Beyond Prostate Cancer: An Androgen Receptor Splice Variant Expression in Multiple Malignancies, Non-Cancer Pathologies, and Development
Abstract
1. Introduction
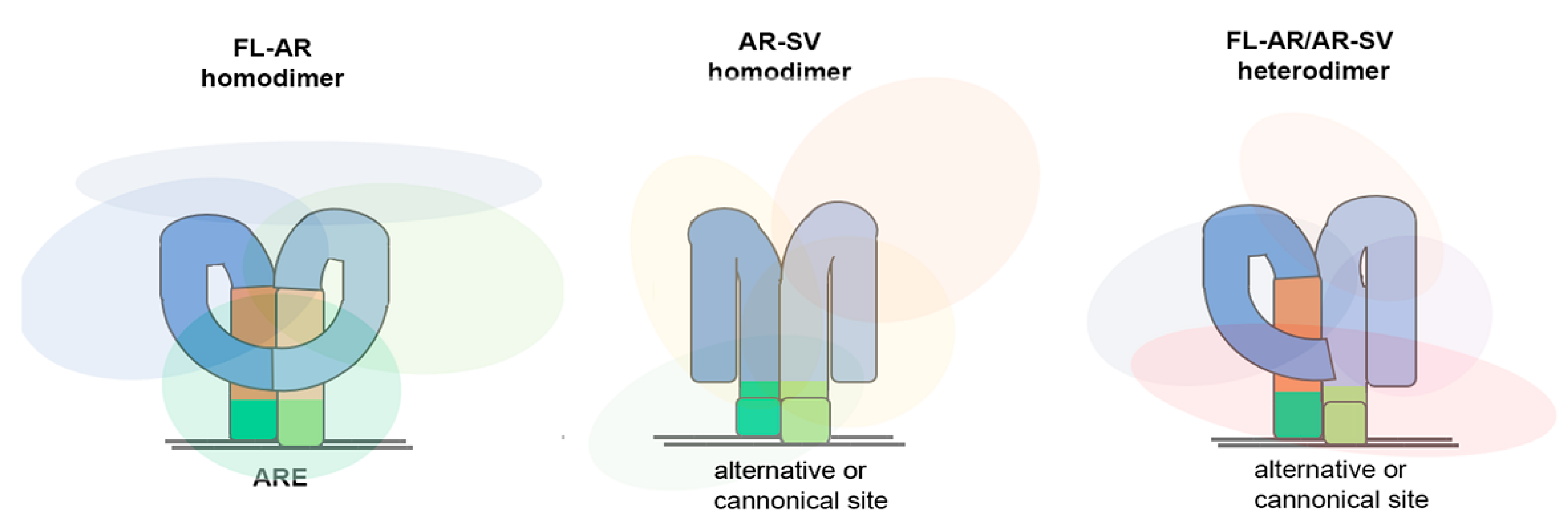
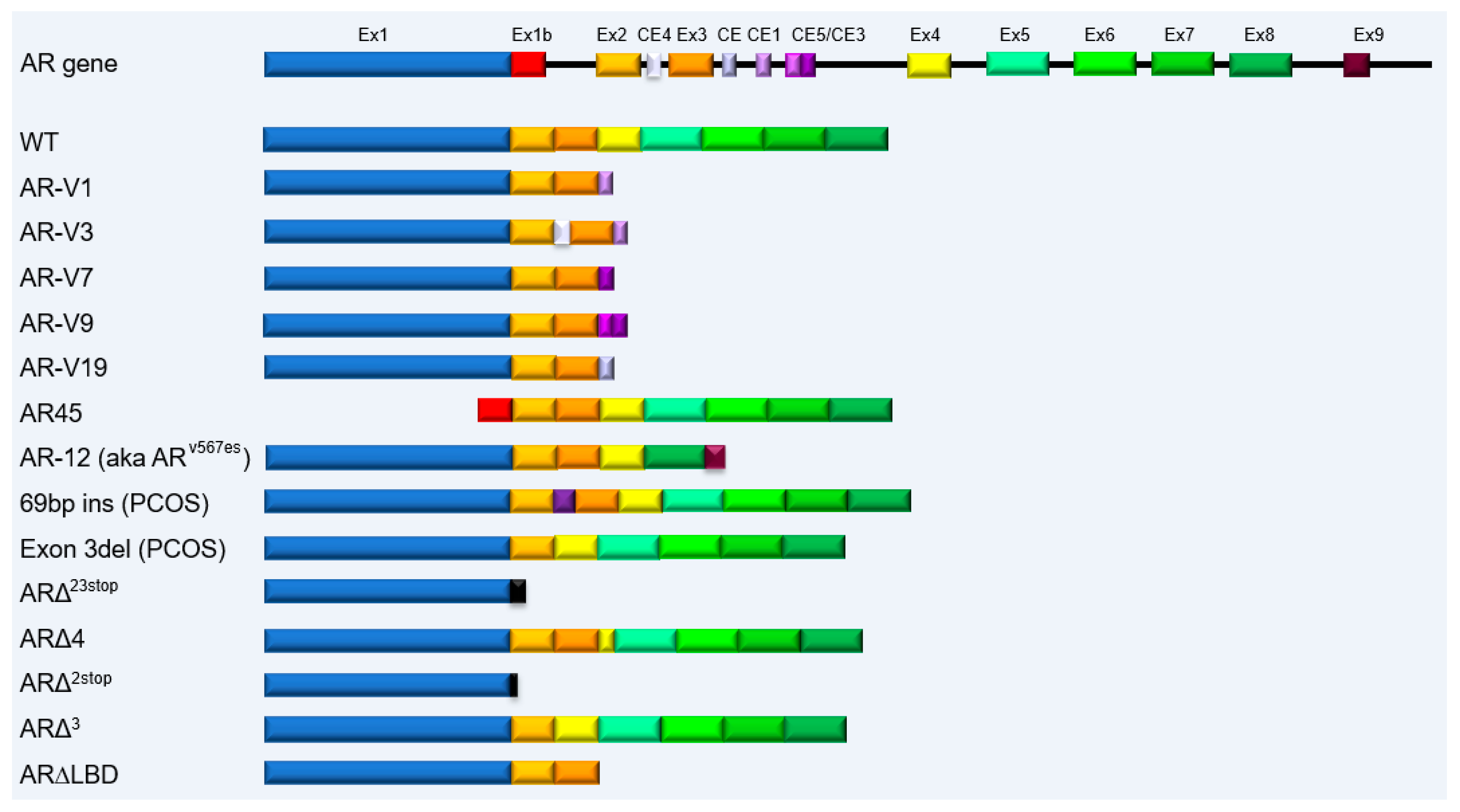
2. AR Splice Variants Expressed in Prostate Cancer
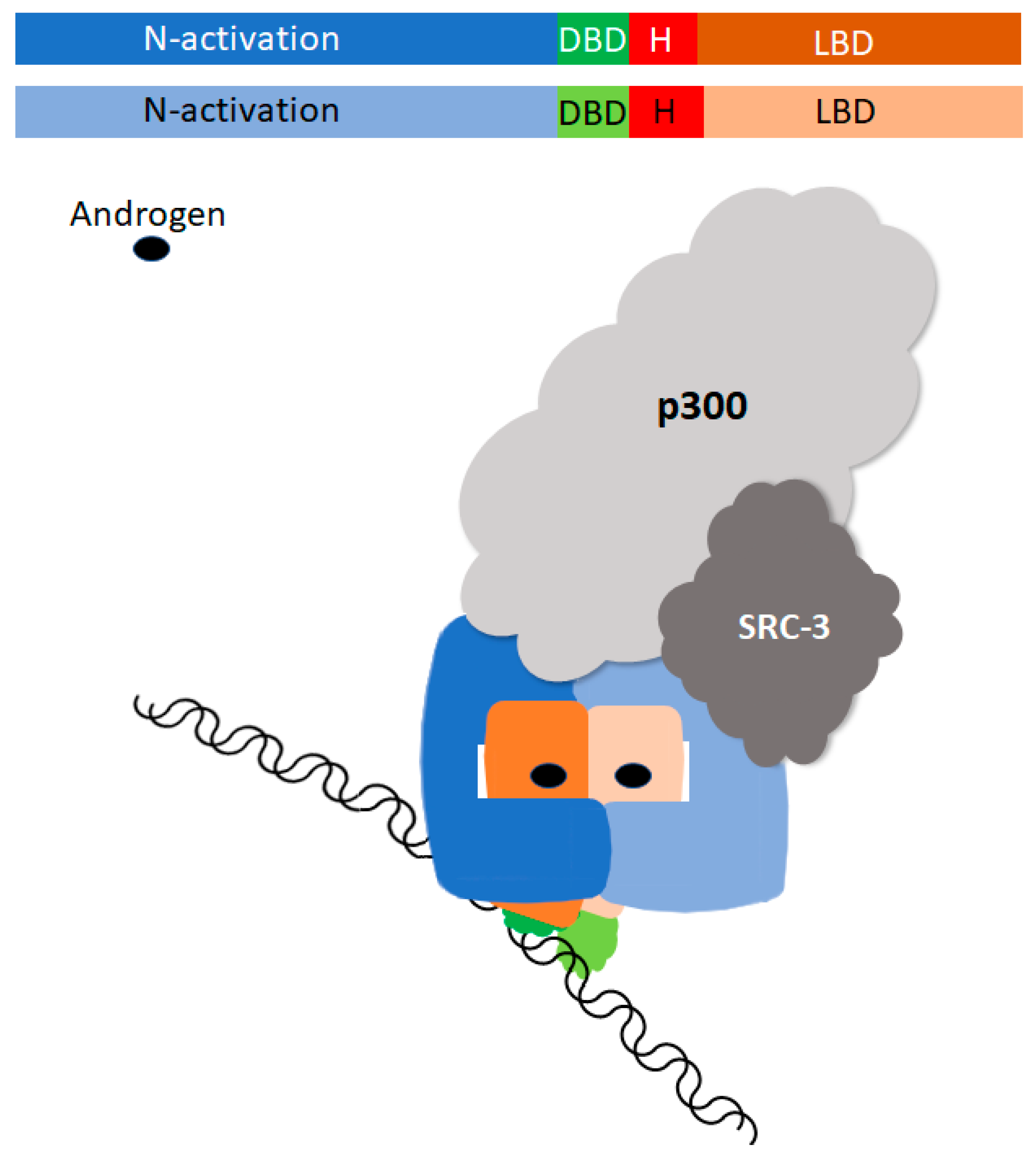
2.1. Classification and Signaling
2.2. Developmental Therapy Targeting Androgen Receptor Splice Variants
3. Sexual Dimorphism of Multiple Cancers
4. Androgen Receptor Splice Variants in Human Malignancies
4.1. Breast Cancer
4.2. Salivary Duct Carcinoma
4.3. Glioblastoma Multiforme
4.4. Renal Cancer
4.5. Bladder Cancers
4.6. Liver Cancer
5. Androgen Receptor Splice Variants in Non-Malignant Cells
5.1. Polycystic Ovarian Syndrome
5.2. Placenta
5.3. Testis
5.4. Neuronal Lipid Raft
6. Androgen Receptor Splice Variants in Immune Cells
6.1. Peripheral Blood Mononuclear Cells
6.2. Fish Immune Cells during Development
6.3. Role of Androgen Receptor Splice Variants in Immunotherapy
7. Implications, Unresolved Questions, and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ipulan-Colet, L.A. Sexual dimorphism through androgen signaling; from external genitalia to muscles. Front. Endocrinol. 2022, 13, 940229. [Google Scholar] [CrossRef]
- Gerald, T.; Raj, G. Testosterone and the Androgen Receptor. Urol. Clin. North Am. 2022, 49, 603–614. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schutz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef]
- Evans, R.M. The nuclear receptor superfamily: A rosetta stone for physiology. Mol. Endocrinol. 2005, 19, 1429–1438. [Google Scholar] [CrossRef]
- Messner, E.A.; Steele, T.M.; Tsamouri, M.M.; Hejazi, N.; Gao, A.C.; Mudryj, M.; Ghosh, P.M. The Androgen Receptor in Prostate Cancer: Effect of Structure, Ligands and Spliced Variants on Therapy. Biomedicines 2020, 8, 422. [Google Scholar] [CrossRef]
- Dahiya, U.R.; Heemers, H.V. Analyzing the Androgen Receptor Interactome in Prostate Cancer: Implications for Therapeutic Intervention. Cells 2022, 11, 936. [Google Scholar] [CrossRef] [PubMed]
- Rey, R.A. The Role of Androgen Signaling in Male Sexual Development at Puberty. Endocrinology 2021, 162, bqaa215. [Google Scholar] [CrossRef] [PubMed]
- Starka, L.; Duskova, M.; Vitku, J. 11-Keto-testosterone and other androgens of adrenal origin. Physiol. Res. 2020, 69, S187–S192. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, D.J.; Hirschberg, A.L.; Bermon, S. Circulating Testosterone as the Hormonal Basis of Sex Differences in Athletic Performance. Endocr. Rev. 2018, 39, 803–829. [Google Scholar] [CrossRef]
- Weikum, E.R.; Liu, X.; Ortlund, E.A. The nuclear receptor superfamily: A structural perspective. Protein Sci. 2018, 27, 1876–1892. [Google Scholar] [CrossRef] [PubMed]
- Migeon, B.R.; Brown, T.R.; Axelman, J.; Migeon, C.J. Studies of the locus for androgen receptor: Localization on the human X chromosome and evidence for homology with the Tfm locus in the mouse. Proc. Natl. Acad. Sci. USA. 1981, 78, 6339–6343. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, A.O.; Faber, P.W.; van Rooij, H.C.; Kuiper, G.G.; Ris, C.; Klaassen, P.; van der Korput, J.A.; Voorhorst, M.M.; van Laar, J.H.; Mulder, E.; et al. The human androgen receptor: Domain structure, genomic organization and regulation of expression. J. Steroid Biochem. 1989, 34, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Veldscholte, J.; Berrevoets, C.A.; Zegers, N.D.; van der Kwast, T.H.; Grootegoed, J.A.; Mulder, E. Hormone-induced dissociation of the androgen receptor-heat-shock protein complex: Use of a new monoclonal antibody to distinguish transformed from nontransformed receptors. Biochemistry 1992, 31, 7422–7430. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yi, P.; Hamilton, R.A.; Shen, H.; Chen, M.; Foulds, C.E.; Mancini, M.A.; Ludtke, S.J.; Wang, Z.; O’Malley, B.W. Structural Insights of Transcriptionally Active, Full-Length Androgen Receptor Coactivator Complexes. Mol. Cell 2020, 79, 812–823.e814. [Google Scholar] [CrossRef]
- Smith, D.F.; Toft, D.O. Minireview: The intersection of steroid receptors with molecular chaperones: Observations and questions. Mol. Endocrinol. 2008, 22, 2229–2240. [Google Scholar] [CrossRef]
- Huggins, C.; Hodges, C.V. Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. J. Urol. 2002, 167, 948–951; discussion 952. [Google Scholar] [CrossRef]
- Magee, D.E.; Singal, R.K. Androgen deprivation therapy: Indications, methods of utilization, side effects and their management. Can. J. Urol. 2020, 27, 11–16. [Google Scholar]
- Shore, N.D.; Antonarakis, E.S.; Cookson, M.S.; Crawford, E.D.; Morgans, A.K.; Albala, D.M.; Hafron, J.; Harris, R.G.; Saltzstein, D.; Brown, G.A.; et al. Optimizing the role of androgen deprivation therapy in advanced prostate cancer: Challenges beyond the guidelines. Prostate 2020, 80, 527–544. [Google Scholar] [CrossRef]
- Yu, E.M.; Aragon-Ching, J.B. Advances with androgen deprivation therapy for prostate cancer. Exp. Opin. Pharmacother. 2022, 23, 1015–1033. [Google Scholar] [CrossRef]
- Roy, S.; Sayyid, R.; Saad, F.; Sun, Y.; Lajkosz, K.; Ong, M.; Klaassen, Z.; Malone, S.; Spratt, D.E.; Wallis, C.J.D.; et al. Addition of Docetaxel to Androgen Receptor Axis-targeted Therapy and Androgen Deprivation Therapy in Metastatic Hormone-sensitive Prostate Cancer: A Network Meta-analysis. Eur. Urol. Oncol. 2022, 5, 494–502. [Google Scholar]
- Yanagisawa, T.; Rajwa, P.; Thibault, C.; Gandaglia, G.; Mori, K.; Kawada, T.; Fukuokaya, W.; Shim, S.R.; Mostafaei, H.; Motlagh, R.S.; et al. Androgen Receptor Signaling Inhibitors in Addition to Docetaxel with Androgen Deprivation Therapy for Metastatic Hormone-sensitive Prostate Cancer: A Systematic Review and Meta-Analysis. Eur. Urol. 2022, 82, 584–598. [Google Scholar]
- Harris, W.P.; Mostaghel, E.A.; Nelson, P.S.; Montgomery, B. Androgen deprivation therapy: Progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat. Clin. Pract. Urol. 2009, 6, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Karantanos, T.; Corn, P.G.; Thompson, T.C. Prostate cancer progression after androgen deprivation therapy: Mechanisms of castrate resistance and novel therapeutic approaches. Oncogene 2013, 32, 5501–5511. [Google Scholar] [PubMed]
- Wadosky, K.M.; Koochekpour, S. Molecular mechanisms underlying resistance to androgen deprivation therapy in prostate cancer. Oncotarget 2016, 7, 64447–64470. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Dehm, S.M.; Sharifi, N. Targeting the Androgen Signaling Axis in Prostate Cancer. J. Clin. Oncol. 2023, JCO2300433. [Google Scholar] [CrossRef]
- Mudryj, M.; Tepper, C.G. On the origins of the androgen receptor low molecular weight species. Horm. Cancer 2013, 4, 259–269. [Google Scholar] [CrossRef]
- Ware, K.E.; Garcia-Blanco, M.A.; Armstrong, A.J.; Dehm, S.M. Biologic and clinical significance of androgen receptor variants in castration resistant prostate cancer. Endocr. Relat. Cancer 2014, 21, T87–T103. [Google Scholar] [CrossRef]
- Bryce, A.H.; Antonarakis, E.S. Androgen receptor splice variant 7 in castration-resistant prostate cancer: Clinical considerations. Int. J. Urol. 2016, 23, 646–653. [Google Scholar] [CrossRef]
- Imamura, Y.; Sadar, M.D. Androgen receptor targeted therapies in castration-resistant prostate cancer: Bench to clinic. Int. J. Urol. 2016, 23, 654–665. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Armstrong, A.J.; Dehm, S.M.; Luo, J. Androgen receptor variant-driven prostate cancer: Clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis. 2016, 19, 231–241. [Google Scholar] [CrossRef]
- Kanayama, M.; Lu, C.; Luo, J.; Antonarakis, E.S. AR Splicing Variants and Resistance to AR Targeting Agents. Cancers 2021, 13, 2563. [Google Scholar] [CrossRef] [PubMed]
- Dehm, S.M.; Schmidt, L.J.; Heemers, H.V.; Vessella, R.L.; Tindall, D.J. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008, 68, 5469–5477. [Google Scholar] [CrossRef]
- Hu, R.; Dunn, T.A.; Wei, S.; Isharwal, S.; Veltri, R.W.; Humphreys, E.; Han, M.; Partin, A.W.; Vessella, R.L.; Isaacs, W.B.; et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009, 69, 16–22. [Google Scholar] [CrossRef]
- Libertini, S.J.; Tepper, C.G.; Rodriguez, V.; Asmuth, D.M.; Kung, H.J.; Mudryj, M. Evidence for calpain-mediated androgen receptor cleavage as a mechanism for androgen independence. Cancer Res. 2007, 67, 9001–9005. [Google Scholar] [CrossRef]
