Investigation of PRKN Mutations in Levodopa-Induced Dyskinesia in Parkinson’s Disease Treatment
Abstract
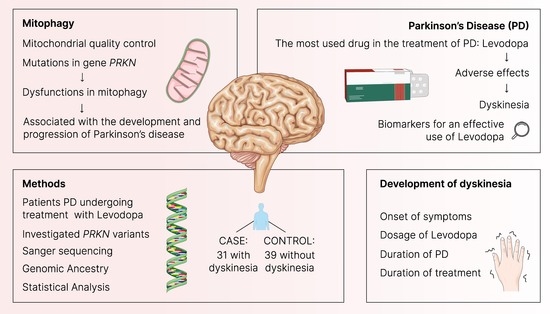
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Selection of Variants
2.3. DNA Extraction and Quantification
2.4. Genotyping
2.5. Genomic Ancestry
2.6. Statistical Analysis
3. Results
3.1. Characterization of the Cohort
3.2. Analysis of Variants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ng, M.Y.W.; Wai, T.; Simonsen, A. Quality Control of the Mitochondrion. Dev. Cell 2021, 56, 881–905. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, H.-H.; Cao, Y.-T.; Zhang, L.-L.; Huang, F.; Yi, C. The Role of Mitochondrial Dynamics and Mitophagy in Carcinogenesis, Metastasis and Therapy. Front. Cell Dev. Biol. 2020, 8, 413. [Google Scholar] [CrossRef]
- Dabravolski, S.A.; Nikiforov, N.G.; Zhuravlev, A.D.; Orekhov, N.A.; Grechko, A.V.; Orekhov, A.N. Role of the MtDNA Mutations and Mitophagy in Inflammaging. Int. J. Mol. Sci. 2022, 23, 1323. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-M.; Jung, Y.-K. A Molecular Approach to Mitophagy and Mitochondrial Dynamics. Mol. Cells 2018, 41, 18–26. [Google Scholar] [CrossRef]
- Park, J.-S.; Davis, R.L.; Sue, C.M. Mitochondrial Dysfunction in Parkinson’s Disease: New Mechanistic Insights and Therapeutic Perspectives. Curr. Neurol. Neurosci. Rep. 2018, 18, 21. [Google Scholar] [CrossRef] [PubMed]
- Pickles, S.; Vigié, P.; Youle, R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018, 28, R170–R185. [Google Scholar] [CrossRef]
- Masaldan, S.; Callegari, S.; Dewson, G. Therapeutic Targeting of Mitophagy in Parkinson’s Disease. Biochem. Soc. Trans. 2022, 50, 783–797. [Google Scholar] [CrossRef]
- Fonseca Cabral, G.; Schaan, A.P.; Cavalcante, G.C.; Sena-dos-Santos, C.; de Souza, T.P.; Souza Port’s, N.M.; dos Santos Pinheiro, J.A.; Ribeiro-dos-Santos, Â.; Vidal, A.F. Nuclear and Mitochondrial Genome, Epigenome and Gut Microbiome: Emerging Molecular Biomarkers for Parkinson’s Disease. Int. J. Mol. Sci. 2021, 22, 9839. [Google Scholar] [CrossRef]
- Liu, J.; Liu, W.; Li, R.; Yang, H. Mitophagy in Parkinson’s Disease: From Pathogenesis to Treatment. Cells 2019, 8, 712. [Google Scholar] [CrossRef]
- Picca, A.; Guerra, F.; Calvani, R.; Romano, R.; Coelho-Júnior, H.J.; Bucci, C.; Marzetti, E. Mitochondrial Dysfunction, Protein Misfolding and Neuroinflammation in Parkinson’s Disease: Roads to Biomarker Discovery. Biomolecules 2021, 11, 1508. [Google Scholar] [CrossRef]
- Li, W.; Fu, Y.; Halliday, G.M.; Sue, C.M. PARK Genes Link Mitochondrial Dysfunction and Alpha-Synuclein Pathology in Sporadic Parkinson’s Disease. Front. Cell Dev. Biol. 2021, 9, 612476. [Google Scholar] [CrossRef]
- Zhang, L.; Dai, L.; Li, D. Mitophagy in Neurological Disorders. J. Neuroinflamm. 2021, 18, 297. [Google Scholar] [CrossRef]
