IFIT2 Depletion Promotes Cancer Stem Cell-like Phenotypes in Oral Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Western Blot Analysis
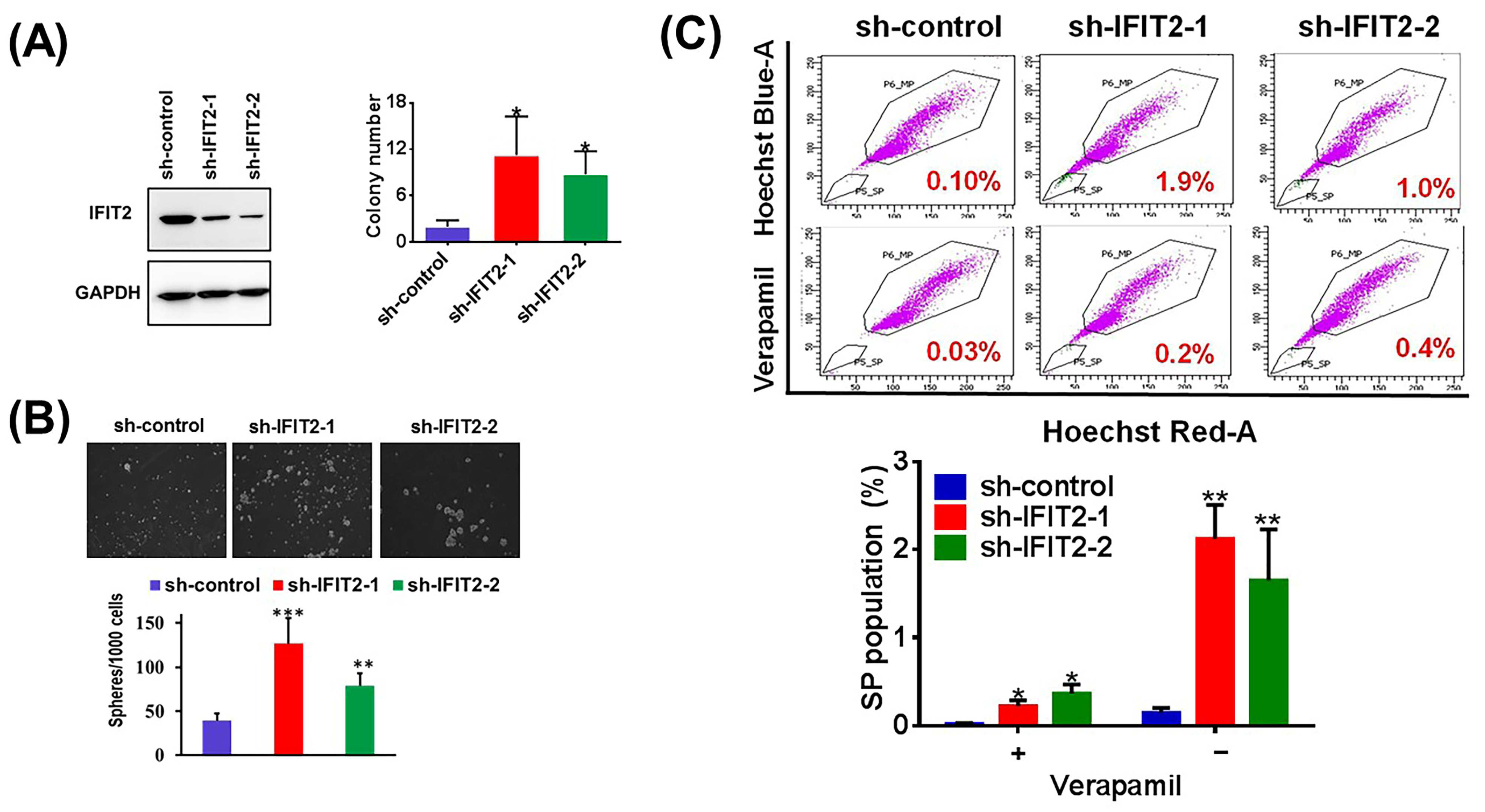
2.3. Anchorage-Independent Growth Assay
2.4. Sphere Formation Assay
2.5. Side Population (SP) Assay
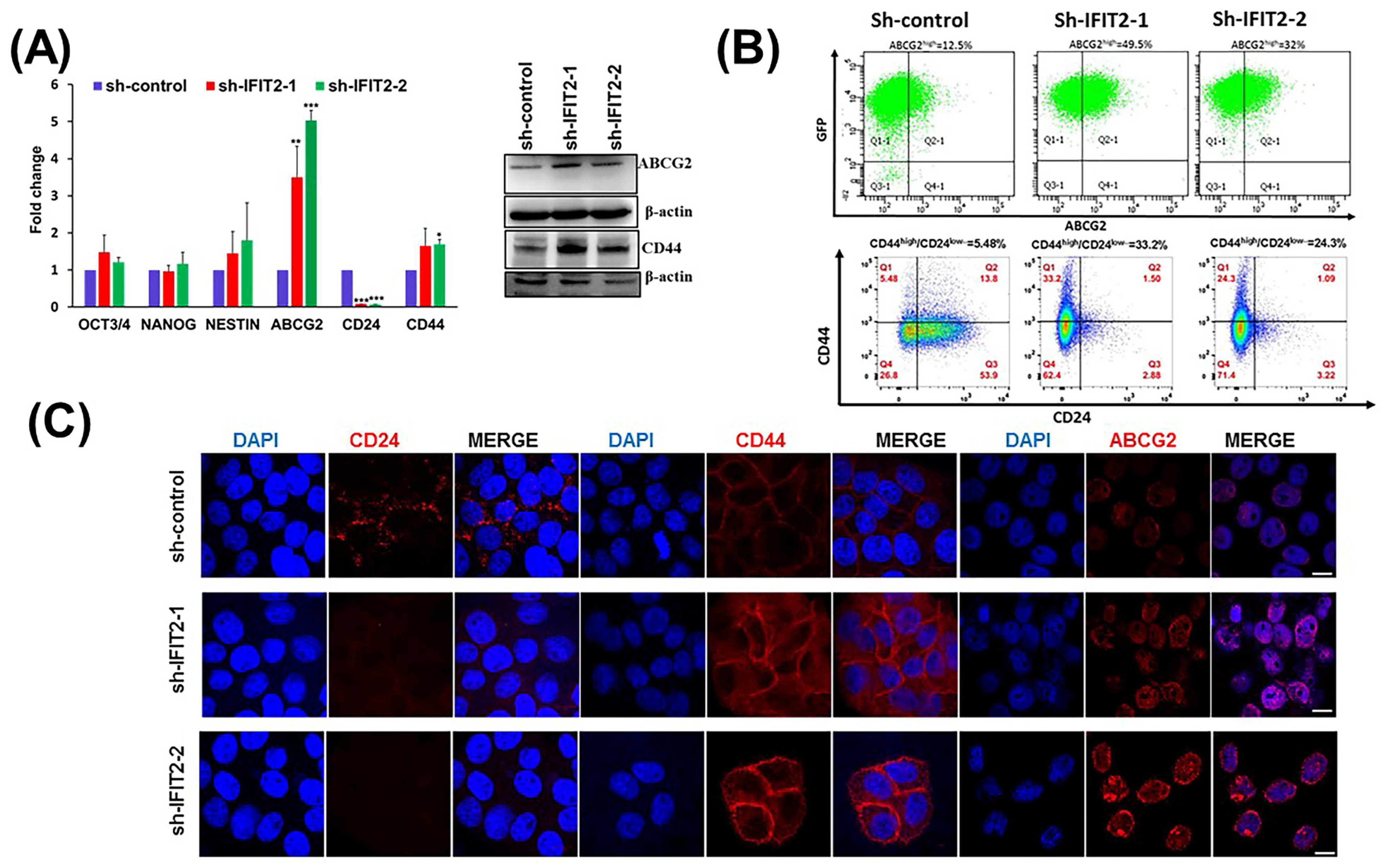
2.6. Quantitative Real-Time PCR (QPCR)
2.7. Flow Cytometry Analysis
2.8. Immunofluorescence and Confocal Microscopy
2.9. Tumorigenicity Assay
2.10. Immunohistochemical Staining
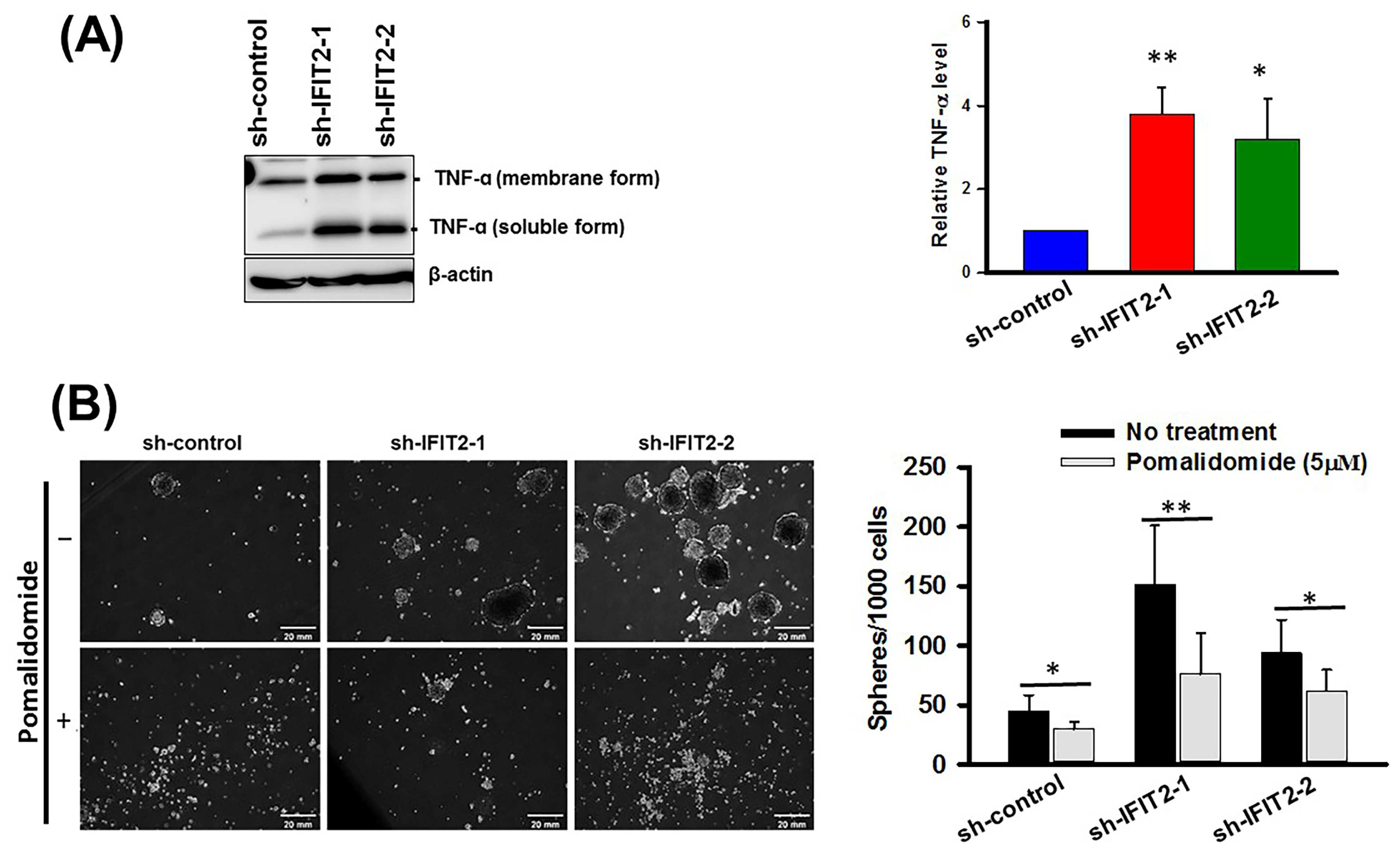
2.11. Enzyme-Linked Immunoassay (ELISA)
2.12. Statistical Analysis
3. Results
3.1. IFIT2 Knockdown Cells Exhibit CSC-like Properties
3.2. Characterization of CSC Markers in IFIT2 Knockdown Cells
3.3. Tumorigenicity in IFIT2 Knockdown Cells
3.4. Clinical Signature of IFIT2 and CSCs in HNC
3.5. Effect of a TNF-α Inhibitor on Spheroid Formation in IFIT2 Knockdown Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shield, K.D.; Ferlay, J.; Jemal, A.; Sankaranarayanan, R.; Chaturvedi, A.K.; Bray, F.; Soerjomataram, I. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J. Clin. 2017, 67, 51–64. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Shingaki, S.; Takada, M.; Sasai, K.; Bibi, R.; Kobayashi, T.; Nomura, T.; Saito, C. Impact of lymph node metastasis on the pattern of failure and survival in oral carcinomas. Am. J. Surg. 2003, 185, 278–284. [Google Scholar] [CrossRef]
- Iyer, N.G.; Tan, D.S.; Tan, V.K.; Wang, W.; Hwang, J.; Tan, N.C.; Sivanandan, R.; Tan, H.K.; Lim, W.T.; Ang, M.K.; et al. Randomized trial comparing surgery and adjuvant radiotherapy versus concurrent chemoradiotherapy in patients with advanced, nonmetastatic squamous cell carcinoma of the head and neck: 10-year update and subset analysis. Cancer 2015, 121, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Jerjes, W.; Upile, T.; Petrie, A.; Riskalla, A.; Hamdoon, Z.; Vourvachis, M.; Karavidas, K.; Jay, A.; Sandison, A.; Thomas, G.J.; et al. Clinicopathological parameters, recurrence, locoregional and distant metastasis in 115 T1-T2 oral squamous cell carcinoma patients. Head Neck Oncol. 2010, 2, 9. [Google Scholar] [CrossRef]