- Guo, Z.; Yang, X.; Sun, F.; Jiang, R.; Linn, D.E.; Chen, H.; Chen, H.; Kong, X.; Melamed, J.; Tepper, C.G.; et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009, 69, 2305–2313. [Google Scholar] [CrossRef]
- Sun, S.; Sprenger, C.C.; Vessella, R.L.; Haugk, K.; Soriano, K.; Mostaghel, E.A.; Page, S.T.; Coleman, I.M.; Nguyen, H.M.; Sun, H.; et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J. Clin. Investig. 2010, 120, 2715–2730. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Attard, G.; Balk, S.P.; Bevan, C.; Burnstein, K.; Cato, L.; Cherkasov, A.; De Bono, J.S.; Dong, Y.; Gao, A.C.; et al. Role of Androgen Receptor Variants in Prostate Cancer: Report from the 2017 Mission Androgen Receptor Variants Meeting. Eur. Urol. 2018, 73, 715–723. [Google Scholar]
- Lu, C.; Luo, J. Decoding the androgen receptor splice variants. Transl. Androl. Urol. 2013, 2, 178–186. [Google Scholar] [PubMed]
- Cato, L.; de Tribolet-Hardy, J.; Lee, I.; Rottenberg, J.T.; Coleman, I.; Melchers, D.; Houtman, R.; Xiao, T.; Li, W.; Uo, T.; et al. ARv7 Represses Tumor-Suppressor Genes in Castration-Resistant Prostate Cancer. Cancer Cell 2019, 35, 401–413.e406. [Google Scholar] [CrossRef]
- Basil, P.; Robertson, M.J.; Bingman, W.E., 3rd.; Dash, A.K.; Krause, W.C.; Shafi, A.A.; Piyarathna, B.; Coarfa, C.; Weigel, N.L. Cistrome and transcriptome analysis identifies unique androgen receptor (AR) and AR-V7 splice variant chromatin binding and transcriptional activities. Sci. Rep. 2022, 12, 5351. [Google Scholar] [CrossRef]
- Cai, L.; Tsai, Y.H.; Wang, P.; Wang, J.; Li, D.; Fan, H.; Zhao, Y.; Bareja, R.; Lu, R.; Wilson, E.M.; et al. ZFX Mediates Non-canonical Oncogenic Functions of the Androgen Receptor Splice Variant 7 in Castrate-Resistant Prostate Cancer. Mol. Cell 2018, 72, 341–354.e346. [Google Scholar] [CrossRef]
- Hu, R.; Lu, C.; Mostaghel, E.A.; Yegnasubramanian, S.; Gurel, M.; Tannahill, C.; Edwards, J.; Isaacs, W.B.; Nelson, P.S.; Bluemn, E.; et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012, 72, 3457–3462. [Google Scholar] [CrossRef]
- Krause, W.C.; Shafi, A.A.; Nakka, M.; Weigel, N.L. Androgen receptor and its splice variant, AR-V7, differentially regulate FOXA1 sensitive genes in LNCaP prostate cancer cells. Int. J. Biochem. Cell Biol. 2014, 54, 49–59. [Google Scholar] [CrossRef]
- Zhang, T.; Armstrong, A.J. Clinical Utility of Circulating Tumor Cells in Advanced Prostate Cancer. Curr. Oncol. Rep. 2016, 18, 3. [Google Scholar] [CrossRef]
- Zhang, T.; Karsh, L.I.; Nissenblatt, M.J.; Canfield, S.E. Androgen Receptor Splice Variant, AR-V7, as a Biomarker of Resistance to Androgen Axis-Targeted Therapies in Advanced Prostate Cancer. Clin. Genitourin. Cancer 2020, 18, 1–10. [Google Scholar] [CrossRef]
- Enikeev, D.; Morozov, A.; Babaevskaya, D.; Bazarkin, A.; Malavaud, B. A Systematic Review of Circulating Tumor Cells Clinical Application in Prostate Cancer Diagnosis. Cancers 2022, 14, 3802. [Google Scholar] [CrossRef]
- Li, Y.; Alsagabi, M.; Fan, D.; Bova, G.S.; Tewfik, A.H.; Dehm, S.M. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011, 71, 2108–2117. [Google Scholar] [CrossRef]
- Tepper, C.G.; Boucher, D.L.; Ryan, P.E.; Ma, A.H.; Xia, L.; Lee, L.F.; Pretlow, T.G.; Kung, H.J. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res. 2002, 62, 6606–6614. [Google Scholar] [PubMed]
- Henzler, C.; Li, Y.; Yang, R.; McBride, T.; Ho, Y.; Sprenger, C.; Liu, G.; Coleman, I.; Lakely, B.; Li, R.; et al. Truncation and constitutive activation of the androgen receptor by diverse genomic rearrangements in prostate cancer. Nat. Commun. 2016, 7, 13668. [Google Scholar] [CrossRef] [PubMed]
- De Laere, B.; van Dam, P.J.; Whitington, T.; Mayrhofer, M.; Diaz, E.H.; Van den Eynden, G.; Vandebroek, J.; Del-Favero, J.; Van Laere, S.; Dirix, L.; et al. Comprehensive Profiling of the Androgen Receptor in Liquid Biopsies from Castration-resistant Prostate Cancer Reveals Novel Intra-AR Structural Variation and Splice Variant Expression Patterns. Eur. Urol. 2017, 72, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Paschalis, A.; Sharp, A.; Welti, J.C.; Neeb, A.; Raj, G.V.; Luo, J.; Plymate, S.R.; de Bono, J.S. Alternative splicing in prostate cancer. Nat. Rev. Clin. Oncol. 2018, 15, 663–675. [Google Scholar] [CrossRef]
- Nadiminty, N.; Tummala, R.; Liu, C.; Lou, W.; Evans, C.P.; Gao, A.C. NF-kappaB2/p52:c-Myc:hnRNPA1 Pathway Regulates Expression of Androgen Receptor Splice Variants and Enzalutamide Sensitivity in Prostate Cancer. Mol. Cancer Ther. 2015, 14, 1884–1895. [Google Scholar] [CrossRef]
- Cao, S.; Zhan, Y.; Dong, Y. Emerging data on androgen receptor splice variants in prostate cancer. Endocr. Relat. Cancer 2016, 23, T199–T210. [Google Scholar] [CrossRef]
- Liu, L.L.; Xie, N.; Sun, S.; Plymate, S.; Mostaghel, E.; Dong, X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 2014, 33, 3140–3150. [Google Scholar] [CrossRef] [PubMed]
- Ferraldeschi, R.; Welti, J.; Powers, M.V.; Yuan, W.; Smyth, T.; Seed, G.; Riisnaes, R.; Hedayat, S.; Wang, H.; Crespo, M.; et al. Second-Generation HSP90 Inhibitor Onalespib Blocks mRNA Splicing of Androgen Receptor Variant 7 in Prostate Cancer Cells. Cancer Res. 2016, 76, 2731–2742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, N.; Huang, J.; Ho, T.T.; Zhu, Z.; Qiu, Z.; Zhou, X.; Bai, C.; Wu, F.; Xu, M.; et al. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget 2016, 7, 15481–15491. [Google Scholar] [CrossRef]
- Adamiecki, R.; Hryniewicz-Jankowska, A.; Ortiz, M.A.; Li, X.; Porter-Hansen, B.A.; Nsouli, I.; Bratslavsky, G.; Kotula, L. In Vivo Models for Prostate Cancer Research. Cancers 2022, 14, 5321. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Chen, H.G.; Li, W.; Yang, X.; Wang, X.; Jiang, R.; Guo, Z.; Chen, H.; Huang, J.; Borowsky, A.D.; et al. Androgen receptor splice variant AR3 promotes prostate cancer via modulating expression of autocrine/paracrine factors. J. Biol. Chem. 2014, 289, 1529–1539. [Google Scholar] [CrossRef]
- Liu, G.; Sprenger, C.; Sun, S.; Epilepsia, K.S.; Haugk, K.; Zhang, X.; Coleman, I.; Nelson, P.S.; Plymate, S. AR variant ARv567es induces carcinogenesis in a novel transgenic mouse model of prostate cancer. Neoplasia 2013, 15, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Zhang, R.; Han, X.; Zhou, J. Targeting the N-terminal domain of the androgen receptor: The effective approach in therapy of CRPC. Eur. J. Med. Chem. 2023, 247, 115077. [Google Scholar] [CrossRef]
- Andersen, R.J.; Mawji, N.R.; Wang, J.; Wang, G.; Haile, S.; Myung, J.K.; Watt, K.; Tam, T.; Yang, Y.C.; Banuelos, C.A.; et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 2010, 17, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Myung, J.K.; Banuelos, C.A.; Fernandez, J.G.; Mawji, N.R.; Wang, J.; Tien, A.H.; Yang, Y.C.; Tavakoli, I.; Haile, S.; Watt, K.; et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J. Clin. Investig. 2013, 123, 2948–2960. [Google Scholar] [CrossRef]
- Brand, L.J.; Olson, M.E.; Ravindranathan, P.; Guo, H.; Kempema, A.M.; Andrews, T.E.; Chen, X.; Raj, G.V.; Harki, D.A.; Dehm, S.M. EPI-001 is a selective peroxisome proliferator-activated receptor-gamma modulator with inhibitory effects on androgen receptor expression and activity in prostate cancer. Oncotarget 2015, 6, 3811–3824. [Google Scholar] [CrossRef]
- Ponnusamy, S.; Coss, C.C.; Thiyagarajan, T.; Watts, K.; Hwang, D.J.; He, Y.; Selth, L.A.; McEwan, I.J.; Duke, C.B.; Pagadala, J.; et al. Novel Selective Agents for the Degradation of Androgen Receptor Variants to Treat Castration-Resistant Prostate Cancer. Cancer Res. 2017, 77, 6282–6298. [Google Scholar] [CrossRef]
- He, Y.; Hwang, D.J.; Ponnusamy, S.; Thiyagarajan, T.; Mohler, M.L.; Narayanan, R.; Miller, D.D. Exploration and Biological Evaluation of Basic Heteromonocyclic Propanamide Derivatives as SARDs for the Treatment of Enzalutamide-Resistant Prostate Cancer. J. Med. Chem. 2021, 64, 11045–11062. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Song, C.H.; Kim, K.J.; Lee, K. A new compound targets the AF-1 of androgen receptor and decreases its activity and protein levels in prostate cancer cells. Am. J. Cancer Res. 2020, 10, 4607–4623. [Google Scholar]
