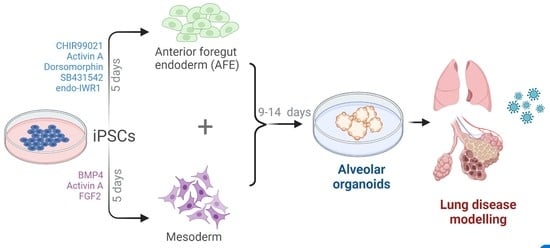
Rapid Generation of Pulmonary Organoids from Induced Pluripotent Stem Cells by Co-Culturing Endodermal and Mesodermal Progenitors for Pulmonary Disease Modelling
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Differentiation
2.2. qRT-PCR
2.3. RNA Sequencing
2.4. Flow Cytometry
2.5. Confocal Imaging and Histology
3. Results
3.1. Differentiation of iPSCs to Endodermal and Mesodermal Progenitors
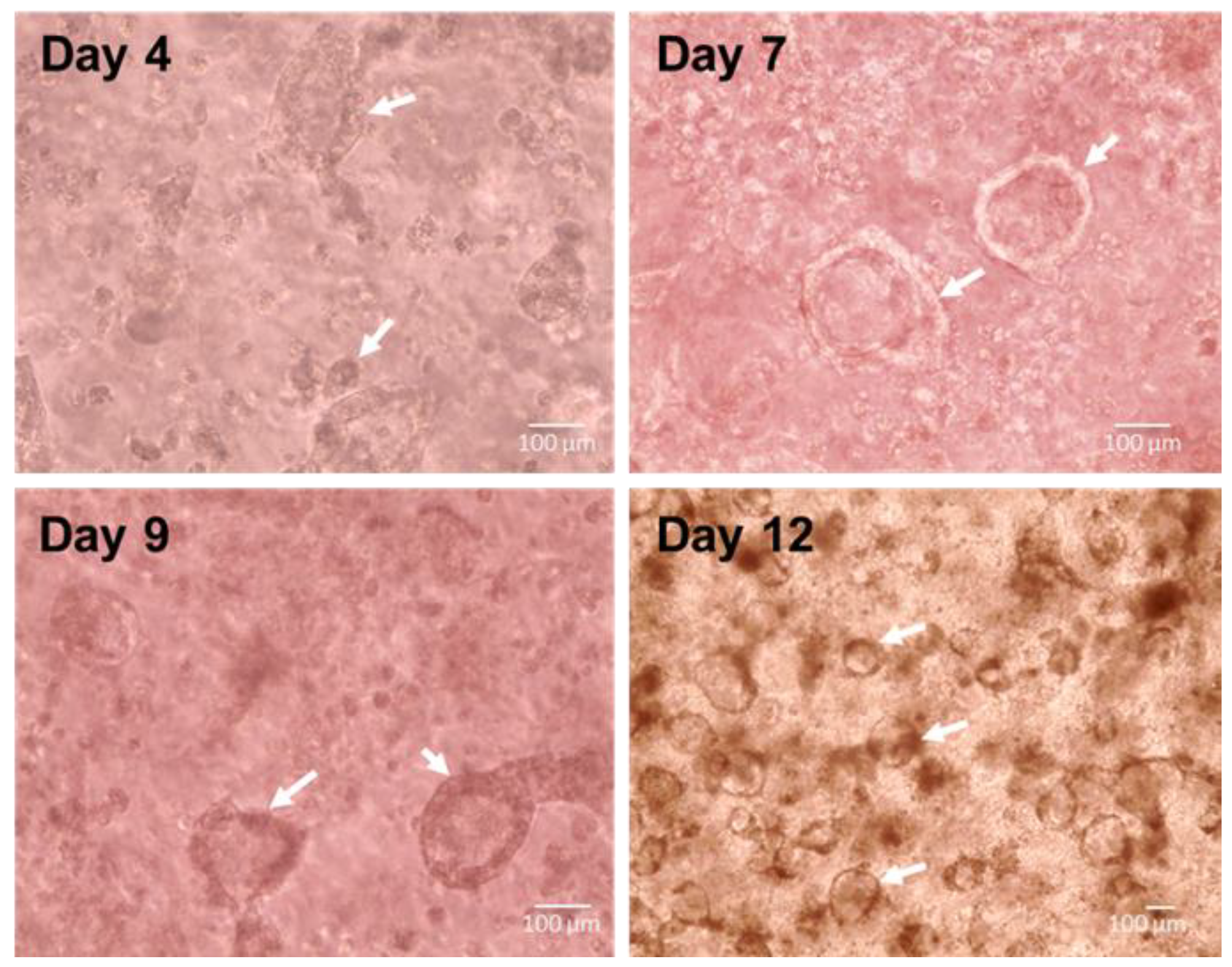
3.2. Spontaneous Pulmonary Organoid Formation by Co-Culture of Endodermal and Mesodermal Progenitors
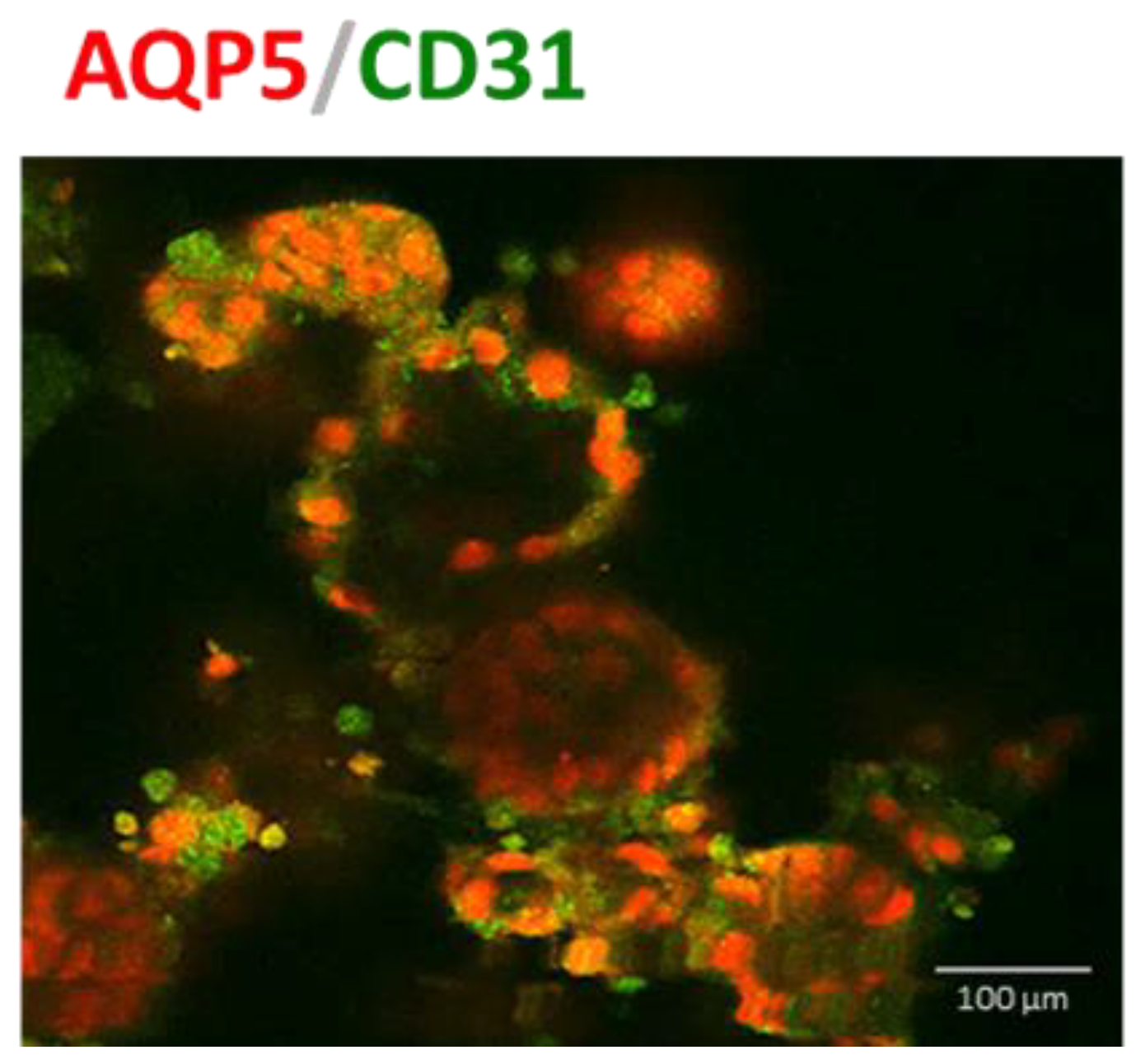
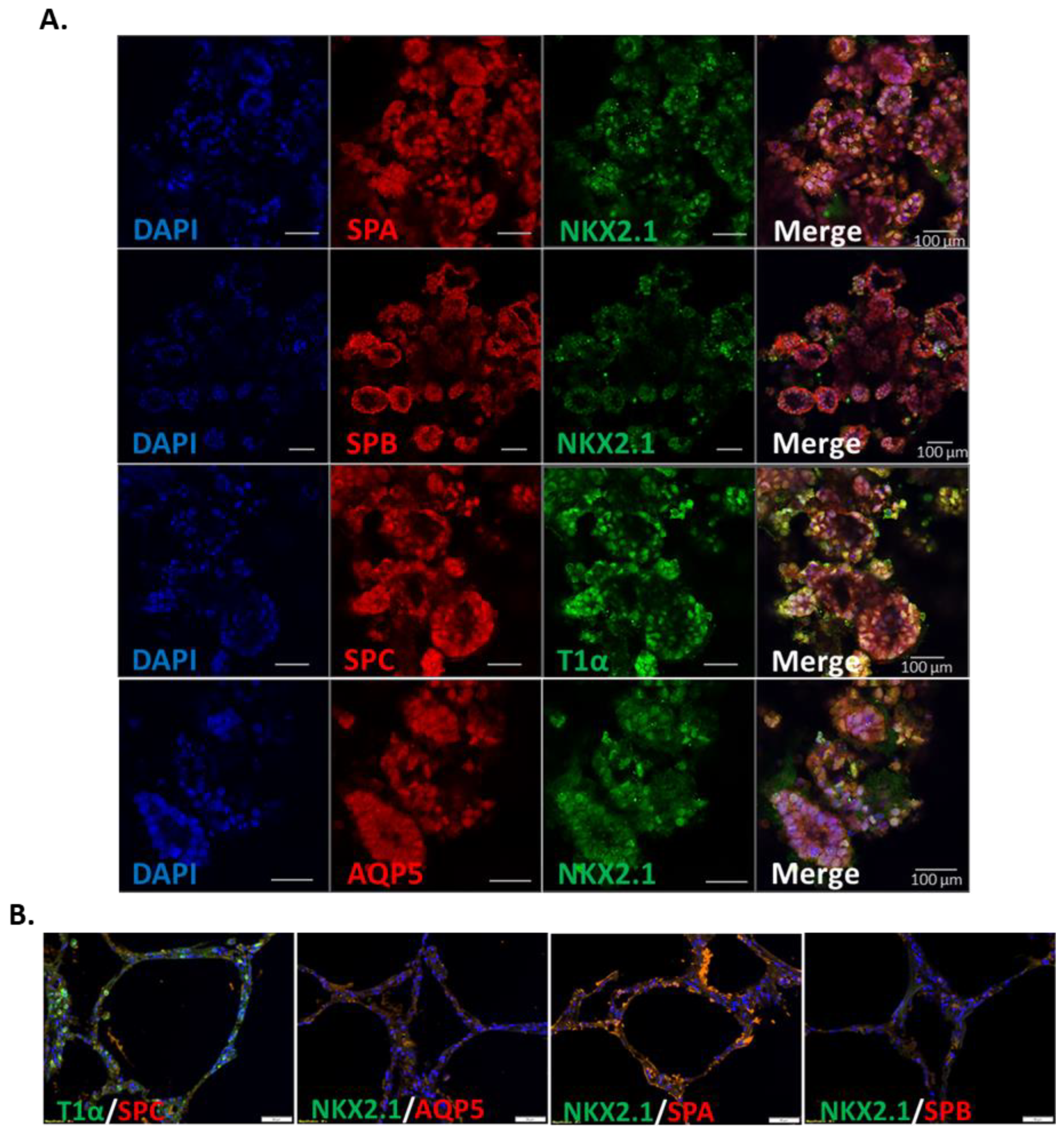
3.3. IPSC-Derived Pulmonary Organoids Express Lung-Specific Marker Genes
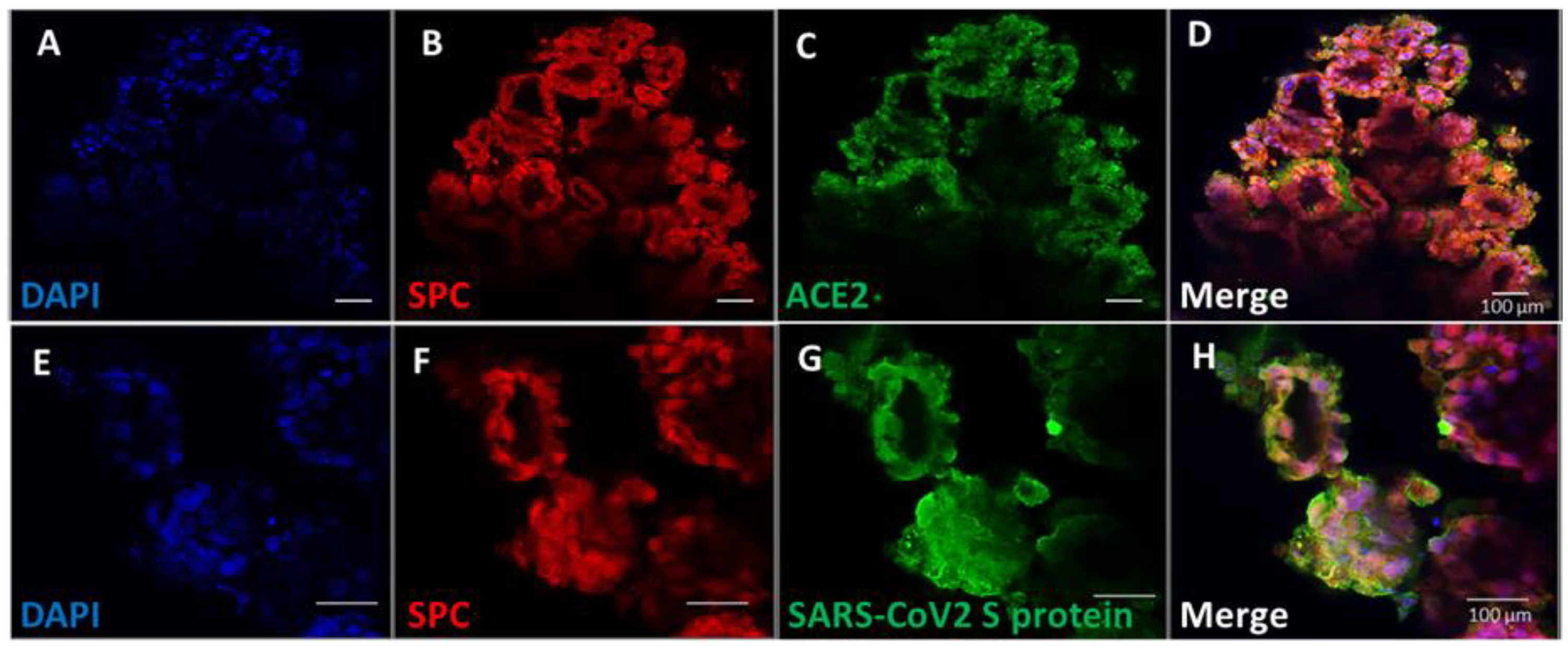
3.4. Pulmonary Organoids Express ACE2 Capable of Binding SARS-CoV-2 Spike Protein
4. Discussion
Limitations of the Study
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Supplemental Information—Detailed Experimental Procedures
Appendix A.1. Cell Culture and Differentiation
Appendix A.2. qRT-PCR
Appendix A.3. RNA Sequencing (RNAseq)
Appendix A.4. Flow Cytometry
Appendix A.5. Binding of SARS-CoV-2 Spike Protein S1 to Organoids
Appendix A.6. 3D Confocal Imaging
Appendix A.7. Histology
Appendix A.8. Statistics and Data Analysis
| Reagent | Supplier | Catalogue Number |
|---|---|---|
| ACE2 primary antibody | R&D Systems | MAB933-SP |
| AQP5 primary antibody | ThermoFisher Scientific | PA5-99403 |
| SFTPA1 primary antibody | Universal Biologics | A3133-50ul |
| SFTPB primary antibody | Biomatik | CAU25609 |
| SFTPC primary antibody | ThermoFisher Scientific | PA571680 |
| T1α primary antibody | ThermoFisher Scientific | BMS1105 |