- Baillie, R.; Tan, S.T.; Itinteang, T. Cancer Stem Cells in Oral Cavity Squamous Cell Carcinoma: A Review. Front. Oncol. 2017, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef]
- Chiou, S.H.; Yu, C.C.; Huang, C.Y.; Lin, S.C.; Liu, C.J.; Tsai, T.H.; Chou, S.H.; Chien, C.S.; Ku, H.H.; Lo, J.F. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin. Cancer Res. 2008, 14, 4085–4095. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Gupta, P.B.; Pastushenko, I.; Skibinski, A.; Blanpain, C.; Kuperwasser, C. Phenotypic Plasticity: Driver of Cancer Initiation, Progression, and Therapy Resistance. Cell Stem Cell 2019, 24, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef]
- Maitland, N.J.; Collins, A.T. Prostate cancer stem cells: A new target for therapy. J. Clin. Oncol. 2008, 26, 2862–2870. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Fujisawa, T.; Husain, S.R.; Puri, R.K. Identification and characterization of cancer stem cells in human head and neck squamous cell carcinoma. BMC Cancer 2014, 14, 173. [Google Scholar] [CrossRef]
- Pohl, A.; Lurje, G.; Kahn, M.; Lenz, H.J. Stem cells in colon cancer. Clin. Color. Cancer 2008, 7, 92–98. [Google Scholar] [CrossRef]
- Batlle, E.; Sancho, E.; Franci, C.; Dominguez, D.; Monfar, M.; Baulida, J.; Garcia De Herreros, A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000, 2, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Cano, A.; Perez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef]
- Baniebrahimi, G.; Mir, F.; Khanmohammadi, R. Cancer stem cells and oral cancer: Insights into molecular mechanisms and therapeutic approaches. Cancer Cell Int. 2020, 20, 113. [Google Scholar] [CrossRef]
- Fensterl, V.; Sen, G.C. The ISG56/IFIT1 gene family. J. Interferon Cytokine Res. 2011, 31, 71–78. [Google Scholar] [CrossRef]
- Diamond, M.S.; Farzan, M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef]