- Goicochea, N.L.; Garnovskaya, M.; Blanton, M.G.; Chan, G.; Weisbart, R.; Lilly, M.B. Development of cell-penetrating bispecific antibodies targeting the N-terminal domain of androgen receptor for prostate cancer therapy. Protein Eng. Des. Sel. 2017, 30, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.B.; Lagas, J.S.; Broestl, L.; Sponagel, J.; Rockwell, N.; Rhee, G.; Rosen, S.F.; Chen, S.; Klein, R.S.; Imoukhuede, P.; et al. Sex differences in cancer mechanisms. Biol. Sex Differ. 2017, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Ramos, C.M.; Quackenbush, J.; DeMeo, D.L. Genome-Wide Sex and Gender Differences in Cancer. Front. Oncol. 2020, 10, 597788. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Clocchiatti, A.; Cora, E.; Zhang, Y.; Dotto, G.P. Sexual dimorphism in cancer. Nat. Rev. Cancer 2016, 16, 330–339. [Google Scholar] [CrossRef]
- Santen, R.J. Assessing individual risk for breast cancer: Role of oestrogens and androgens. Breast Cancer Res. 2008, 10, S10. [Google Scholar] [CrossRef]
- Santen, R.J. Menopausal hormone therapy and breast cancer. J. Steroid Biochem. Mol. Biol. 2014, 142, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Xiang, L.; Li, T.; Bai, Z.; Hallmarks, C. Biomarkers and Breast Cancer Molecular Subtypes. J. Cancer 2016, 7, 1281–1294. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.E.; Clements, J.A.; Birrell, S.N.; Tilley, W.D. Prostate-specific antigen and gross cystic disease fluid protein-15 are co-expressed in androgen receptor-positive breast tumours. Br. J. Cancer 1998, 78, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Kuenen-Boumeester, V.; Van der Kwast, T.H.; van Putten, W.L.; Claassen, C.; van Ooijen, B.; Henzen-Logmans, S.C. Immunohistochemical determination of androgen receptors in relation to oestrogen and progesterone receptors in female breast cancer. Int. J. Cancer 1992, 52, 581–584. [Google Scholar] [CrossRef]
- Agoff, S.N.; Swanson, P.E.; Linden, H.; Hawes, S.E.; Lawton, T.J. Androgen receptor expression in estrogen receptor-negative breast cancer. Immunohistochemical, clinical, and prognostic associations. Am. J. Clin. Pathol. 2003, 120, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Kaaks, R.; Rinaldi, S.; Key, T.J.; Berrino, F.; Peeters, P.H.; Biessy, C.; Dossus, L.; Lukanova, A.; Bingham, S.; Khaw, K.T.; et al. Postmenopausal serum androgens, oestrogens and breast cancer risk: The European prospective investigation into cancer and nutrition. Endocr. Relat. Cancer 2005, 12, 1071–1082. [Google Scholar] [CrossRef]
- Secreto, G.; Toniolo, P.; Pisani, P.; Recchione, C.; Cavalleri, A.; Fariselli, G.; Totis, A.; Di Pietro, S.; Berrino, F. Androgens and breast cancer in premenopausal women. Cancer Res. 1989, 49, 471–476. [Google Scholar]
- Grattarola, R. The Premenstrual Endometrial Pattern of Women with Breast Cancer. A Study of Progestational Activity. Cancer 1964, 17, 1119–1122. [Google Scholar] [CrossRef]
- Somboonporn, W.; Davis, S.R. Postmenopausal testosterone therapy and breast cancer risk. Maturitas 2004, 49, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Key, T.; Appleby, P.; Barnes, I.; Reeves, G.; Endogenous, H.; Collaborative, G.B.C. Endogenous sex hormones and breast cancer in postmenopausal women: Reanalysis of nine prospective studies. J. Natl. Cancer Inst. 2002, 94, 606–616. [Google Scholar]
- Secreto, G.; Zumoff, B. Abnormal production of androgens in women with breast cancer. Anticancer Res. 1994, 14, 2113–2117. [Google Scholar] [PubMed]
- You, C.-P.; Tsoi, H.; Man, E.P.S.; Leung, M.-H.; Khoo, U.S. Modulating the Activity of Androgen Receptor for Treating Breast Cancer. Int. J. Mol. Sci. 2022, 23, 15342. [Google Scholar]
- Brys, M.; Wojcik, M.; Romanowicz-Makowska, H.; Krajewska, W.M. Androgen receptor status in female breast cancer: RT-PCR and Western blot studies. J. Cancer Res. Clin. Oncol. 2002, 128, 85–90. [Google Scholar]
- Bieche, I.; Parfait, B.; Tozlu, S.; Lidereau, R.; Vidaud, M. Quantitation of androgen receptor gene expression in sporadic breast tumors by real-time RT-PCR: Evidence that MYC is an AR-regulated gene. Carcinogenesis 2001, 22, 1521–1526. [Google Scholar] [CrossRef]
- Soreide, J.A.; Lea, O.A.; Varhaug, J.E.; Skarstein, A.; Kvinnsland, S. Androgen receptors in operable breast cancer: Relation to other steroid hormone receptors, correlations to prognostic factors and predictive value for effect of adjuvant tamoxifen treatment. Eur. J. Surg. Oncol. 1992, 18, 112–118. [Google Scholar]
- Basile, D.; Cinausero, M.; Iacono, D.; Pelizzari, G.; Bonotto, M.; Vitale, M.G.; Gerratana, L.; Puglisi, F. Androgen receptor in estrogen receptor positive breast cancer: Beyond expression. Cancer Treat. Rev. 2017, 61, 15–22. [Google Scholar] [CrossRef]
- Hickey, T.E.; Robinson, J.L.; Carroll, J.S.; Tilley, W.D. Minireview: The androgen receptor in breast tissues: Growth inhibitor, tumor suppressor, oncogene? Mol. Endocrinol. 2012, 26, 1252–1267. [Google Scholar] [CrossRef] [PubMed]
- Hickey, T.E.; Selth, L.A.; Chia, K.M.; Laven-Law, G.; Milioli, H.H.; Roden, D.; Jindal, S.; Hui, M.; Finlay-Schultz, J.; Ebrahimie, E.; et al. The androgen receptor is a tumor suppressor in estrogen receptor-positive breast cancer. Nat. Med. 2021, 27, 310–320. [Google Scholar] [CrossRef]
- Glaser, R.L.; Dimitrakakis, C. Rapid response of breast cancer to neoadjuvant intramammary testosterone-anastrozole therapy: Neoadjuvant hormone therapy in breast cancer. Menopause 2014, 21, 673–678. [Google Scholar] [CrossRef]
- Glaser, R.L.; York, A.E.; Dimitrakakis, C. Subcutaneous testosterone-letrozole therapy before and concurrent with neoadjuvant breast chemotherapy: Clinical response and therapeutic implications. Menopause 2017, 24, 859–864. [Google Scholar] [CrossRef]
- Boni, C.; Pagano, M.; Panebianco, M.; Bologna, A.; Sierra, N.M.; Gnoni, R.; Formisano, D.; Bisagni, G. Therapeutic activity of testosterone in metastatic breast cancer. Anticancer Res. 2014, 34, 1287–1290. [Google Scholar]
- Bluemn, E.G.; Coleman, I.M.; Lucas, J.M.; Coleman, R.T.; Hernandez-Lopez, S.; Tharakan, R.; Bianchi-Frias, D.; Dumpit, R.F.; Kaipainen, A.; Corella, A.N.; et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017, 32, 474–489.e476. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.A.; D’Amato, N.C.; Gu, H.; Babbs, B.; Wulfkuhle, J.; Petricoin, E.F.; Gallagher, I.; Dong, T.; Torkko, K.; Liu, B.; et al. Synergy between Androgen Receptor Antagonism and Inhibition of mTOR and HER2 in Breast Cancer. Mol. Cancer Ther. 2017, 16, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Han, J.; Liang, X.; Sun, S.; Jiang, Y.; Xia, B.; Niu, M.; Li, D.; Zhang, J.; Wang, S.; et al. Androgen Receptor Expression and Bicalutamide Antagonize Androgen Receptor Inhibit beta-Catenin Transcription Complex in Estrogen Receptor-Negative Breast Cancer. Cell Physiol. Biochem. 2017, 43, 2212–2225. [Google Scholar] [CrossRef] [PubMed]
- Barton, V.N.; D’Amato, N.C.; Gordon, M.A.; Lind, H.T.; Spoelstra, N.S.; Babbs, B.L.; Heinz, R.E.; Elias, A.; Jedlicka, P.; Jacobsen, B.M.; et al. Multiple molecular subtypes of triple-negative breast cancer critically rely on androgen receptor and respond to enzalutamide in vivo. Mol. Cancer Ther. 2015, 14, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Chia, K.M.; Liu, J.; Francis, G.D.; Naderi, A. A feedback loop between androgen receptor and ERK signaling in estrogen receptor-negative breast cancer. Neoplasia 2011, 13, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Chen, Y.; Lim, E.; Wimberly, H.; Bailey, S.T.; Imai, Y.; Rimm, D.L.; Liu, X.S.; Brown, M. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell 2011, 20, 119–131. [Google Scholar] [CrossRef]
- Giovannelli, P.; Di Donato, M.; Auricchio, F.; Castoria, G.; Migliaccio, A. Androgens Induce Invasiveness of Triple Negative Breast Cancer Cells Through AR/Src/PI3-K Complex Assembly. Sci. Rep. 2019, 9, 4490. [Google Scholar] [CrossRef]
- Park, S.; Koo, J.S.; Kim, M.S.; Park, H.S.; Lee, J.S.; Lee, J.S.; Kim, S.I.; Park, B.W.; Lee, K.S. Androgen receptor expression is significantly associated with better outcomes in estrogen receptor-positive breast cancers. Ann. Oncol. 2011, 22, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
- Asano, Y.; Kashiwagi, S.; Onoda, N.; Kurata, K.; Morisaki, T.; Noda, S.; Takashima, T.; Ohsawa, M.; Kitagawa, S.; Hirakawa, K. Clinical verification of sensitivity to preoperative chemotherapy in cases of androgen receptor-expressing positive breast cancer. Br. J. Cancer 2016, 114, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Tsvetkova, V.; Griguolo, G.; Miglietta, F.; Mantiero, M.; Tasca, G.; Cumerlato, E.; Giorgi, C.A.; Giarratano, T.; Faggioni, G.; et al. Androgen Receptor Expression and Association with Distant Disease-Free Survival in Triple Negative Breast Cancer: Analysis of 263 Patients Treated With Standard Therapy for Stage I–III Disease. Front. Oncol. 2019, 9, 452. [Google Scholar] [PubMed]