| TTF1 primary antibody | Caltag Medsystems | H00007080-M01-100ug |
| Donkey anti Mouse A488 secondary antibody | Abcam | ab150105 |
| Donkey anti Rabbit A594 secondary antibody | Abcam | ab150076 |
| FOXA2 e660 conjugated antibody | eBioscience | 50-4778 |
| GATA4 A647 conjugated antibody | Bioss | bs-1778R-A647 |
| PDGFRa A594 conjugated antibody | Bioss | bs-0231R-A594 |
| SOX17 PerCP CY5.5 conjugated antibody | BD Pharmingen | 562387 |
| TBX1 A488 conjugated antibody | Bioss | bs-8257R-A488 |
| VEGFR2 PE conjugated antibody | Bioss | bs-10412R-PE |
| FOXA2 primer | BioRad | qHsaCID0014658 |
| GATA4 primer | BioRad | qHsaCID0012121 |
| PDGFRa primer | BioRad | qHsaCID0007202 |
| SOX17 primer | BioRad | qHsaCED0046246 |
| TBX1 primer | BioRad | qHsaCID0013392 |
| VEGFR2 primer | BioRad | qHsaCID0006310 |
| B2M | Forward: 5′ GAGGCTATCCAGCGTACTCCA 3′ | Reverse: 5′ CGGCAGGCATACTCATC TTTT 3′ |
| GAPDH | Forward: 5′ CATGTTCGTCATGGGTG TGAACCA 3′ | Reverse: 5′ ATGGCATGGACTGTGGT CATGAGT 3′ |
| Recombinant SARS-CoV-2 Spike S1 protein | R&D Systems | 10522-CV |
| SARS-CoV-2 Spike S1 Subunit Antibody | R&D Systems | MAB105403-SP |
Appendix B. Supplemental Information—Supplemental Figures

Appendix C. Gene Ontology Terms of Lung-Development-Related Gene Sets
References
- Mitchell, A.; Drinnan, C.T.; Jensen, T.; Finck, C. Production of high purity alveolar-like cells from iPSCs through depletion of uncommitted cells after AFE induction. Differentiation 2017, 96, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Morrisey, E.E.; Hogan, B.L.M. Preparing for the First Breath: Genetic and Cellular Mechanisms in Lung Development. Dev. Cell 2010, 18, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Green, M.D.; Chen, A.; Nostro, M.