- Pidugu, V.K.; Pidugu, H.B.; Wu, M.M.; Liu, C.J.; Lee, T.C. Emerging Functions of Human IFIT Proteins in Cancer. Front. Mol. Biosci. 2019, 6, 148. [Google Scholar] [CrossRef]
- Zhou, X.; Michal, J.J.; Zhang, L.; Ding, B.; Lunney, J.K.; Liu, B.; Jiang, Z. Interferon induced IFIT family genes in host antiviral defense. Int. J. Biol. Sci. 2013, 9, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Terenzi, F.; White, C.; Pal, S.; Williams, B.R.; Sen, G.C. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J. Virol. 2007, 81, 8656–8665. [Google Scholar] [CrossRef] [PubMed]
- Pidugu, V.K.; Wu, M.M.; Yen, A.H.; Pidugu, H.B.; Chang, K.W.; Liu, C.J.; Lee, T.C. IFIT1 and IFIT3 promote oral squamous cell carcinoma metastasis and contribute to the anti-tumor effect of gefitinib via enhancing p-EGFR recycling. Oncogene 2019, 38, 3232–3247. [Google Scholar] [CrossRef] [PubMed]
- Lo, U.G.; Bao, J.; Cen, J.; Yeh, H.C.; Luo, J.; Tan, W.; Hsieh, J.T. Interferon-induced IFIT5 promotes epithelial-to-mesenchymal transition leading to renal cancer invasion. Am. J. Clin. Exp. Urol. 2019, 7, 31–45. [Google Scholar] [PubMed]
- Zhao, Y.; Altendorf-Hofmann, A.; Pozios, I.; Camaj, P.; Daberitz, T.; Wang, X.; Niess, H.; Seeliger, H.; Popp, F.; Betzler, C.; et al. Elevated interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) is a poor prognostic marker in pancreatic ductal adenocarcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Koh, S.Y.; Moon, J.Y.; Unno, T.; Cho, S.K. Baicalein Suppresses Stem Cell-Like Characteristics in Radio- and Chemoresistant MDA-MB-231 Human Breast Cancer Cells through Up-Regulation of IFIT2. Nutrients 2019, 11, 624. [Google Scholar] [CrossRef]
- Chen, L.; Zhai, W.; Zheng, X.; Xie, Q.; Zhou, Q.; Tao, M.; Zhu, Y.; Wu, C.; Jiang, J. Decreased IFIT2 Expression Promotes Gastric Cancer Progression and Predicts Poor Prognosis of the Patients. Cell. Physiol. Biochem. 2018, 45, 15–25. [Google Scholar] [CrossRef]