- Chan, G.H.-F. The Role of Genetic Data in Selecting Device-Aided Therapies in Patients with Advanced Parkinson’s Disease: A Mini-Review. Front. Aging Neurosci. 2022, 14, 895430. [Google Scholar] [CrossRef]
- Miller, S.; Muqit, M.M.K. Therapeutic Approaches to Enhance PINK1/Parkin Mediated Mitophagy for the Treatment of Parkinson’s Disease. Neurosci. Lett. 2019, 705, 7–13. [Google Scholar] [CrossRef]
- Sliter, D.A.; Martinez, J.; Hao, L.; Chen, X.; Sun, N.; Fischer, T.D.; Burman, J.L.; Li, Y.; Zhang, Z.; Narendra, D.P.; et al. Parkin and PINK1 Mitigate STING-Induced Inflammation. Nature 2018, 561, 258–262. [Google Scholar] [CrossRef]
- Reich, S.G.; Savitt, J.M. Parkinson’s Disease. Med. Clin. N. Am. 2019, 103, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, J.; Tan, E.K. Parkinson’s Disease: Etiopathogenesis and Treatment. J. Neurol. Neurosurg. Psychiatry 2020, 91, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.K.; Tanner, C.M.; Brundin, P. Parkinson Disease Epidemiology, Pathology, Genetics, and Pathophysiology. Clin. Geriatr. Med. 2020, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Aarsland, D.; Batzu, L.; Halliday, G.M.; Geurtsen, G.J.; Ballard, C.; Ray Chaudhuri, K.; Weintraub, D. Parkinson Disease-Associated Cognitive Impairment. Nat. Rev. Dis. Primers 2021, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Brakedal, B.; Toker, L.; Haugarvoll, K.; Tzoulis, C. A Nationwide Study of the Incidence, Prevalence and Mortality of Parkinson’s Disease in the Norwegian Population. NPJ Park. Dis. 2022, 8, 19. [Google Scholar] [CrossRef]
- Bandopadhyay, R.; Mishra, N.; Rana, R.; Kaur, G.; Ghoneim, M.M.; Alshehri, S.; Mustafa, G.; Ahmad, J.; Alhakamy, N.A.; Mishra, A. Molecular Mechanisms and Therapeutic Strategies for Levodopa-Induced Dyskinesia in Parkinson’s Disease: A Perspective through Preclinical and Clinical Evidence. Front. Pharmacol. 2022, 13, 805388. [Google Scholar] [CrossRef]
- Scarduzio, M.; Hess, E.J.; Standaert, D.G.; Eskow Jaunarajs, K.L. Striatal Synaptic Dysfunction in Dystonia and Levodopa-Induced Dyskinesia. Neurobiol. Dis. 2022, 166, 105650. [Google Scholar] [CrossRef]
- Xiao, B.; Kuruvilla, J.; Tan, E.-K. Mitophagy and Reactive Oxygen Species Interplay in Parkinson’s Disease. NPJ Park. Dis. 2022, 8, 135. [Google Scholar] [CrossRef]
- Goiran, T.; Eldeeb, M.A.; Zorca, C.E.; Fon, E.A. Hallmarks and Molecular Tools for the Study of Mitophagy in Parkinson’s Disease. Cells 2022, 11, 2097. [Google Scholar] [CrossRef]
- Silvian, L.F. PINK1/Parkin Pathway Activation for Mitochondrial Quality Control–Which Is the Best Molecular Target for Therapy? Front. Aging Neurosci. 2022, 14, 890823. [Google Scholar] [CrossRef]
- Alter, A.; Fava, V.M.; Huong, N.T.; Singh, M.; Orlova, M.; Van Thuc, N.; Katoch, K.; Thai, V.H.; Ba, N.N.; Abel, L.; et al. Linkage Disequilibrium Pattern and Age-At-Diagnosis Are Critical for Replicating Genetic Associations across Ethnic Groups in Leprosy. Hum. Genet. 2012, 132, 107–116. [Google Scholar] [CrossRef]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single Nucleotide Polymorphisms and Cancer Susceptibility. Oncotarget 2017, 8, 110635–110649. [Google Scholar] [CrossRef]