-C.; d’Souza, S.L.; Schaniel, C.; Lemischka, I.R.; Gouon-Evans, V.; Keller, G.; Snoeck, H.-W. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.X.L.; Islam, M.N.; O’Neill, J.; Hu, Z.; Yang, Y.-G.; Chen, Y.-W.; Mumau, M.; Green, M.D.; Vunjak-Novakovic, G.; Bhattacharya, J.; et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014, 32, 84–91. [Google Scholar] [CrossRef]
- Hayden, P.J.; Harbell, J.W. Special review series on 3D organotypic culture models: Introduction and historical perspective. In Vitro Cell. Dev. Biol.-Anim. 2021, 57, 95–103. [Google Scholar] [CrossRef]
- Tran, F.; Klein, C.; Arlt, A.; Imm, S.; Knappe, E.; Simmons, A.; Rosenstiel, P.; Seibler, P. Stem Cells and Organoid Technology in Precision Medicine in Inflammation: Are We There Yet? Front. Immunol. 2020, 11, 573562. [Google Scholar] [CrossRef]
- Ohata, K.; Ott, H.C. Human-scale lung regeneration based on decellularized matrix scaffolds as a biologic platform. Surg. Today 2020, 50, 633–643. [Google Scholar] [CrossRef]
- Gotoh, S.; Ito, I.; Nagasaki, T.; Yamamoto, Y.; Konishi, S.; Korogi, Y.; Matsumoto, H.; Muro, S.; Hirai, T.; Funato, M.; et al. Generation of Alveolar Epithelial Spheroids via Isolated Progenitor Cells from Human Pluripotent Stem Cells. Stem Cell Rep. 2014, 3, 394–403. [Google Scholar] [CrossRef]
- Barkauskas, C.E.; Cronce, M.J.; Rackley, C.R.; Bowie, E.J.; Keene, D.R.; Stripp, B.R.; Randell, S.H.; Noble, P.W.; Hogan, B.L.M. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Investig. 2013, 123, 3025–3036. [Google Scholar] [CrossRef]
- Dye, B.R.; Hill, D.R.; Ferguson, M.A.H.; Tsai, Y.-H.; Nagy, M.S.; Dyal, R.; Wells, J.M.; Mayhew, C.N.; Nattiv, R.; Klein, O.D.; et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 2015, 4, e05098. [Google Scholar] [CrossRef]