- Su, W.; Xiao, W.; Chen, L.; Zhou, Q.; Zheng, X.; Ju, J.; Jiang, J.; Wang, Z. Decreased IFIT2 Expression In Human Non-Small-Cell Lung Cancer Tissues Is Associated With Cancer Progression And Poor Survival Of The Patients. OncoTargets Ther. 2019, 12, 8139–8149. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.C.; Liu, C.J.; Chang, K.W.; Lee, T.C. Depleting IFIT2 mediates atypical PKC signaling to enhance the migration and metastatic activity of oral squamous cell carcinoma cells. Oncogene 2013, 32, 3686–3697. [Google Scholar] [CrossRef] [PubMed]
- Regmi, P.; Lai, K.C.; Liu, C.J.; Lee, T.C. SAHA Overcomes 5-FU Resistance in IFIT2-Depleted Oral Squamous Cell Carcinoma Cells. Cancers 2020, 12, 3527. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.C.; Chang, K.W.; Liu, C.J.; Kao, S.Y.; Lee, T.C. IFN-induced protein with tetratricopeptide repeats 2 inhibits migration activity and increases survival of oral squamous cell carcinoma. Mol. Cancer Res. 2008, 6, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, N.; Liu, S.; Jiang, M.; Wan, J.; Zhang, Y.; Wan, L.; Xie, C.; Le, A. Overexpression of IFIT2 inhibits the proliferation of chronic myeloid leukemia cells by regulating the BCRABL/AKT/mTOR pathway. Int. J. Mol. Med. 2020, 45, 1187–1194. [Google Scholar] [PubMed]
- Chen, L.; Liu, S.; Xu, F.; Kong, Y.; Wan, L.; Zhang, Y.; Zhang, Z. Inhibition of Proteasome Activity Induces Aggregation of IFIT2 in the Centrosome and Enhances IFIT2-Induced Cell Apoptosis. Int. J. Biol. Sci. 2017, 13, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Wang, Y.; Ma, Z.; Yang, R.; Liang, S.; Zhang, M.; Song, S.; Li, S.; Liu, G.; Fan, D.; et al. MicroRNA-645, up-regulated in human adencarcinoma of gastric esophageal junction, inhibits apoptosis by targeting tumor suppressor IFIT2. BMC Cancer 2014, 14, 633. [Google Scholar] [CrossRef] [PubMed]
- Ohsugi, T.; Yamaguchi, K.; Zhu, C.; Ikenoue, T.; Furukawa, Y. Decreased expression of interferon-induced protein 2 (IFIT2) by Wnt/beta-catenin signaling confers anti-apoptotic properties to colorectal cancer cells. Oncotarget 2017, 8, 100176–100186. [Google Scholar] [CrossRef]