- Isola, J.J. Immunohistochemical demonstration of androgen receptor in breast cancer and its relationship to other prognostic factors. J. Pathol. 1993, 170, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Arce-Salinas, C.; Riesco-Martinez, M.C.; Hanna, W.; Bedard, P.; Warner, E. Complete Response of Metastatic Androgen Receptor-Positive Breast Cancer to Bicalutamide: Case Report and Review of the Literature. J. Clin. Oncol. 2016, 34, e21–e24. [Google Scholar]
- Gucalp, A.; Tolaney, S.; Isakoff, S.J.; Ingle, J.N.; Liu, M.C.; Carey, L.A.; Blackwell, K.; Rugo, H.; Nabell, L.; Forero, A.; et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin. Cancer Res. 2013, 19, 5505–5512. [Google Scholar] [CrossRef]
- Traina, T.A.; Miller, K.; Yardley, D.A.; Eakle, J.; Schwartzberg, L.S.; O’Shaughnessy, J.; Gradishar, W.; Schmid, P.; Winer, E.; Kelly, C.; et al. Enzalutamide for the Treatment of Androgen Receptor-Expressing Triple-Negative Breast Cancer. J. Clin. Oncol. 2018, 36, 884–890. [Google Scholar] [CrossRef]
- Hoeferlin, L.A.; Chalfant, C.E.; Park, M.A. Challenges in the Treatment of Triple Negative and HER2-Overexpressing Breast Cancer. J. Surg. Sci. 2013, 1, 3–7. [Google Scholar]
- Hu, D.G.; Hickey, T.E.; Irvine, C.; Wijayakumara, D.D.; Lu, L.; Tilley, W.D.; Selth, L.A.; Mackenzie, P.I. Identification of androgen receptor splice variant transcripts in breast cancer cell lines and human tissues. Horm. Cancer 2014, 5, 61–71. [Google Scholar]
- Hickey, T.E.; Irvine, C.M.; Dvinge, H.; Tarulli, G.A.; Hanson, A.R.; Ryan, N.K.; Pickering, M.A.; Birrell, S.N.; Hu, D.G.; Mackenzie, P.I.; et al. Expression of androgen receptor splice variants in clinical breast cancers. Oncotarget 2015, 6, 44728–44744. [Google Scholar] [CrossRef]
- Aceto, N.; Bardia, A.; Wittner, B.S.; Donaldson, M.C.; O’Keefe, R.; Engstrom, A.; Bersani, F.; Zheng, Y.; Comaills, V.; Niederhoffer, K.; et al. AR Expression in Breast Cancer CTCs Associates with Bone Metastases. Mol. Cancer Res. 2018, 16, 720–727. [Google Scholar] [CrossRef]
- Kasimir-Bauer, S.; Keup, C.; Hoffmann, O.; Hauch, S.; Kimmig, R.; Bittner, A.K. Circulating Tumor Cells Expressing the Prostate Specific Membrane Antigen (PSMA) Indicate Worse Outcome in Primary, Non-Metastatic Triple-Negative Breast Cancer. Front. Oncol. 2020, 10, 1658. [Google Scholar] [PubMed]
- Ferguson, D.C.; Mata, D.A.; Tay, T.K.; Traina, T.A.; Gucalp, A.; Chandarlapaty, S.; D’Alfonso, T.M.; Brogi, E.; Mullaney, K.; Ladanyi, M.; et al. Androgen receptor splice variant-7 in breast cancer: Clinical and pathologic correlations. Mod. Pathol. 2022, 35, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Mitani, Y.; Rao, P.H.; Maity, S.N.; Lee, Y.C.; Ferrarotto, R.; Post, J.C.; Licitra, L.; Lippman, S.M.; Kies, M.S.; Weber, R.S.; et al. Alterations associated with androgen receptor gene activation in salivary duct carcinoma of both sexes: Potential therapeutic ramifications. Clin. Cancer Res. 2014, 20, 6570–6581.e814. [Google Scholar]
- Dalin, M.G.; Desrichard, A.; Katabi, N.; Makarov, V.; Walsh, L.A.; Lee, K.W.; Wang, Q.; Armenia, J.; West, L.; Dogan, S.; et al. Comprehensive Molecular Characterization of Salivary Duct Carcinoma Reveals Actionable Targets and Similarity to Apocrine Breast Cancer. Clin. Cancer Res. 2016, 22, 4623–4633. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.H. Salivary duct carcinoma: New developments—Morphological variants including pure in situ high grade lesions; proposed molecular classification. Head Neck Pathol. 2013, 7, S48–S58. [Google Scholar] [PubMed]
- Di Palma, S.; Simpson, R.H.; Marchio, C.; Skalova, A.; Ungari, M.; Sandison, A.; Whitaker, S.; Parry, S.; Reis-Filho, J.S. Salivary duct carcinomas can be classified into luminal androgen receptor-positive, HER2 and basal-like phenotypes. Histopathology 2012, 61, 629–643. [Google Scholar] [CrossRef]
- Chiosea, S.I.; Williams, L.; Griffith, C.C.; Thompson, L.D.; Weinreb, I.; Bauman, J.E.; Luvison, A.; Roy, S.; Seethala, R.R.; Nikiforova, M.N. Molecular characterization of apocrine salivary duct carcinoma. Am. J. Surg. Pathol. 2015, 39, 744–752. [Google Scholar] [CrossRef]
- Luk, P.P.; Weston, J.D.; Yu, B.; Selinger, C.I.; Ekmejian, R.; Eviston, T.J.; Lum, T.; Gao, K.; Boyer, M.; O’Toole, S.A.; et al. Salivary duct carcinoma: Clinicopathologic features, morphologic spectrum, and somatic mutations. Head Neck 2016, 38, E1838–E1847. [Google Scholar]
- Williams, L.; Thompson, L.D.; Seethala, R.R.; Weinreb, I.; Assaad, A.M.; Tuluc, M.; Din, N.U.; Purgina, B.; Lai, C.; Griffith, C.C.; et al. Salivary duct carcinoma: The predominance of apocrine morphology, prevalence of histologic variants and androgen receptor expression. Am. J. Surg. Pathol. 2015, 39, 705–713. [Google Scholar]
- Locati, L.D.; Quattrone, P.; Bossi, P.; Marchiano, A.V.; Cantu, G.; Licitra, L. A complete remission with androgen-deprivation therapy in a recurrent androgen receptor-expressing adenocarcinoma of the parotid gland. Ann. Oncol. 2003, 14, 1327–1328. [Google Scholar] [CrossRef]
- Jaspers, H.C.; Verbist, B.M.; Schoffelen, R.; Mattijssen, V.; Slootweg, P.J.; van der Graaf, W.T.; van Herpen, C.M. Androgen receptor-positive salivary duct carcinoma: A disease entity with promising new treatment options. J. Clin. Oncol. 2011, 29, e473–e476. [Google Scholar] [PubMed]
- Boon, E.; van Boxtel, W.; Buter, J.; de Jong, R.J.B.; van Es, R.J.J.; Bel, M.; Fiets, E.; Oosting, S.F.; Slingerland, M.; Hoeben, A.; et al. Androgen deprivation therapy for androgen receptor-positive advanced salivary duct carcinoma: A nationwide case series of 35 patients in The Netherlands. Head Neck 2018, 40, 605–613. [Google Scholar] [PubMed]
- van Boxtel, W.; Locati, L.D.; van Engen-van Grunsven, A.C.H.; Bergamini, C.; Jonker, M.A.; Fiets, E.; Cavalieri, S.; Tooten, S.; Bos, E.; Quattrone, P.; et al. Adjuvant androgen deprivation therapy for poor-risk, androgen receptor-positive salivary duct carcinoma. Eur. J. Cancer 2019, 110, 62–70. [Google Scholar] [CrossRef]
- Fushimi, C.; Tada, Y.; Takahashi, H.; Nagao, T.; Ojiri, H.; Masubuchi, T.; Matsuki, T.; Miura, K.; Kawakita, D.; Hirai, H.; et al. A prospective phase II study of combined androgen blockade in patients with androgen receptor-positive metastatic or locally advanced unresectable salivary gland carcinoma. Ann. Oncol. 2018, 29, 979–984. [Google Scholar] [PubMed]
- Kawakita, D.; Nagao, T.; Takahashi, H.; Kano, S.; Honma, Y.; Hirai, H.; Saigusa, N.; Akazawa, K.; Tani, K.; Ojiri, H.; et al. Survival benefit of HER2-targeted or androgen deprivation therapy in salivary duct carcinoma. Ther. Adv. Med. Oncol. 2022, 14, 17588359221119538. [Google Scholar] [CrossRef]
- Ho, A.L.; Foster, N.R.; Zoroufy, A.J.; Campbell, J.D.; Worden, F.; Price, K.; Adkins, D.; Bowles, D.W.; Kang, H.; Burtness, B.; et al. Phase II Study of Enzalutamide for Patients with Androgen Receptor-Positive Salivary Gland Cancers (Alliance A091404). J. Clin. Oncol. 2022, 40, 4240–4249. [Google Scholar]
- Yang, R.K.; Zhao, P.; Lu, C.; Luo, J.; Hu, R. Expression pattern of androgen receptor and AR-V7 in androgen-deprivation therapy-naive salivary duct carcinomas. Hum. Pathol. 2019, 84, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Gargano, S.M.; Senarathne, W.; Feldman, R.; Florento, E.; Stafford, P.; Swensen, J.; Vranic, S.; Gatalica, Z. Novel therapeutic targets in salivary duct carcinoma uncovered by comprehensive molecular profiling. Cancer Med. 2019, 8, 7322–7329. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro-Oncology 2020, 22, iv1–iv96. [Google Scholar]
- Gittleman, H.; Ostrom, Q.T.; Stetson, L.C.; Waite, K.; Hodges, T.R.; Wright, C.H.; Wright, J.; Rubin, J.B.; Berens, M.E.; Lathia, J.; et al. Sex is an important prognostic factor for glioblastoma but not for nonglioblastoma. Neurooncol. Pract. 2019, 6, 451–462. [Google Scholar] [CrossRef]
- Broestl, L.; Warrington, N.M.; Grandison, L.; Abou-Antoun, T.; Tung, O.; Shenoy, S.; Tallman, M.M.; Rhee, G.; Yang, W.; Sponagel, J.; et al. Gonadal sex patterns p21-induced cellular senescence in mouse and human glioblastoma. Commun. Biol. 2022, 5, 781. [Google Scholar] [CrossRef]
- Yu, X.; Jiang, Y.; Wei, W.; Cong, P.; Ding, Y.; Xiang, L.; Wu, K. Androgen receptor signaling regulates growth of glioblastoma multiforme in men. Tumour Biol. 2015, 36, 967–972. [Google Scholar] [CrossRef]