- Dye, B.R.; Dedhia, P.H.; Miller, A.J.; Nagy, M.S.; White, E.S.; Shea, L.D.; Spence, J.R. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. eLife 2016, 5, e19732. [Google Scholar] [CrossRef] [PubMed]
- Leeman, K.T.; Pessina, P.; Lee, J.H.; Kim, C.F. Mesenchymal Stem Cells Increase Alveolar Differentiation in Lung Progenitor Organoid Cultures. Sci. Rep. 2019, 9, 6479. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gordon, J.; Manley, N.R.; Litingtung, Y.; Chiang, C. Bmp4 is required for tracheal formation: A novel mouse model for tracheal agenesis. Dev. Biol. 2008, 322, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, C.; Liu, Y.-H.; Xing, Y.; Hu, L.; Borok, Z.; Kwong, K.Y.C.; Minoo, P. Mesodermal deletion of transforming growth factor-beta receptor II disrupts lung epithelial morphogenesis: Cross-talk between TGF-beta and Sonic hedgehog pathways. J. Biol. Chem. 2008, 283, 36257–36264. [Google Scholar] [CrossRef] [PubMed]
- Min, H.; Danilenko, D.M.; Scully, S.A.; Bolon, B.; Ring, B.D.; Tarpley, J.E.; DeRose, M.; Simonet, W.S. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998, 12, 3156–3161. [Google Scholar] [CrossRef]
- Ackerman, K.G.; Wang, J.; Luo, L.; Fujiwara, Y.; Orkin, S.H.; Beier, D.R. Gata4 is necessary for normal pulmonary lobar development. Am. J. Respir. Cell Mol. Biol. 2007, 36, 391–397. [Google Scholar] [CrossRef]
- Golzio, C.; Havis, E.; Daubas, P.; Nuel, G.; Babarit, C.; Munnich, A.; Vekemans, M.; Zaffran, S.; Lyonnet, S.; Etchevers, H.C. ISL1 directly regulates FGF10 transcription during human cardiac outflow formation. PLoS ONE 2012, 7, e30677. [Google Scholar] [CrossRef]