- Lai, K.C.; Liu, C.J.; Lin, T.J.; Mar, A.C.; Wang, H.H.; Chen, C.W.; Hong, Z.X.; Lee, T.C. Blocking TNF-alpha inhibits angiogenesis and growth of IFIT2-depleted metastatic oral squamous cell carcinoma cells. Cancer Lett. 2016, 370, 207–215. [Google Scholar] [CrossRef]
- Du, F.; Liu, H.; Lu, Y.; Zhao, X.; Fan, D. Epithelial-to-Mesenchymal Transition: Liaison between Cancer Metastasis and Drug Resistance. Crit. Rev. Oncog. 2017, 22, 275–282. [Google Scholar] [CrossRef]
- Niess, H.; Camaj, P.; Renner, A.; Ischenko, I.; Zhao, Y.; Krebs, S.; Mysliwietz, J.; Jackel, C.; Nelson, P.J.; Blum, H.; et al. Side population cells of pancreatic cancer show characteristics of cancer stem cells responsible for resistance and metastasis. Target Oncol. 2015, 10, 215–227. [Google Scholar] [CrossRef]
- Hamburger, A.W.; Salmon, S.E. Primary bioassay of human tumor stem cells. Science 1977, 197, 461–463. [Google Scholar] [CrossRef]
- Song, J.; Chang, I.; Chen, Z.; Kang, M.; Wang, C.Y. Characterization of side populations in HNSCC: Highly invasive, chemoresistant and abnormal Wnt signaling. PLoS ONE 2010, 5, e11456. [Google Scholar] [CrossRef]
- Greve, B.; Kelsch, R.; Spaniol, K.; Eich, H.T.; Gotte, M. Flow cytometry in cancer stem cell analysis and separation. Cytometry A 2012, 81, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Hirschmann-Jax, C.; Foster, A.E.; Wulf, G.G.; Nuchtern, J.G.; Jax, T.W.; Gobel, U.; Goodell, M.A.; Brenner, M.K. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc. Natl. Acad. Sci. USA 2004, 101, 14228–14233. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, W.; Jiang, Q.; Gong, M.; Chen, R.; Wu, H.; Han, R.; Chen, Y.; Han, D. Lipopolysaccharide-induced testicular dysfunction and epididymitis in mice: A critical role of tumor necrosis factor alphadagger. Biol. Reprod. 2019, 100, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Steele, R.; Sowadski, M.; Crawford, S.E.; Varvares, M.; Ray, R.B. Identification of molecular signature of head and neck cancer stem-like cells. Sci. Rep. 2015, 5, 7819. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, D.S.; Tevis, K.M.; Blessing, W.A.; Colson, Y.L.; Zaman, M.H.; Grinstaff, M.W. Breast Cancer Spheroids Reveal a Differential Cancer Stem Cell Response to Chemotherapeutic Treatment. Sci. Rep. 2017, 7, 10382. [Google Scholar] [CrossRef]