- Bao, D.; Cheng, C.; Lan, X.; Xing, R.; Chen, Z.; Zhao, H.; Sun, J.; Wang, Y.; Niu, C.; Zhang, B.; et al. Regulation of p53wt glioma cell proliferation by androgen receptor-mediated inhibition of small VCP/p97-interacting protein expression. Oncotarget 2017, 8, 23142–23154. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lozano, D.C.; Velazquez-Vazquez, D.E.; Del Moral-Morales, A.; Camacho-Arroyo, I. Dihydrotestosterone Induces Proliferation, Migration and Invasion of Human Glioblastoma Cell Lines. Onco Targets Ther. 2020, 13, 8813–8823. [Google Scholar] [CrossRef] [PubMed]
- Verzat, C.; Delisle, M.B.; Courriere, P.; Hollande, E. Influence of host sex on the growth of a human glioblastoma line in athymic mice. Neuropathol. Appl. Neurobiol. 1990, 16, 141–151. [Google Scholar] [CrossRef]
- Chen, T.C.; Chuang, J.Y.; Ko, C.Y.; Kao, T.J.; Yang, P.Y.; Yu, C.H.; Liu, M.S.; Hu, S.L.; Tsai, Y.T.; Chan, H.; et al. AR ubiquitination induced by the curcumin analog suppresses growth of temozolomide-resistant glioblastoma through disrupting GPX4-Mediated redox homeostasis. Redox Biol. 2020, 30, 101413. [Google Scholar] [CrossRef]
- Rodriguez-Lozano, D.C.; Pina-Medina, A.G.; Hansberg-Pastor, V.; Bello-Alvarez, C.; Camacho-Arroyo, I. Testosterone Promotes Glioblastoma Cell Proliferation, Migration and Invasion Through Androgen Receptor Activation. Front. Endocrinol. 2019, 10, 16. [Google Scholar] [CrossRef]
- Orozco, M.; Valdez, R.A.; Ramos, L.; Cabeza, M.; Segovia, J.; Romano, M.C. Dutasteride combined with androgen receptor antagonists inhibit glioblastoma U87 cell metabolism, proliferation and invasion capacity: Androgen regulation. Steroids 2020, 164, 108733. [Google Scholar] [PubMed]
- Zalcman, N.; Canello, T.; Ovadia, H.; Charbit, H.; Zelikovitch, B.; Mordechai, A.; Fellig, Y.; Rabani, S.; Shahar, T.; Lossos, A.; et al. Androgen receptor: A potential therapeutic target for glioblastoma. Oncotarget 2018, 9, 19980–19993. [Google Scholar] [CrossRef]
- Scelo, G.; Li, P.; Chanudet, E.; Muller, D.C. Variability of Sex Disparities in Cancer Incidence over 30 Years: The Striking Case of Kidney Cancer. Eur. Urol. Focus 2018, 4, 586–590. [Google Scholar]
- Peired, A.J.; Campi, R.; Angelotti, M.L.; Antonelli, G.; Conte, C.; Lazzeri, E.; Becherucci, F.; Calistri, L.; Serni, S.; Romagnani, P. Sex and Gender Differences in Kidney Cancer: Clinical and Experimental Evidence. Cancers 2021, 13, 4588. [Google Scholar] [CrossRef]
- Langner, C.; Ratschek, M.; Rehak, P.; Schips, L.; Zigeuner, R. Steroid hormone receptor expression in renal cell carcinoma: An immunohistochemical analysis of 182 tumors. J. Urol. 2004, 171, 611–614. [Google Scholar]
- Zhu, G.; Liang, L.; Li, L.; Dang, Q.; Song, W.; Yeh, S.; He, D.; Chang, C. The expression and evaluation of androgen receptor in human renal cell carcinoma. Urology 2014, 83, 510.e19–510.e24. [Google Scholar] [CrossRef]
- Ha, Y.S.; Lee, G.T.; Modi, P.; Kwon, Y.S.; Ahn, H.; Kim, W.J.; Kim, I.Y. Increased Expression of Androgen Receptor mRNA in Human Renal Cell Carcinoma Cells is Associated with Poor Prognosis in Patients with Localized Renal Cell Carcinoma. J. Urol. 2015, 194, 1441–1448. [Google Scholar] [CrossRef]
- Yuan, P.; Ge, Y.; Liu, X.; Wang, S.; Ye, Z.; Xu, H.; Chen, Z. The Association of Androgen Receptor Expression with Renal Cell Carcinoma Risk: A Systematic Review and Meta-Analysis. Pathol. Oncol. Res. 2020, 26, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.J.; Kang, M.J.; Kim, K.M.; Bae, J.S.; Park, H.S.; Moon, W.S.; Chung, M.J.; Lee, H.; Lee, D.G.; Jang, K.Y. Acetylation status of P53 and the expression of DBC1, SIRT1 and androgen receptor are associated with survival in clear cell renal cell carcinoma patients. Pathology 2013, 45, 574–580. [Google Scholar] [CrossRef]
- Pak, S.; Kim, W.; Kim, Y.; Song, C.; Ahn, H. Dihydrotestosterone promotes kidney cancer cell proliferation by activating the STAT5 pathway via androgen and glucocorticoid receptors. J. Cancer Res. Clin. Oncol. 2019, 145, 2293–2301. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.T.; Han, C.S.; Kwon, Y.S.; Patel, R.; Modi, P.K.; Kwon, S.J.; Faiena, I.; Patel, N.; Singer, E.A.; Ahn, H.J.; et al. Intracrine androgen biosynthesis in renal cell carcinoma. Br. J. Cancer 2017, 116, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Makhov, P.; Joshi, S.; Ghatalia, P.; Kutikov, A.; Uzzo, R.G.; Kolenko, V.M. Resistance to Systemic Therapies in Clear Cell Renal Cell Carcinoma: Mechanisms and Management Strategies. Mol. Cancer Ther. 2018, 17, 1355–1364. [Google Scholar]
- Muglia, V.F.; Prando, A. Renal cell carcinoma: Histological classification and correlation with imaging findings. Radiol. Bras. 2015, 48, 166–174. [Google Scholar]
- Bialek, J.; Piwonka, M.; Kawan, F.; Fornara, P.; Theil, G. Differential Expression of the Androgen Receptor, Splice Variants and Relaxin 2 in Renal Cancer. Life 2021, 11, 731. [Google Scholar] [CrossRef] [PubMed]
- Foersch, S.; Schindeldecker, M.; Keith, M.; Tagscherer, K.E.; Fernandez, A.; Stenzel, P.J.; Pahernik, S.; Hohenfellner, M.; Schirmacher, P.; Roth, W.; et al. Prognostic relevance of androgen receptor expression in renal cell carcinomas. Oncotarget 2017, 8, 78545–78555. [Google Scholar] [CrossRef]
- Neschadim, A.; Summerlee, A.J.; Silvertown, J.D. Targeting the relaxin hormonal pathway in prostate cancer. Int. J. Cancer 2015, 137, 2287–2295. [Google Scholar] [CrossRef]
- Samuel, C.S.; Hewitson, T.D. Relaxin and the progression of kidney disease. Curr. Opin. Nephrol. Hypertens. 2009, 18, 9–14. [Google Scholar] [CrossRef]
- Samuel, C.S.; Zhao, C.; Bond, C.P.; Hewitson, T.D.; Amento, E.P.; Summers, R.J. Relaxin-1-deficient mice develop an age-related progression of renal fibrosis. Kidney Int. 2004, 65, 2054–2064. [Google Scholar] [CrossRef] [PubMed]
- Schafer, E.J.; Jemal, A.; Wiese, D.; Sung, H.; Kratzer, T.B.; Islami, F.; Dahut, W.L.; Knudsen, K.E. Disparities and Trends in Genitourinary Cancer Incidence and Mortality in the USA. Eur. Urol. 2022, 84, 117–126. [Google Scholar] [CrossRef]
- Laor, E.; Schiffman, Z.J.; Braunstein, J.D.; Reid, R.E.; Tolia, B.M.; Koss, L.G.; Freed, S.Z. Androgen receptors in bladder tumors. Urology 1985, 25, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yin, Y.; Stemler, K.; Humphrey, P.; Kibel, A.S.; Mysorekar, I.U.; Ma, L. Constitutive beta-catenin activation induces male-specific tumorigenesis in the bladder urothelium. Cancer Res. 2013, 73, 5914–5925. [Google Scholar] [CrossRef]
- Terada, S.; Suzuki, N.; Uchide, K.; Akasofu, K.; Nishida, E. Effect of testosterone on the development of bladder tumors and calculi in female rats. Gynecol. Obstet. Investig. 1992, 34, 105–110. [Google Scholar] [CrossRef]
- Hsu, J.W.; Hsu, I.; Xu, D.; Miyamoto, H.; Liang, L.; Wu, X.R.; Shyr, C.R.; Chang, C. Decreased tumorigenesis and mortality from bladder cancer in mice lacking urothelial androgen receptor. Am. J. Pathol. 2013, 182, 1811–1820. [Google Scholar]
- Wang, C.S.; Li, C.C.; Juan, Y.S.; Wu, W.J.; Lee, H.Y. 5alpha-reductase inhibitors impact prognosis of urothelial carcinoma. BMC Cancer 2020, 20, 872. [Google Scholar] [CrossRef]
- Katleba, K.; Lombard, A.P.; Tsamouri, M.M.; Baek, H.B.; Nishida, K.S.; Libertini, S.J.; Platero, A.J.; Ma, A.H.; Pan, C.X.; Ghosh, P.M.; et al. Depletion of androgen receptor low molecular weight isoform reduces bladder tumor cell viability and induces apoptosis. Cancer Lett. 2021, 504, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Katleba, K.D.; Tsamouri, M.M.; Jathal, M.; Baek, H.B.; Armenta, R.B.; Tepper, C.G.; Cortopassi, G.; Ghosh, P.M.; Mudryj, M. Androgen receptor-dependent regulation of metabolism in high grade bladder cancer cells. Sci. Rep. 2023, 13, 1762. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.L.; Lai, H.C.; Yeh, S.; Cai, X.; Chang, C. Androgen receptor roles in hepatocellular carcinoma, fatty liver, cirrhosis and hepatitis. Endocr. Relat. Cancer 2014, 21, R165–R182. [Google Scholar] [CrossRef]
- Kanda, T.; Jiang, X.; Yokosuka, O. Androgen receptor signaling in hepatocellular carcinoma and pancreatic cancers. World J. Gastroenterol. 2014, 20, 9229–9236. [Google Scholar] [PubMed]
- Montgomery, E.J.; Xing, E.; Campbell, M.J.; Li, P.K.; Blachly, J.S.; Tsung, A.; Coss, C.C. Constitutively Active Androgen Receptor in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2022, 23, 13768. [Google Scholar] [CrossRef]