- Gong, H.; Yan, Y.; Fang, B.; Xue, Y.; Yin, P.; Li, L.; Zhang, G.; Sun, X.; Chen, Z.; Ma, H.; et al. Knockdown of Nucleosome Assembly Protein 1-Like 1 Induces Mesoderm Formation and Cardiomyogenesis Via Notch Signaling in Murine-Induced Pluripotent Stem Cells. Stem Cell 2014, 32, 1759–1773. [Google Scholar] [CrossRef]
- Chen, Y.-W.; Huang, S.X.; de Carvalho, A.L.R.T.; Ho, S.-H.; Islam, M.N.; Volpi, S.; Notarangelo, L.D.; Ciancanelli, M.; Casanova, J.-L.; Bhattacharya, J.; et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017, 19, 542–549. [Google Scholar] [CrossRef]
- Suezawa, T.; Kanagaki, S.; Moriguchi, K.; Masui, A.; Nakao, K.; Toyomoto, M.; Tamai, K.; Mikawa, R.; Hirai, T.; Murakami, K.; et al. Disease modeling of pulmonary fibrosis using human pluripotent stem cell-derived alveolar organoids. Stem Cell Rep. 2021, 16, 2973–2987. [Google Scholar] [CrossRef]
- Han, Y.; Duan, X.; Yang, L.; Nilsson-Payant, B.E.; Wang, P.; Duan, F.; Tang, X.; Yaron, T.M.; Zhang, T.; Uhl, S.; et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021, 589, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Surendran, H.; Nandakumar, S.; Pal, R. Human Induced Pluripotent Stem Cell-Derived Lung Epithelial System for SARS-CoV-2 Infection Modeling and Its Potential in Drug Repurposing. Stem Cells Dev. 2020, 29, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.K.; Wang, S.; Smith, D.; Carlin, A.F.; Rana, T.M. Revealing Tissue-Specific SARS-CoV-2 Infection and Host Responses using Human Stem Cell-Derived Lung and Cerebral Organoids. Stem Cell Rep. 2021, 16, 437–445. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, N.; Fan, S.; Zhao, L.; Song, W.; Gong, Y.; Shen, Q.; Zhang, C.; Ren, P.; Lin, C.; et al. Establishment of human distal lung organoids for SARS-CoV-2 infection. Cell Discov. 