- Morata-Tarifa, C.; Jimenez, G.; Garcia, M.A.; Entrena, J.M.; Grinan-Lison, C.; Aguilera, M.; Picon-Ruiz, M.; Marchal, J.A. Low adherent cancer cell subpopulations are enriched in tumorigenic and metastatic epithelial-to-mesenchymal transition-induced cancer stem-like cells. Sci. Rep. 2016, 6, 18772. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Zhou, Y.; Zhai, B.; Liao, J.; Xu, W.; Zhang, R.; Li, J.; Zhang, Y.; Chen, L.; Qian, H.; et al. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol. 2011, 11, 71. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, Y.; Mao, L.; Zhang, Z.; Chen, W. Side population in oral squamous cell carcinoma possesses tumor stem cell phenotypes. Cancer Lett. 2009, 277, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Yanamoto, S.; Kawasaki, G.; Yamada, S.; Yoshitomi, I.; Kawano, T.; Yonezawa, H.; Rokutanda, S.; Naruse, T.; Umeda, M. Isolation and characterization of cancer stem-like side population cells in human oral cancer cells. Oral Oncol. 2011, 47, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Todoroki, K.; Ogasawara, S.; Akiba, J.; Nakayama, M.; Naito, Y.; Seki, N.; Kusukawa, J.; Yano, H. CD44v3+/CD24− cells possess cancer stem cell-like properties in human oral squamous cell carcinoma. Int. J. Oncol. 2016, 48, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Ghuwalewala, S.; Ghatak, D.; Das, P.; Dey, S.; Sarkar, S.; Alam, N.; Panda, C.K.; Roychoudhury, S. CD44(high)CD24(low) molecular signature determines the Cancer Stem Cell and EMT phenotype in Oral Squamous Cell Carcinoma. Stem Cell Res 2016, 16, 405–417. [Google Scholar] [CrossRef]