- Ma, W.L.; Hsu, C.L.; Wu, M.H.; Wu, C.T.; Wu, C.C.; Lai, J.J.; Jou, Y.S.; Chen, C.W.; Yeh, S.; Chang, C. Androgen receptor is a new potential therapeutic target for the treatment of hepatocellular carcinoma. Gastroenterology 2008, 135, 947–955.e5. [Google Scholar]
- Wu, M.H.; Ma, W.L.; Hsu, C.L.; Chen, Y.L.; Ou, J.H.; Ryan, C.K.; Hung, Y.C.; Yeh, S.; Chang, C. Androgen receptor promotes hepatitis B virus-induced hepatocarcinogenesis through modulation of hepatitis B virus RNA transcription. Sci. Transl. Med. 2010, 2, 32ra35. [Google Scholar] [CrossRef]
- Liu, W.C.; Liu, Q.Y. Molecular mechanisms of gender disparity in hepatitis B virus-associated hepatocellular carcinoma. World J. Gastroenterol. 2014, 20, 6252–6261. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Chen, P.J.; Yeh, S.H. Gender disparity in chronic hepatitis B: Mechanisms of sex hormones. J. Gastroenterol. Hepatol. 2015, 30, 1237–1245. [Google Scholar] [CrossRef]
- Kalra, M.; Mayes, J.; Assefa, S.; Kaul, A.K.; Kaul, R. Role of sex steroid receptors in pathobiology of hepatocellular carcinoma. World J. Gastroenterol. 2008, 14, 5945–5961. [Google Scholar]
- Zhang, H.; Li, X.X.; Yang, Y.; Zhang, Y.; Wang, H.Y.; Zheng, X.F.S. Significance and mechanism of androgen receptor overexpression and androgen receptor/mechanistic target of rapamycin cross-talk in hepatocellular carcinoma. Hepatology 2018, 67, 2271–2286. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Lopez, S.; Diaz-Bethencourt, D.; Concepcion-Massip, T.; de Basoa, M.C.M.-F.; Plata-Bello, A.; Gonzalez-Rodriguez, A.; Perez-Hernandez, F.; Plata-Bello, J. The androgen receptor expression and its activity have different relationships with prognosis in hepatocellular carcinoma. Sci. Rep. 2020, 10, 22046. [Google Scholar] [CrossRef] [PubMed]
- Rajaram, P.; Rivera, A.; Muthima, K.; Olveda, N.; Muchalski, H.; Chen, Q.H. Second-Generation Androgen Receptor Antagonists as Hormonal Therapeutics for Three Forms of Prostate Cancer. Molecules 2020, 25, 2448. [Google Scholar] [CrossRef]
- Grimaldi, C.; Bleiberg, H.; Gay, F.; Messner, M.; Rougier, P.; Kok, T.C.; Cirera, L.; Cervantes, A.; De Greve, J.; Paillot, B.; et al. Evaluation of antiandrogen therapy in unresectable hepatocellular carcinoma: Results of a European Organization for Research and Treatment of Cancer multicentric double-blind trial. J. Clin. Oncol. 1998, 16, 411–417. [Google Scholar]
- Dauki, A.M.; Blachly, J.S.; Kautto, E.A.; Ezzat, S.; Abdel-Rahman, M.H.; Coss, C.C. Transcriptionally Active Androgen Receptor Splice Variants Promote Hepatocellular Carcinoma Progression. Cancer Res. 2020, 80, 561–575. [Google Scholar] [CrossRef]
- Lee, S.E.; Alcedo, K.P.; Kim, H.J.; Snider, N.T. Alternative Splicing in Hepatocellular Carcinoma. Cell Mol. Gastroenterol. Hepatol. 2020, 10, 699–712. [Google Scholar] [CrossRef]
- Wang, F.; Pan, J.; Liu, Y.; Meng, Q.; Lv, P.; Qu, F.; Ding, G.L.; Klausen, C.; Leung, P.C.; Chan, H.C.; et al. Alternative splicing of the androgen receptor in polycystic ovary syndrome. Proc. Natl. Acad. Sci. USA 2015, 112, 4743–4748. [Google Scholar] [CrossRef]
- Marin-Aguilera, M.; Jimenez, N.; Reig, O.; Montalbo, R.; Verma, A.K.; Castellano, G.; Mengual, L.; Victoria, I.; Pereira, M.V.; Mila-Guasch, M.; et al. Androgen Receptor and Its Splicing Variant 7 Expression in Peripheral Blood Mononuclear Cells and in Circulating Tumor Cells in Metastatic Castration-Resistant Prostate Cancer. Cells 2020, 9, 203. [Google Scholar] [CrossRef]
- Meakin, A.S.; Saif, Z.; Tuck, A.R.; Clifton, V.L. Human placental androgen receptor variants: Potential regulators of male fetal growth. Placenta 2019, 80, 18–26. [Google Scholar] [CrossRef]
- Meakin, A.S.; Morrison, J.L.; Bradshaw, E.L.; Holman, S.L.; Saif, Z.; Gatford, K.L.; Wallace, M.J.; Bischof, R.J.; Moss, T.J.M.; Clifton, V.L. Identification of placental androgen receptor isoforms in a sheep model of maternal allergic asthma. Placenta 2021, 104, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Laurentino, S.S.; Pinto, P.I.; Tomas, J.; Cavaco, J.E.; Sousa, M.; Barros, A.; Power, D.M.; Canario, A.V.; Socorro, S. Identification of androgen receptor variants in testis from humans and other vertebrates. Andrologia 2013, 45, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Wu, Y.S.; Zhao, J.; Li, W. AR3 messenger ribonucleic acid expression and its functional implication in human primary testicular failure. Andrologia 2014, 46, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Garza-Contreras, J.; Duong, P.; Snyder, B.D.; Schreihofer, D.A.; Cunningham, R.L. Presence of Androgen Receptor Variant in Neuronal Lipid Rafts. eNeuro 2017, 4, ENEURO.0109-17.2017. [Google Scholar] [CrossRef]
- Sanchez-Hernandez, M.; Arizcun, M.; Garcia-Alcazar, A.; Sarropoulou, E.; Mulero, V.; Garcia-Ayala, A. Fish granulocytes express a constitutively active androgen receptor variant. Dev. Comp. Immunol. 2014, 45, 115–122. [Google Scholar] [CrossRef]
- Ben-Batalla, I.; Vargas-Delgado, M.E.; von Amsberg, G.; Janning, M.; Loges, S. Influence of Androgens on Immunity to Self and Foreign: Effects on Immunity and Cancer. Front. Immunol. 2020, 11, 1184. [Google Scholar] [CrossRef]
- Dodd, K.C.; Menon, M. Sex bias in lymphocytes: Implications for autoimmune diseases. Front. Immunol. 2022, 13, 945762. [Google Scholar] [CrossRef]
- Kronzer, V.L.; Bridges, S.L., Jr.; Davis, J.M., 3rd. Why women have more autoimmune diseases than men: An evolutionary perspective. Evol. Appl. 2021, 14, 629–633. [Google Scholar] [CrossRef]
- Lee, T.P.; Chiang, B.L. Sex differences in spontaneous versus induced animal models of autoimmunity. Autoimmun. Rev. 2012, 11, A422–A429. [Google Scholar] [CrossRef]
- Brown, M.A.; Su, M.A. An Inconvenient Variable: Sex Hormones and Their Impact on T Cell Responses. J. Immunol. 2019, 202, 1927–1933. [Google Scholar] [CrossRef]
- Bereshchenko, O.; Bruscoli, S.; Riccardi, C. Glucocorticoids, Sex Hormones and Immunity. Front. Immunol. 2018, 9, 1332. [Google Scholar] [CrossRef]
- Fitzpatrick, F.; Lepault, F.; Homo-Delarche, F.; Bach, J.F.; Dardenne, M. Influence of castration, alone or combined with thymectomy, on the development of diabetes in the nonobese diabetic mouse. Endocrinology 1991, 129, 1382–1390. [Google Scholar] [CrossRef] [PubMed]
- Fox, H.S. Androgen treatment prevents diabetes in nonobese diabetic mice. J. Exp. Med. 1992, 175, 1409–1412. [Google Scholar] [CrossRef] [PubMed]
- Liva, S.M.; Voskuhl, R.R. Testosterone acts directly on CD4+ T lymphocytes to increase IL-10 production. J. Immunol. 2001, 167, 2060–2067. [Google Scholar] [CrossRef]
- Qu, F.; Xie, W.; Nakabayashi, M.; Zhang, H.; Jeong, S.H.; Wang, X.; Komura, K.; Sweeney, C.J.; Sartor, O.; Lee, G.M.; et al. Association of AR-V7 and Prostate-Specific Antigen RNA Levels in Blood with Efficacy of Abiraterone Acetate and Enzalutamide Treatment in Men with Prostate Cancer. Clin. Cancer Res. 2017, 23, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Dapas, M.; Dunaif, A. Deconstructing a Syndrome: Genomic Insights into PCOS Causal Mechanisms and Classification. Endocr. Rev. 2022, 43, 927–965. [Google Scholar] [CrossRef]
- Conway, G.; Dewailly, D.; Diamanti-Kandarakis, E.; Escobar-Morreale, H.F.; Franks, S.; Gambineri, A.; Kelestimur, F.; Macut, D.; Micic, D.; Pasquali, R.; et al. The polycystic ovary syndrome: A position statement from the European Society of Endocrinology. Eur. J. Endocrinol. 2014, 171, P1–P29. [Google Scholar]
- Norman, R.J.; Dewailly, D.; Legro, R.S.; Hickey, T.E. Polycystic ovary syndrome. Lancet 2007, 370, 685–697. [Google Scholar] [CrossRef]
- Walters, K.A. Role of androgens in normal and pathological ovarian function. Reproduction 2015, 149, R193–R218. [Google Scholar] [CrossRef]
- Walters, K.A.; Handelsman, D.J. Androgen receptor splice variants and polycystic ovary syndrome: Cause or effect? Asian J. Androl. 2016, 18, 442–443. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.C.; Wang, P.H.; Yeh, S.; Wang, R.S.; Xie, C.; Xu, Q.; Zhou, X.; Chao, H.T.; Tsai, M.Y.; Chang, C. Subfertility and defective folliculogenesis in female mice lacking androgen receptor. Proc. Natl. Acad. Sci. USA 2004, 101, 11209–11214. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Hammes, S.R. Granulosa cell-specific androgen receptors are critical regulators of ovarian development and function. Mol. Endocrinol. 2010, 24, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.A.; Antoine, H.J.; Pall, M.; Taylor, K.D.; Azziz, R.; Goodarzi, M.O. Association of androgen receptor CAG repeat polymorphism and polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2008, 93, 1939–1945. [Google Scholar] [CrossRef]
