peptide Inhibitors of SARS-CoV-2. ACS Nano 2020, 14, 5143–5147. [Google Scholar] [CrossRef]
- Ling, R.; Dai, Y.; Huang, B.; Huang, W.; Yu, J.; Lu, X.; Jiang, Y. In Silico Design of Antiviral Peptides Targeting the Spike Protein of SARS-CoV-2. Peptides 2020, 130, 170328. [Google Scholar] [CrossRef]
- Larue, R.C.; Xing, E.; Kenney, A.D.; Zhang, Y.; Tuazon, J.A.; Li, J.; Yount, J.S.; Li, P.-K.; Sharma, A. Rationally Designed ACE2-Derived Peptides Inhibit SARS-CoV-2. Bioconjugate Chem. 2021, 32, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Morse, J.S.; Lalonde, T.; Xu, S.; Liu, W.R. Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV. ChemBioChem 2020, 21, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Pei, P.; Qin, H.; Chen, J.; Wang, F.; He, C.; He, S.; Hong, B.; Liu, K.; Qiao, R.; Fan, H.; et al. Computational Design of Ultrashort Peptide Inhibitors of the Receptor-Binding Domain of the SARS-CoV-2 S Protein. Brief. Bioinform. 2021, 22, bbab243. [Google Scholar] [CrossRef]
- Abouhajar, F.; Chaudhuri, R.; Valiulis, S.N.; Stuart, D.D.; Malinick, A.S.; Xue, M.; Cheng, Q. Label-Free Analysis of Binding and Inhibition of SARS-CoV-19 Spike Proteins to ACE2 Receptor with ACE2-Derived Peptides by Surface Plasmon Resonance. ACS Appl. Bio Mater. 2022, 6, 182–190. [Google Scholar] [CrossRef]
- Chitsike, L.; Krstenansky, J.; Duerksen-Hughes, P.J. ACE2 : S1 RBD Interaction-Targeted Peptides and Small Molecules as Potential COVID-19 Therapeutics. Adv. Pharmacol. Pharm. Sci. 2021, 2021, 1828792. [Google Scholar] [CrossRef]
- Karoyan, P.; Vieillard, V.; Gómez-Morales, L.; Odile, E.; Guihot, A.; Luyt, C.-E.; Denis, A.; Grondin, P.; Lequin, O. Human ACE2 Peptide-Mimics Block SARS-CoV-2 Pulmonary Cells Infection. Commun. Biol. 2021, 4, 197. [Google Scholar] [CrossRef]
- Kolaskar, A.S.; Tongaonkar, P.C. A Semi-Empirical Method for Prediction of Antigenic Determinants on Protein Antigens. FEBS Lett. 1990, 276, 172–174. [Google Scholar] [CrossRef]
- Chan, K.K.; Dorosky, D.; Sharma, P.; Abbasi, S.A.; Dye, J.M.; Kranz, D.M.; Herbert, A.S.; Procko, E. Engineering Human ACE2 to Optimize Binding to the Spike Protein of SARS Coronavirus 2. Science 2020, 369, 1261–1265. [Google Scholar] [CrossRef]
- Chopra, A.; Shukri, A.H.; Adhikary, H.; Lukinović, V.; Hoekstra, M.; Cowpland, M.; Biggar, K.K. A Peptide Array Pipeline for the Development of Spike-ACE2 Interaction Inhibitors. Peptides 2022, 158, 170898. [Google Scholar] [CrossRef]
- Rajpoot, S.; Ohishi, T.; Kumar, A.; Pan, Q.; Banerjee, S.; Zhang, K.Y.J.; Baig, M.S. A Novel Therapeutic Peptide Blocks SARS-CoV-2 Spike Protein Binding with Host Cell ACE2 Receptor. Drugs R D 2021, 21, 273–283. [Google Scholar] [CrossRef]
- Odolczyk, N.; Klim, J.; Podsiadła-Białoskórska, M.; Winiewska-Szajewska, M.; Szolajska, E.; Zielenkiewicz, U.; Poznański, J.; Zielenkiewicz, P. Improvement of Native Structure-Based Peptides as Efficient Inhibitors of Protein-Protein Interactions of SARS-CoV-2 Spike Protein and Human ACE2. Front. Mol. Biosci. 2022, 9, 983014. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.A. Clinical Trials of Repurposed Antivirals for SARS-CoV-2. Antimicrob. Agents Chemother. 2020, 64, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Spinner, C.D.; Gottlieb, R.L.; Criner, G.J.; Arribas López, J.R.; Cattelan, A.M.; Soriano Viladomiu, A.; Ogbuagu, O.; Malhotra, P.; Mullane, K.M.; Castagna, A.; et al. Effect of Remdesivir vs Standard Care on Clinical Status at 11 Days in Patients With Moderate COVID-19: A Randomized Clinical Trial. JAMA 2020, 324, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Baig, M.H.; Ahmad, K.; Saeed, M.; Alharbi, A.M.; Barreto, G.E.; Ashraf, G.M.; Choi, I. Peptide Based Therapeutics and Their Use for the Treatment of Neurodegenerative and Other Diseases. Biomed. Pharmacother. 2018, 103, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Heydari, H.; Golmohammadi, R.; Mirnejad, R.; Tebyanian, H.; Fasihi-Ramandi, M.; Moosazadeh Moghaddam, M. Antiviral Peptides against Coronaviridae Family: A Review. Peptides 2021, 139, 170526. [Google Scholar] [CrossRef] [PubMed]