- Tsai, L.L.; Hu, F.W.; Lee, S.S.; Yu, C.H.; Yu, C.C.; Chang, Y.C. Oct4 mediates tumor initiating properties in oral squamous cell carcinomas through the regulation of epithelial-mesenchymal transition. PLoS ONE 2014, 9, e87207. [Google Scholar] [CrossRef]
- Vesuna, F.; Lisok, A.; Kimble, B.; Raman, V. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia 2009, 11, 1318–1328. [Google Scholar] [CrossRef]
- Ye, P.; Nadkarni, M.A.; Hunter, N. Regulation of E-cadherin and TGF-beta3 expression by CD24 in cultured oral epithelial cells. Biochem. Biophys. Res. Commun. 2006, 349, 229–235. [Google Scholar] [CrossRef]
- Schneeberger, E.E.; Lynch, R.D. The tight junction: A multifunctional complex. Am. J. Physiol. Cell Physiol. 2004, 286, C1213–C1228. [Google Scholar] [CrossRef]
- Bhat-Nakshatri, P.; Appaiah, H.; Ballas, C.; Pick-Franke, P.; Goulet, R., Jr.; Badve, S.; Srour, E.F.; Nakshatri, H. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24− phenotype. BMC Cancer 2010, 10, 411. [Google Scholar] [CrossRef]
- Techasen, A.; Namwat, N.; Loilome, W.; Bungkanjana, P.; Khuntikeo, N.; Puapairoj, A.; Jearanaikoon, P.; Saya, H.; Yongvanit, P. Tumor necrosis factor-alpha (TNF-alpha) stimulates the epithelial-mesenchymal transition regulator Snail in cholangiocarcinoma. Med. Oncol. 2012, 29, 3083–3091. [Google Scholar] [CrossRef]
- Zhang, L.; Jiao, M.; Wu, K.; Li, L.; Zhu, G.; Wang, X.; He, D.; Wu, D. TNF-alpha induced epithelial mesenchymal transition increases stemness properties in renal cell carcinoma cells. Int. J. Clin. Exp. Med. 2014, 7, 4951–4958. [Google Scholar] [PubMed]
- Li, J.; Zha, X.M.; Wang, R.; Li, X.D.; Xu, B.; Xu, Y.J.; Yin, Y.M. Regulation of CD44 expression by tumor necrosis factor-alpha and its potential role in breast cancer cell migration. Biomed. Pharmacother. 2012, 66, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Hong, H.S.; Liu, Z.X.; Kim, R.H.; Kang, M.K.; Park, N.H.; Shin, K.H. TNFα enhances cancer stem cell-like phenotype via Notch-Hes1 activation in oral squamous cell carcinoma cells. Biochem. Biophys. Res. Commun. 2012, 424, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Reich, N.C. A death-promoting role for ISG54/IFIT2. J. Interferon Cytokine Res. 2013, 33, 199–205. [Google Scholar] [CrossRef]