- Sartori, P.V.U. Mapeamento Fino das Sequências Exônicas de Genes de Susceptibilidade do Hospedeiro à Hanseníase. Ph.D. Thesis, Pontifical Catholic University of Paraná, Curitiba, Brazil, 2019. [Google Scholar]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989; ISBN 9780879693732. [Google Scholar]
- Ferraz, R.S.; Silva, C.S.; Cavalcante, G.C.; de Queiroz, N.N.M.; Felício, K.M.; Felício, J.S.; Ribeiro-dos-Santos, Â. Variants in the VDR Gene May Influence 25(OH)D Levels in Type 1 Diabetes Mellitus in a Brazilian Population. Nutrients 2022, 14, 1010. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2014, 7, 539. [Google Scholar] [CrossRef]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A New Bioinformatics Analysis Tools Framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.P.C.; Ribeiro-Rodrigues, E.M.; Ribeiro-dos-Santos, Â.K.C.; Pereira, R.; Gusmão, L.; Amorim, A.; Guerreiro, J.F.; Zago, M.A.; Matte, C.; Hutz, M.H.; et al. Assessing Individual Interethnic Admixture and Population Substructure Using a 48-Insertion-Deletion (INSEL) Ancestry-Informative Marker (AIM) Panel. Hum. Mutat. 2010, 31, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Ramos, B.R.d.A.; D’Elia, M.P.B.; Amador, M.A.T.; Santos, N.P.C.; Santos, S.E.B.; da Cruz Castelli, E.; Witkin, S.S.; Miot, H.A.; Miot, L.D.B.; da Silva, M.G. Neither Self-Reported Ethnicity nor Declared Family Origin Are Reliable Indicators of Genomic Ancestry. Genetica 2016, 144, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Andrade, R.B.; Amador, M.A.T.; Cavalcante, G.C.; Leitão, L.P.C.; Fernandes, M.R.; Modesto, A.A.C.; Moreira, F.C.; Khayat, A.S.; Assumpção, P.P.; Ribeiro-dos-Santos, Â.; et al. Estimating Asian Contribution to the Brazilian Population: A New Application of a Validated Set of 61 Ancestry Informative Markers. G3 Genes Genomes Genet. 2018, 8, 3577–3582. [Google Scholar] [CrossRef]
- JASP Team. JASP, version 0.16.3; Computer Software. 2022.
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2014. Available online: http://www.R-project.org/ (accessed on 12 November 2022).
- Ebersbach, G.; Baas, H.; Csoti, I.; Müngersdorf, M.; Deuschl, G. Scales in Parkinson’s Disease. J. Neurol. 2006, 253 (Suppl. 4), IV32–IV35. [Google Scholar] [CrossRef]
- Regnault, A.; Boroojerdi, B.; Meunier, J.; Bani, M.; Morel, T.; Cano, S. Does the MDS-UPDRS Provide the Precision to Assess Progression in Early Parkinson’s Disease? Learnings from the Parkinson’s Progression Marker Initiative Cohort. J. Neurol. 2019, 266, 1927–1936. [Google Scholar] [CrossRef]
- Cavalcante, G.C.; de Moraes, M.R.; Valente, C.M.D.; Silva, C.S.; Modesto, A.A.C.; de Assumpção, P.B.; de Assumpção, P.P.; Santos, S.; Ribeiro-dos-Santos, Â. Investigation of INDEL Variants in Apoptosis: The Relevance to Gastric Cancer. BMC Med. Genet. 2020, 21, 207. [Google Scholar] [CrossRef]