- Peng, C.Y.; Long, X.Y.; Lu, G.X. Association of AR rs6152G/A gene polymorphism with susceptibility to polycystic ovary syndrome in Chinese women. Reprod. Fertil. Dev. 2010, 22, 881–885. [Google Scholar] [CrossRef]
- Escobar, J.C.; Patel, S.S.; Beshay, V.E.; Suzuki, T.; Carr, B.R. The human placenta expresses CYP17 and generates androgens de novo. J. Clin. Endocrinol. Metab. 2011, 96, 1385–1392. [Google Scholar] [CrossRef]
- Sathishkumar, K.; Balakrishnan, M.; Chinnathambi, V.; Chauhan, M.; Hankins, G.D.; Yallampalli, C. Fetal sex-related dysregulation in testosterone production and their receptor expression in the human placenta with preeclampsia. J. Perinatol. 2012, 32, 328–335. [Google Scholar] [CrossRef]
- Fornes, R.; Maliqueo, M.; Hu, M.; Hadi, L.; Jimenez-Andrade, J.M.; Ebefors, K.; Nystrom, J.; Labrie, F.; Jansson, T.; Benrick, A.; et al. The effect of androgen excess on maternal metabolism, placental function and fetal growth in obese dams. Sci. Rep. 2017, 7, 8066. [Google Scholar] [CrossRef]
- Cleys, E.R.; Halleran, J.L.; Enriquez, V.A.; da Silveira, J.C.; West, R.C.; Winger, Q.A.; Anthony, R.V.; Bruemmer, J.E.; Clay, C.M.; Bouma, G.J. Androgen receptor and histone lysine demethylases in ovine placenta. PLoS ONE 2015, 10, e0117472. [Google Scholar] [CrossRef][Green Version]
- Khatri, P.; Hoffmann, B.; Schuler, G. Androgen receptor is widely expressed in bovine placentomes and up-regulated during differentiation of bovine trophoblast giant cells. Placenta 2013, 34, 416–423. [Google Scholar] [CrossRef]
- Dobbins, T.A.; Sullivan, E.A.; Roberts, C.L.; Simpson, J.M. Australian national birthweight percentiles by sex and gestational age, 1998–2007. Med. J. Aust. 2012, 197, 291–294. [Google Scholar] [CrossRef]
- Kumar, S.; Gordon, G.H.; Abbott, D.H.; Mishra, J.S. Androgens in maternal vascular and placental function: Implications for preeclampsia pathogenesis. Reproduction 2018, 156, R155–R167. [Google Scholar] [CrossRef] [PubMed]
- McWhorter, E.S.; West, R.C.; Russ, J.E.; Ali, A.; Winger, Q.A.; Bouma, G.J. LIN28B regulates androgen receptor in human trophoblast cells through Let-7c. Mol. Reprod. Dev. 2019, 86, 1086–1093. [Google Scholar] [CrossRef]
- Zhou, X. Roles of androgen receptor in male and female reproduction: Lessons from global and cell-specific androgen receptor knockout (ARKO) mice. J. Androl. 2010, 31, 235–243. [Google Scholar]
- Cunha, G.R.; Cao, M.; Aksel, S.; Derpinghaus, A.; Baskin, L.S. Mouse-human species differences in early testicular development and its implications. Differentiation 2023, 129, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Li, Z.F.; Yang, W.X. What Does Androgen Receptor Signaling Pathway in Sertoli Cells During Normal Spermatogenesis Tell Us? Front. Endocrinol. 2022, 13, 838858. [Google Scholar] [CrossRef] [PubMed]
- Elliott, D.J.; Grellscheid, S.N. Alternative RNA splicing regulation in the testis. Reproduction 2006, 132, 811–819. [Google Scholar] [CrossRef]
- Ogawa, S.; Inoue, S.; Watanabe, T.; Orimo, A.; Hosoi, T.; Ouchi, Y.; Muramatsu, M. Molecular cloning and characterization of human estrogen receptor betacx: A potential inhibitor ofestrogen action in human. Nucleic Acids Res. 1998, 26, 3505–3512. [Google Scholar] [CrossRef]
- Inoue, S.; Ogawa, S.; Horie, K.; Hoshino, S.; Goto, W.; Hosoi, T.; Tsutsumi, O.; Muramatsu, M.; Ouchi, Y. An estrogen receptor beta isoform that lacks exon 5 has dominant negative activity on both ERalpha and ERbeta. Biochem. Biophys. Res. Commun. 2000, 279, 814–819. [Google Scholar] [CrossRef]
- Hirata, S.; Shoda, T.; Kato, J.; Hoshi, K. Novel isoforms of the mRNA for human female sex steroid hormone receptors. J. Steroid Biochem. Mol. Biol. 2002, 83, 25–30. [Google Scholar] [CrossRef]
- Saunders, P.T.; Millar, M.R.; Macpherson, S.; Irvine, D.S.; Groome, N.P.; Evans, L.R.; Sharpe, R.M.; Scobie, G.A. ERbeta1 and the ERbeta2 splice variant (ERbetacx/beta2) are expressed in distinct cell populations in the adult human testis. J. Clin. Endocrinol. Metab. 2002, 87, 2706–2715. [Google Scholar]
- Scobie, G.A.; Macpherson, S.; Millar, M.R.; Groome, N.P.; Romana, P.G.; Saunders, P.T. Human oestrogen receptors: Differential expression of ER alpha and beta and the identification of ER beta variants. Steroids 2002, 67, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Shoda, T.; Hirata, S.; Kato, J.; Hoshi, K. Cloning of the novel isoform of the estrogen receptor beta cDNA (ERbeta isoform M cDNA) from the human testicular cDNA library. J. Steroid Biochem. Mol. Biol. 2002, 82, 201–208. [Google Scholar] [CrossRef]
- Aschim, E.L.; Saether, T.; Wiger, R.; Grotmol, T.; Haugen, T.B. Differential distribution of splice variants of estrogen receptor beta in human testicular cells suggests specific functions in spermatogenesis. J. Steroid Biochem. Mol. Biol. 2004, 92, 97–106. [Google Scholar] [CrossRef]
- Ahrens-Fath, I.; Politz, O.; Geserick, C.; Haendler, B. Androgen receptor function is modulated by the tissue-specific AR45 variant. FEBS J. 2005, 272, 74–84. [Google Scholar] [CrossRef]
- Zhu, Y.; Dalrymple, S.L.; Coleman, I.; Zheng, S.L.; Xu, J.; Hooper, J.E.; Antonarakis, E.S.; De Marzo, A.M.; Meeker, A.K.; Nelson, P.S.; et al. Role of androgen receptor splice variant-7 (AR-V7) in prostate cancer resistance to 2nd-generation androgen receptor signaling inhibitors. Oncogene 2020, 39, 6935–6949. [Google Scholar] [CrossRef]
- Quigley, C.A.; Evans, B.A.; Simental, J.A.; Marschke, K.B.; Sar, M.; Lubahn, D.B.; Davies, P.; Hughes, I.A.; Wilson, E.M.; French, F.S. Complete androgen insensitivity due to deletion of exon C of the androgen receptor gene highlights the functional importance of the second zinc finger of the androgen receptor in vivo. Mol. Endocrinol. 1992, 6, 1103–1112. [Google Scholar]
- Kritzer, M.F. Selective colocalization of immunoreactivity for intracellular gonadal hormone receptors and tyrosine hydroxylase in the ventral tegmental area, substantia nigra, and retrorubral fields in the rat. J. Comp. Neurol. 1997, 379, 247–260. [Google Scholar] [CrossRef]
- Hatzoglou, A.; Kampa, M.; Kogia, C.; Charalampopoulos, I.; Theodoropoulos, P.A.; Anezinis, P.; Dambaki, C.; Papakonstanti, E.A.; Stathopoulos, E.N.; Stournaras, C.; et al. Membrane androgen receptor activation induces apoptotic regression of human prostate cancer cells in vitro and in vivo. J. Clin. Endocrinol. Metab. 2005, 90, 893–903. [Google Scholar] [CrossRef][Green Version]
- Aguila, S.; Castillo-Briceno, P.; Sanchez, M.; Cabas, I.; Garcia-Alcazar, A.; Meseguer, J.; Mulero, V.; Garcia-Ayala, A. Specific and non-overlapping functions of testosterone and 11-ketotestosterone in the regulation of professional phagocyte responses in the teleost fish gilthead seabream. Mol. Immunol. 2013, 53, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Kritzer, M.F.; Creutz, L.M. Region and sex differences in constituent dopamine neurons and immunoreactivity for intracellular estrogen and androgen receptors in mesocortical projections in rats. J. Neurosci. 2008, 28, 9525–9535. [Google Scholar] [CrossRef]
- Sarkey, S.; Azcoitia, I.; Garcia-Segura, L.M.; Garcia-Ovejero, D.; DonCarlos, L.L. Classical androgen receptors in non-classical sites in the brain. Horm. Behav. 2008, 53, 753–764. [Google Scholar] [CrossRef]
- Papakonstanti, E.A.; Kampa, M.; Castanas, E.; Stournaras, C. A rapid, nongenomic, signaling pathway regulates the actin reorganization induced by activation of membrane testosterone receptors. Mol. Endocrinol. 2003, 17, 870–881. [Google Scholar] [CrossRef]
- Gorczynska, E.; Handelsman, D.J. Androgens rapidly increase the cytosolic calcium concentration in Sertoli cells. Endocrinology 1995, 136, 2052–2059. [Google Scholar] [CrossRef]
- Benten, W.P.; Lieberherr, M.; Stamm, O.; Wrehlke, C.; Guo, Z.; Wunderlich, F. Testosterone signaling through internalizable surface receptors in androgen receptor-free macrophages. Mol. Biol. Cell 1999, 10, 3113–3123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Estrada, M.; Espinosa, A.; Muller, M.; Jaimovich, E. Testosterone stimulates intracellular calcium release and mitogen-activated protein kinases via a G protein-coupled receptor in skeletal muscle cells. Endocrinology 2003, 144, 3586–3597. [Google Scholar] [CrossRef] [PubMed]