- Nepali, K.; Sharma, R.; Sharma, S.; Thakur, A.; Liou, J.-P. Beyond the Vaccines: A Glance at the Small Molecule and Peptide-Based Anti-COVID19 Arsenal. J. Biomed. Sci. 2022, 29, 65. [Google Scholar] [CrossRef]
- Dahal, A.; Sonju, J.J.; Kousoulas, K.G.; Jois, S.D. Peptides and Peptidomimetics as Therapeutic Agents for COVID-19. Pept. Sci. 2022, 114, e24245. [Google Scholar] [CrossRef]
- Schütz, D.; Ruiz-Blanco, Y.B.; Münch, J.; Kirchhoff, F.; Sanchez-Garcia, E.; Müller, J.A. Peptide and Peptide-Based Inhibitors of SARS-CoV-2 Entry. Adv. Drug Deliv. Rev. 2020, 167, 47–65. [Google Scholar] [CrossRef]
- Panchal, D.; Kataria, J.; Patel, K.; Crowe, K.; Pai, V.; Azizogli, A.-R.; Kadian, N.; Sanyal, S.; Roy, A.; Dodd-o, J.; et al. Peptide-Based Inhibitors for SARS-CoV-2 and SARS-CoV. Adv. Ther. 2021, 4, 2100104. [Google Scholar] [CrossRef]
- Shah, J.N.; Guo, G.-Q.; Krishnan, A.; Ramesh, M.; Katari, N.K.; Shahbaaz, M.; Abdellattif, M.H.; Singh, S.K.; Dua, K. Peptides-Based Therapeutics: Emerging Potential Therapeutic Agents for COVID-19. Therapies 2022, 77, 319–328. [Google Scholar] [CrossRef]
- Maas, M.N.; Hintzen, J.C.J.; Löffler, P.M.G.; Mecinović, J. Targeting SARS-CoV-2 Spike Protein by Stapled hACE2 Peptides. Chem. Commun. 2021, 57, 3283–3286. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Dawber, R.S.; Zhang, P.; Walko, M.; Wilson, A.J.; Wang, X. Peptide-Based Inhibitors of Protein–Protein Interactions: Biophysical, Structural and Cellular Consequences of Introducing a Constraint. Chem. Sci. 2021, 12, 5977–5993. [Google Scholar] [CrossRef] [PubMed]
- Lucana, M.C.; Arruga, Y.; Petrachi, E.; Roig, A.; Lucchi, R.; Oller-Salvia, B. Protease-Resistant Peptides for Targeting and Intracellular Delivery of Therapeutics. Pharmaceutics 2021, 13, 2065. [Google Scholar] [CrossRef] [PubMed]
- Rossino, G.; Marchese, E.; Galli, G.; Verde, F.; Finizio, M.; Serra, M.; Linciano, P.; Collina, S. Peptides as Therapeutic Agents: Challenges and Opportunities in the Green Transition Era. Molecules 2023, 28, 7165. [Google Scholar] [CrossRef]
- Lee, M.F.; Poh, C.L. Strategies to Improve the Physicochemical Properties of Peptide-Based Drugs. Pharm. Res. 2023, 40, 617–632. [Google Scholar] [CrossRef]
- de Vries, R.D.; Schmitz, K.S.; Bovier, F.T.; Predella, C.; Khao, J.; Noack, D.; Haagmans, B.L.; Herfst, S.; Stearns, K.N.; Drew-Bear, J.; et al. Intranasal Fusion Inhibitory Lipopeptide Prevents Direct-Contact SARS-CoV-2 Transmission in Ferrets. Science 2021, 371, 1379–1382. [Google Scholar] [CrossRef]
- Oliveira, E.H.; Monteleone-Cassiano, A.C.; Tavares, L.; Santos, J.C.; Lima, T.M.; Gomes, G.F.; Tanaka, P.P.; Monteiro, C.J.; Munuera, M.; Batah, S.S.; et al. A Mimetic Peptide of ACE2 Protects against SARS-CoV-2 Infection and Decreases Pulmonary Inflammation Related to COVID-19. Antivir. Res. 2024, 229, 105968. [Google Scholar] [CrossRef]
- Fan, H.; He, S.; Han, P.; Hong, B.; Liu, K.; Li, M.; Wang, S.; Tong, Y. Cepharanthine: A Promising Old Drug against SARS-CoV-2. Adv. Biol. 2022, 6, 2200148. [Google Scholar] [CrossRef]
- Hijikata, A.; Shionyu-Mitsuyama, C.; Nakae, S.; Shionyu, M.; Ota, M.; Kanaya, S.; Hirokawa, T.; Nakajima, S.; Watashi, K.; Shirai, T. Evaluating Cepharanthine Analogues as Natural Drugs against SARS-CoV-2. FEBS Open Bio 2022, 12, 285–294. [Google Scholar] [CrossRef]
- Williams, B.A.; Jones, C.H.; Welch, V.; True, J.M. Outlook of Pandemic Preparedness in a Post-COVID-19 World. npj Vaccines 2023, 8, 178. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).