- Terron, A.; Bal-Price, A.; Paini, A.; Monnet-Tschudi, F.; Bennekou, S.H.; Leist, M.; Schildknecht, S. An Adverse Outcome Pathway for Parkinsonian Motor Deficits Associated with Mitochondrial Complex I Inhibition. Arch. Toxicol. 2017, 92, 41–82. [Google Scholar] [CrossRef]
- Grünewald, A.; Kumar, K.R.; Sue, C.M. New Insights into the Complex Role of Mitochondria in Parkinson’s Disease. Prog. Neurobiol. 2019, 177, 73–93. [Google Scholar] [CrossRef]
- Hely, M.A.; Morris, J.G.L.; Reid, W.G.J.; Trafficante, R. Sydney Multicenter Study of Parkinson’s Disease: Non-L-Dopa-Responsive Problems Dominate at 15 Years. Mov. Disord. 2005, 20, 190–199. [Google Scholar] [CrossRef]
- Lücking, C.B.; Dürr, A.; Bonifati, V.; Vaughan, J.; De Michele, G.; Gasser, T.; Harhangi, B.S.; Meco, G.; Denèfle, P.; Wood, N.W.; et al. Association between Early-Onset Parkinson’s Disease and Mutations in the Parkin Gene. N. Engl. J. Med. 2000, 342, 1560–1567. [Google Scholar] [CrossRef]
- Sassone, J.; Valtorta, F.; Ciammola, A. Early Dyskinesias in Parkinson’s Disease Patients with Parkin Mutation: A Primary Corticostriatal Synaptopathy? Front. Neurosci. 2019, 13, 273. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Alphonsa, T.A.; Husain, R.A.; Ahmed, S.S.; Subramaniyan, K.; Kumar, S. Association of Rs1801582 and Rs1801334 PARK2 Polymorphisms with Risk of Parkinson’s Disease: A Case-Control Study in South India and Meta-Analysis. Meta Gene 2016, 10, 32–38. [Google Scholar] [CrossRef]
- Filatova, E.; Shadrina, M.I.; Fedotova, E.Y.; Ivanova-Smolenskaya, I.A.; Illarioshkin, S.N.; Limborska, S.A.; Slominsky, P.A. Analysis of Known Point Mutations and SNPs in Genes Responsible for Monogenic Parkinson’s Disease in Russian Patients. Adv. Park. Dis. 2013, 2, 28–30. [Google Scholar] [CrossRef][Green Version]
- Ku, S.; Glass, G.A. Age of Parkinson’s Disease Onset as a Predictor for the Development of Dyskinesia. Mov. Disord. 2010, 25, 1177–1182. [Google Scholar] [CrossRef]
- Kwon, D.K.; Kwatra, M.; Wang, J.; Ko, H.S. Levodopa-Induced Dyskinesia in Parkinson’s Disease: Pathogenesis and Emerging Treatment Strategies. Cells 2022, 11, 3736. [Google Scholar] [CrossRef]
- Warren Olanow, C.; Kieburtz, K.; Rascol, O.; Poewe, W.; Schapira, A.H.; Emre, M.; Nissinen, H.; Leinonen, M.; Stocchi, F. Factors Predictive of the Development of Levodopa-Induced Dyskinesia and Wearing-off in Parkinson’s Disease. Mov. Disord. 2013, 28, 1064–1071. [Google Scholar] [CrossRef]


| Variant | Alleles | MAF |
|---|---|---|
| rs9458609 | A>G | 0.30678 |
| rs1446954927 | A>G | 0.00007 |
| rs1562397973 | G>A | 0.000007 |
| rs1188406822 | G>A | 0.000007 |
| rs897123960 | T>C | 0.000014 |
| rs1469130556 | A>G | 0.00007 |
| rs1037288938 | G>A | 0.000021 |
| rs1453267522 | T>G | 0.000014 |
| rs6935164 | A>G | 0.497471 |
| rs1460805773 | T>G | 0.000007 |