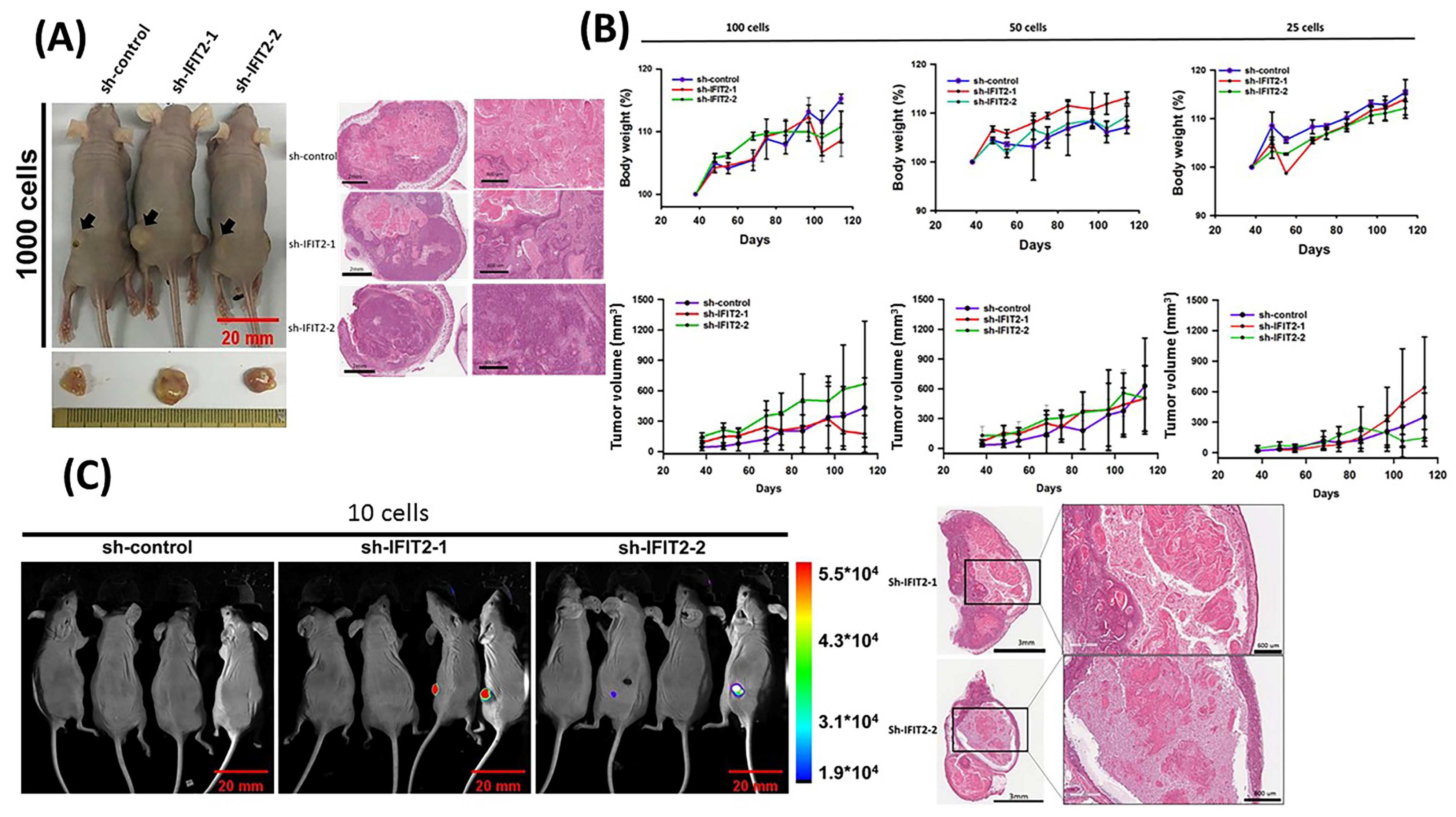

| Cell Number | Subline | Latency (Days) | Tumor Incidence (%) |
|---|---|---|---|
| 100,000 | sh-control | 21 | 100 (10/10) |
| sh-IFIT2-1 | 21 | 100 (10/10) | |
| sh-IFIT2-2 | 21 | 100 (10/10) | |
| 50,000 | sh-control | 21 | 100 (10/10) |
| sh-IFIT2-1 | 21 | 100 (10/10) | |
| sh-IFIT2-2 | 21 | 100 (10/10) | |
| 10,000 | sh-control | 30 | 100 (10/10) |
| sh-IFIT2-1 | 28 | 100 (10/10) | |
| sh-IFIT2-2 | 28 | 100 (10/10) | |
| 1000 | sh-control | 36 | 100 (10/10) |
| sh-IFIT2-1 | 28 | 100 (10/10) | |
| sh-IFIT2-2 | 32 | 100 (10/10) | |
| 100 | sh-control | 55 | 60 (6/10) |
| sh-IFIT2-1 | 48 | 70 (7/10) | |
| sh-IFIT2-2 | 48 | 80 (8/10) | |
| 50 | sh-control | 68 | 50 (5/10) |
| sh-IFIT2-1 | 48 | 70 (7/10) | |
| sh-IFIT2-2 | 48 | 70 (7/10) | |
| 25 | sh-control | 75 | 16.7 (1/6) |
| sh-IFIT2-1 | 68 | 66.7 (4/6) | |
| sh-IFIT2-2 | 68 | 50 (3/6) | |
| 10 | sh-control | 114 | 0 (0/8) |
| sh-IFIT2-1 | 114 | 25 (2/8) | |
| sh-IFIT2-2 | 114 | 25 (2/8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, K.-C.; Regmi, P.; Liu, C.-J.; Lo, J.-F.; Lee, T.-C. IFIT2 Depletion Promotes Cancer Stem Cell-like Phenotypes in Oral Cancer. Biomedicines 2023, 11, 896. https://doi.org/10.3390/biomedicines11030896
Lai K-C, Regmi P, Liu C-J, Lo J-F, Lee T-C. IFIT2 Depletion Promotes Cancer Stem Cell-like Phenotypes in Oral Cancer. Biomedicines. 2023; 11(3):896. https://doi.org/10.3390/biomedicines11030896
Chicago/Turabian StyleLai, Kuo-Chu, Prabha Regmi, Chung-Ji Liu, Jeng-Fan Lo, and Te-Chang Lee. 2023. "IFIT2 Depletion Promotes Cancer Stem Cell-like Phenotypes in Oral Cancer" Biomedicines 11, no. 3: 896. https://doi.org/10.3390/biomedicines11030896
APA StyleLai, K.-C., Regmi, P., Liu, C.-J., Lo, J.-F., & Lee, T.-C. (2023). IFIT2 Depletion Promotes Cancer Stem Cell-like Phenotypes in Oral Cancer. Biomedicines, 11(3), 896. https://doi.org/10.3390/biomedicines11030896