- Estrada, M.; Uhlen, P.; Ehrlich, B.E. Ca2+ oscillations induced by testosterone enhance neurite outgrowth. J. Cell Sci. 2006, 119, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Holmes, S.; Abbassi, B.; Su, C.; Singh, M.; Cunningham, R.L. Oxidative stress defines the neuroprotective or neurotoxic properties of androgens in immortalized female rat dopaminergic neuronal cells. Endocrinology 2013, 154, 4281–4292. [Google Scholar] [CrossRef]
- Steinsapir, J.; Socci, R.; Reinach, P. Effects of androgen on intracellular calcium of LNCaP cells. Biochem. Biophys. Res. Commun. 1991, 179, 90–96. [Google Scholar] [CrossRef]
- Weiss, B.; Faus, H.; Haendler, B. Phylogenetic conservation of the androgen receptor AR45 variant form in placental mammals. Gene 2007, 399, 105–111. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Chen, K.; Haendler, B.; McDonald, T.V.; Bian, J.S. Stimulation of N-terminal truncated isoform of androgen receptor stabilizes human ether-a-go-go-related gene-encoded potassium channel protein via activation of extracellular signal regulated kinase 1/2. Endocrinology 2008, 149, 5061–5069. [Google Scholar] [CrossRef] [PubMed]
- Schuppe, E.R.; Miles, M.C.; Fuxjager, M.J. Evolution of the androgen receptor: Perspectives from human health to dancing birds. Mol. Cell Endocrinol. 2020, 499, 110577. [Google Scholar] [CrossRef] [PubMed]
- Blazquez, M.; Piferrer, F. Sea bass (Dicentrarchus labrax) androgen receptor: cDNA cloning, tissue-specific expression and mRNA levels during early development and sex differentiation. Mol. Cell Endocrinol. 2005, 237, 37–48. [Google Scholar] [CrossRef]
- de Waal, P.P.; Wang, D.S.; Nijenhuis, W.A.; Schulz, R.W.; Bogerd, J. Functional characterization and expression analysis of the androgen receptor in zebrafish (Danio rerio) testis. Reproduction 2008, 136, 225–234. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Mulero, V.; Meseguer, J.; Ayala, A.G. An overview of cell renewal in the testis throughout the reproductive cycle of a seasonal breeding teleost, the gilthead seabream (Sparus aurata L.). Biol. Reprod. 2005, 72, 593–601. [Google Scholar] [CrossRef]
- Liarte, S.; Chaves-Pozo, E.; Garcia-Alcazar, A.; Mulero, V.; Meseguer, J.; Garcia-Ayala, A. Testicular involution prior to sex change in gilthead seabream is characterized by a decrease in DMRT1 gene expression and by massive leukocyte infiltration. Reprod. Biol. Endocrinol. 2007, 5, 20. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Pelegrin, P.; Mulero, V.; Meseguer, J.; Ayala, A.G. A role for acidophilic granulocytes in the testis of the gilthead seabream (Sparus aurata L., Teleostei). J. Endocrinol. 2003, 179, 165–174. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Mulero, V.; Meseguer, J.; Ayala, A.G. Professional phagocytic granulocytes of the bony fish gilthead seabream display functional adaptation to testicular microenvironment. J. Leukoc. Biol. 2005, 78, 345–351. [Google Scholar] [CrossRef]
- Lin, P.Y.; Sun, L.; Thibodeaux, S.R.; Ludwig, S.M.; Vadlamudi, R.K.; Hurez, V.J.; Bahar, R.; Kious, M.J.; Livi, C.B.; Wall, S.R.; et al. B7-H1-dependent sex-related differences in tumor immunity and immunotherapy responses. J. Immunol. 2010, 185, 2747–2753. [Google Scholar] [CrossRef]
- Conforti, F.; Pala, L.; Bagnardi, V.; De Pas, T.; Martinetti, M.; Viale, G.; Gelber, R.D.; Goldhirsch, A. Cancer immunotherapy efficacy and patients’ sex: A systematic review and meta-analysis. Lancet Oncol. 2018, 19, 737–746. [Google Scholar] [PubMed]
- Wang, S.; Cowley, L.A.; Liu, X.S. Sex Differences in Cancer Immunotherapy Efficacy, Biomarkers and Therapeutic Strategy. Molecules 2019, 24, 3214. [Google Scholar] [CrossRef] [PubMed]
- Conforti, F.; Pala, L.; Bagnardi, V.; Viale, G.; De Pas, T.; Pagan, E.; Pennacchioli, E.; Cocorocchio, E.; Ferrucci, P.F.; De Marinis, F.; et al. Sex-Based Heterogeneity in Response to Lung Cancer Immunotherapy: A Systematic Review and Meta-Analysis. J. Natl. Cancer Inst. 2019, 111, 772–781. [Google Scholar] [CrossRef]
- Gamat, M.; McNeel, D.G. Androgen deprivation and immunotherapy for the treatment of prostate cancer. Endocr. Relat. Cancer 2017, 24, T297–T310. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Piulats, J.M.; Gross-Goupil, M.; Goh, J.; Ojamaa, K.; Hoimes, C.J.; Vaishampayan, U.; Berger, R.; Sezer, A.; Alanko, T.; et al. Pembrolizumab for Treatment-Refractory Metastatic Castration-Resistant Prostate Cancer: Multicohort, Open-Label Phase II KEYNOTE-199 Study. J. Clin. Oncol. 2020, 38, 395–405. [Google Scholar]
- Bishop, J.L.; Sio, A.; Angeles, A.; Roberts, M.E.; Azad, A.A.; Chi, K.N.; Zoubeidi, A. PD-L1 is highly expressed in Enzalutamide resistant prostate cancer. Oncotarget 2015, 6, 234–242. [Google Scholar] [CrossRef]
- Mercader, M.; Bodner, B.K.; Moser, M.T.; Kwon, P.S.; Park, E.S.; Manecke, R.G.; Ellis, T.M.; Wojcik, E.M.; Yang, D.; Flanigan, R.C.; et al. T cell infiltration of the prostate induced by androgen withdrawal in patients with prostate cancer. Proc. Natl. Acad. Sci. USA 2001, 98, 14565–14570. [Google Scholar] [CrossRef] [PubMed]
- Kissick, H.T.; Sanda, M.G.; Dunn, L.K.; Pellegrini, K.L.; On, S.T.; Noel, J.K.; Arredouani, M.S. Androgens alter T-cell immunity by inhibiting T-helper 1 differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, 9887–9892. [Google Scholar] [CrossRef]
- Graff, J.N.; Alumkal, J.J.; Drake, C.G.; Thomas, G.V.; Redmond, W.L.; Farhad, M.; Cetnar, J.P.; Ey, F.S.; Bergan, R.C.; Slottke, R.; et al. Early evidence of anti-PD-1 activity in enzalutamide-resistant prostate cancer. Oncotarget 2016, 7, 52810–52817. [Google Scholar] [CrossRef]
- Graff, J.N.; Beer, T.M.; Alumkal, J.J.; Slottke, R.E.; Redmond, W.L.; Thomas, G.V.; Thompson, R.F.; Wood, M.A.; Koguchi, Y.; Chen, Y.; et al. A phase II single-arm study of pembrolizumab with enzalutamide in men with metastatic castration-resistant prostate cancer progressing on enzalutamide alone. J. Immunother. Cancer 2020, 8, e000642. [Google Scholar] [CrossRef]

| SVs Detected | Biological Processes | Known/Putative Targets | References | |
|---|---|---|---|---|
| Prostate Cancer | V1–V14 (and others) | AR signaling, cell cycle, mTORC1, and OX/PHOS | UBE2C, KLK3, EDN2, ETS2, SRD5A1, ORM1 BIRC3, FKBP5, and HES1 | [37,39,40,42] |
| Breast Cancer | V1, V3, V7, V45, V9, V15, V16, V17, and V18 | immune function, signaling, and cell movement | multiple | [109,110,111,112,113] |
| Salivary Duct Carcinoma | V7 | viability/ proliferation | unknown | [128,129] |
| Glioblastoma Multiforme | V7 | unknown | unknown | [140] |
| Renal Carcinoma | V1, V3, V4, and V7 | expression correlates with RLN2 | unknown | [152] |
| Bladder Cancer | V1, V7, and V19 | cell cycle, mTOR, OXPHOS, ribosome, mitochondria, and HIF1a | mTOR and FKBP5 | [163,164] |
| Liver Cancer | V1, V3, and V7 | migration and invasion; mTOR signaling | SNA12 | [177] |
| Polycystic Ovarian Syndrome | 69 bp ins between exons 2 and 3, exon 3 deletion | limits conversion of androgens to estrogen; fails to upregulate ratios of estradiol to total testosterone | CYP17A1 | [188] |
| Peripheral Blood Mononuclear Cells | V7 | response to AA/ENZ and taxanes | [229] | |
| Placenta | V1, V7 and AR45 | unknown | unknown | [196,199] |
| Testis | AR∆2Stop, AR∆223Stop, AR∆3, and AR∆4 | spermatogenesis | unknown | [211,214] |
| Neuronal Lipid Rafts | AR45 | interacts with GPCR proteins Gαq and Gαo | unknown | [226] |
| Fish Granulocytes (functional equivalent of mammalian neutrophils) | AR∆LBD | immunocompetance | [230] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katleba, K.D.; Ghosh, P.M.; Mudryj, M. Beyond Prostate Cancer: An Androgen Receptor Splice Variant Expression in Multiple Malignancies, Non-Cancer Pathologies, and Development. Biomedicines 2023, 11, 2215. https://doi.org/10.3390/biomedicines11082215
Katleba KD, Ghosh PM, Mudryj M. Beyond Prostate Cancer: An Androgen Receptor Splice Variant Expression in Multiple Malignancies, Non-Cancer Pathologies, and Development. Biomedicines. 2023; 11(8):2215. https://doi.org/10.3390/biomedicines11082215
Chicago/Turabian StyleKatleba, Kimberley D., Paramita M. Ghosh, and Maria Mudryj. 2023. "Beyond Prostate Cancer: An Androgen Receptor Splice Variant Expression in Multiple Malignancies, Non-Cancer Pathologies, and Development" Biomedicines 11, no. 8: 2215. https://doi.org/10.3390/biomedicines11082215
APA StyleKatleba, K. D., Ghosh, P. M., & Mudryj, M. (2023). Beyond Prostate Cancer: An Androgen Receptor Splice Variant Expression in Multiple Malignancies, Non-Cancer Pathologies, and Development. Biomedicines, 11(8), 2215. https://doi.org/10.3390/biomedicines11082215