| rs972410489 | G>C | 0.000007 |
| rs1372726016 | G>A | 0.000007 |
| rs187267734 | C>A | 0.000007 |
| rs182928580 | G>A | 0.003313 |
| Variable | Case | Control | p-Value |
|---|---|---|---|
| n | 31 | 39 | |
| Age of onset of symptoms, years a | 47.5 ± 1.68 | 55.3 ± 1.55 | 0.001 |
| Sex, % male/female b | 74.2/25.8 | 61.5/38.5 | 0.263 |
| European ancestry c | 0.643 ± 0.036 | 0.588 ± 0.035 | 0.309 |
| Native American ancestry c | 0.214 ± 0.031 | 0.259 ± 0.033 | 0.350 |
| African ancestry c | 0.143 ± 0.029 | 0.154 ± 0.031 | 0.692 |
| Family history, % yes/no b | 17.2/82.8 | 30.6/69.4 | 0.215 |
| Primary Symptom, % tremor/others b | 48.4/51.6 | 66.7/33.3 | 0.123 |
| Duration of PD d | 10.0 ± 0.92 | 6.9 ± 0.62 | 0.0037 |
| Duration of treatment d | 8.1 ± 0.68 | 4.97 ± 0.51 | 8.1 × 10−5 |
| LEDD d | 1066.1 ± 94.7 | 540.2 ± 44.7 | 1.8 × 10−6 |
| UPDRS d | 73.2 ± 6.5 | 64.3 ± 4.3 | 0.370 |
| Variant | Genotype | Case (%) | Control (%) | p-Value a | OR (95%CI) b |
|---|---|---|---|---|---|
| rs9458609 | n = 29 | n = 36 | |||
| AA | 16 (55.2) | 17 (47.2) | 0.773 | 0.817 (0.207–3.229) | |
| AG | 9 (31.0) | 17 (47.2) | 0.807 | 0.847 (0.224–3.203) | |
| GG | 4 (13.8) | 2 (5.6) | 0.330 | 3.306 (0.299–36.601) | |
| A = 0.7/G = 0.3 | A = 0.7/G = 0.3 | ||||
| rs6935164 | n = 28 | n = 30 | |||
| AA | 6 (21.4) | 13 (43.3) | 0.062 | 0.229 (0.049–1.079) | |
| AG | 12 (42.8) | 10 (33.3) | 0.293 | 2.320 (0.483–11.148) | |
| GG | 10 (35.8) | 7 (23.4) | 0.287 | 2.333 (0.491–11.080) | |
| A = 0.4/G = 0.6 | A = 0.6/G = 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bispo, A.G.; Silva, C.S.; Sena-dos-Santos, C.; Dalledone Moura, D.; Koshimoto, B.H.B.; Santos-Lobato, B.L.; Ribeiro-dos-Santos, Â.; Cavalcante, G.C. Investigation of PRKN Mutations in Levodopa-Induced Dyskinesia in Parkinson’s Disease Treatment. Biomedicines 2023, 11, 2230. https://doi.org/10.3390/biomedicines11082230
Bispo AG, Silva CS, Sena-dos-Santos C, Dalledone Moura D, Koshimoto BHB, Santos-Lobato BL, Ribeiro-dos-Santos Â, Cavalcante GC. Investigation of PRKN Mutations in Levodopa-Induced Dyskinesia in Parkinson’s Disease Treatment. Biomedicines. 2023; 11(8):2230. https://doi.org/10.3390/biomedicines11082230
Chicago/Turabian StyleBispo, Ana Gabrielle, Caio S. Silva, Camille Sena-dos-Santos, Dafne Dalledone Moura, Brenda Hanae Bentes Koshimoto, Bruno Lopes Santos-Lobato, Ândrea Ribeiro-dos-Santos, and Giovanna C. Cavalcante. 2023. "Investigation of PRKN Mutations in Levodopa-Induced Dyskinesia in Parkinson’s Disease Treatment" Biomedicines 11, no. 8: 2230. https://doi.org/10.3390/biomedicines11082230
APA StyleBispo, A. G., Silva, C. S., Sena-dos-Santos, C., Dalledone Moura, D., Koshimoto, B. H. B., Santos-Lobato, B. L., Ribeiro-dos-Santos, Â., & Cavalcante, G. C. (2023). Investigation of PRKN Mutations in Levodopa-Induced Dyskinesia in Parkinson’s Disease Treatment. Biomedicines, 11(8), 2230. https://doi.org/10.3390/biomedicines11082230