2021, 7, 108. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Alieva, M.; Wellens, L.M.; Ariese, H.C.R.; Jamieson, P.R.; Vonk, A.M.; Amatngalim, G.D.; Hu, H.; Oost, K.C.; Snippert, H.J.G.; et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019, 14, 1756–1771. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, J.; Zhao, S.; Han, L.; Yang, H.; Lin, Y.; Rong, Z. Long-Term Engraftment Promotes Differentiation of Alveolar Epithelial Cells from Human Embryonic Stem Cell Derived Lung Organoids. Stem Cells Dev. 2018, 27, 1339–1349. [Google Scholar] [CrossRef]
- Wang, R.; McCauley, K.B.; Kotton, D.N.; Hawkins, F. Differentiation of human airway-organoids from induced pluripotent stem cells (iPSCs). Methods Cell Biol. 2020, 159, 95–114. [Google Scholar]
- Kelleher, J.; Dickinson, A.; Cain, S.; Hu, Y.; Bates, N.; Harvey, A.; Ren, J.; Zhang, W.; Moreton, F.C.; Muir, K.W.; et al. Patient-Specific iPSC Model of a Genetic Vascular Dementia Syndrome Reveals Failure of Mural Cells to Stabilize Capillary Structures. Stem Cell Rep. 2019, 13, 817–831. [Google Scholar] [CrossRef]
- Porotto, M.; Ferren, M.; Chen, Y.-W.; Siu, Y.; Makhsous, N.; Rima, B.; Briese, T.; Greninger, A.L.; Snoeck, H.-W.; Moscona, A. Authentic Modeling of Human Respiratory Virus Infection in Human Pluripotent Stem Cell-Derived Lung Organoids. mBio 2019, 10, e00723-19. [Google Scholar] [CrossRef]
- Ackermann, M.; Kempf, H.; Hetzel, M.; Hesse, C.; Hashtchin, A.R.; Brinkert, K.; Schott, J.W.; Haake, K.; Kuhnel, M.P.; Glage, S.; et al. Bioreactor-based mass production of human iPSC-derived macrophages enables immunotherapies against bacterial airway infections. Nat. Commun. 2018, 9, 5088. [Google Scholar] [CrossRef] [PubMed]
- Trump, L.R.; Nayak, R.C.; Singh, A.K.; Emberesh, S.; Wellendorf, A.M.; Lutzko, C.M.; Cancelas, J.A. Neutrophils Derived from Genetically Modified Human Induced Pluripotent Stem Cells Circulate and Phagocytose Bacteria In Vivo. Stem Cells Transl. Med. 2019, 8, 557–567. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitchell, A.; Yu, C.; Zhao, X.; Pearmain, L.; Shah, R.; Hanley, K.P.; Felton, T.; Wang, T. Rapid Generation of Pulmonary Organoids from Induced Pluripotent Stem Cells by Co-Culturing Endodermal and Mesodermal Progenitors for Pulmonary Disease Modelling. Biomedicines 2023, 11, 1476. https://doi.org/10.3390/biomedicines11051476
Mitchell A, Yu C, Zhao X, Pearmain L, Shah R, Hanley KP, Felton T, Wang T. Rapid Generation of Pulmonary Organoids from Induced Pluripotent Stem Cells by Co-Culturing Endodermal and Mesodermal Progenitors for Pulmonary Disease Modelling. Biomedicines. 2023; 11(5):1476. https://doi.org/10.3390/biomedicines11051476
Chicago/Turabian StyleMitchell, Adam, Chaowen Yu, Xiangjun Zhao, Laurence Pearmain, Rajesh Shah, Karen Piper Hanley, Timothy Felton, and Tao Wang. 2023. "Rapid Generation of Pulmonary Organoids from Induced Pluripotent Stem Cells by Co-Culturing Endodermal and Mesodermal Progenitors for Pulmonary Disease Modelling" Biomedicines 11, no. 5: 1476. https://doi.org/10.3390/biomedicines11051476
APA StyleMitchell, A., Yu, C., Zhao, X., Pearmain, L., Shah, R., Hanley, K. P., Felton, T., & Wang, T. (2023). Rapid Generation of Pulmonary Organoids from Induced Pluripotent Stem Cells by Co-Culturing Endodermal and Mesodermal Progenitors for Pulmonary Disease Modelling. Biomedicines, 11(5), 1476. https://doi.org/10.3390/biomedicines11051476