Recoding of Nonsense Mutation as a Pharmacological Strategy
Abstract
1. Introduction
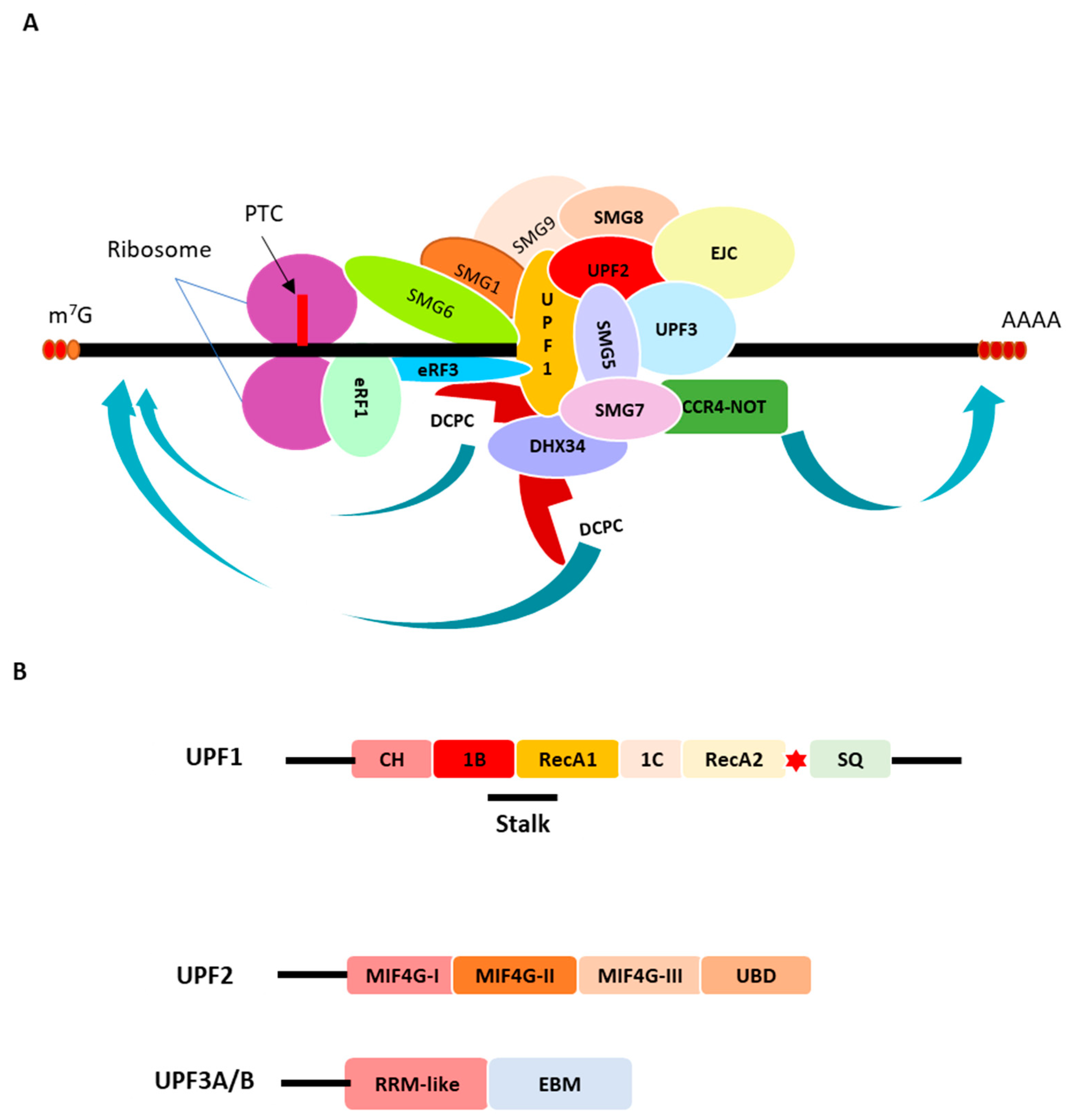
2. The Nonsense-Mediated mRNA Decay (NMD) Machinery
The Codon Effects
3. Nonsense-Mediated mRNA Decay (NMD) Inhibition by Drugs
3.1. Second Generation of Aminoglycosides Used for Pharmacological Therapy
3.2. Drugs That Induce Readthrough of PTCs by Non-Aminoglycoside Compounds
3.3. Combinatorial Drug Therapy
3.4. Challenges Associated with Drug-Stimulated Nonsense Suppression Studies
3.5. Development of NMD Inhibitors: New Pharmacological Perspectives
4. Conclusions and Discussion
5. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Campofelice, A.; Lentini, L.; Di Leonardo, A.; Melfi, R.; Tutone, M.; Pace, A.; Pibiri, I. Strategies against Nonsense: Oxadiazoles as Translational Readthrough-Inducing Drugs (TRIDs). Int. J. Mol. Sci. 2019, 20, 3329. [Google Scholar] [CrossRef]
- Antonarakis, S.E.; Cooper, D.N. Human Gene Mutation in Inherited Disease: Molecular Mechanisms and Clinical Consequences. In Emery Rimoin’s Princ Pract Med Genet, 6th ed.; Rimoin, D., Pyeritz, R., Bruce, K., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; Chapter 7; pp. 1–48. [Google Scholar]
- Peltz, S.W.; Morsy, M.; Welch, E.M.; Jacobson, A. Ataluren as an Agent for Therapeutic Nonsense Suppression. Annu. Rev. Med. 2013, 64, 407–425. [Google Scholar] [CrossRef]
- Lejeune, F. Nonsense-Mediated mRNA Decay, a Finely Regulated Mechanism. Biomedicines 2022, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Loudon, J.A. Repurposing Amlexanox as a ‘Run the Red Light Cure-All’ with Read-Through—A ‘No-Nonsense’ Approach to Personalised Medicine. J. Bioanal. Biomed. 2013, 5, 79–96. [Google Scholar] [CrossRef]
- Ng, M.Y.; Zhang, H.; Weil, A.; Singh, V.; Jamiolkowski, R.; Baradaran-Heravi, A.; Roberge, M.; Jacobson, A.; Friesen, W.; Welch, E.; et al. New in Vitro Assay Measuring Direct Interaction of Nonsense Suppressors with the Eukaryotic Protein Synthesis Machinery. ACS Med. Chem. Lett. 2018, 9, 1285–1291. [Google Scholar] [CrossRef]
- Ardiçli, D.; Haliloǧlu, G.; Alikaşifoǧlu, M.; Topaloǧlu, H. Diagnostic Pathway to Nonsense Mutation Dystrophinopathy: A Tertiary-Center, Retrospective Experience. Neuropediatrics 2019, 50, 41–45. [Google Scholar] [CrossRef]
- Li, N.; Wang, L.; Sun, X.; Lu, Z.; Suo, X.; Li, J.; Peng, J.; Peng, R. A novel mutation in VRK1 associated with distal spinal muscular atrophy. J. Hum. Genet. 2019, 64, 215–219. [Google Scholar] [CrossRef]
- Li, K.; Turner, A.N.; Chen, M.; Brosius, S.N.; Schoeb, T.R.; Messiaen, L.M.; Bedwell, D.M.; Zinn, K.R.; Anastasaki, C.; Gutmann, D.H.; et al. Mice with missense and nonsense NF1 mutations display divergent phenotypes compared with human neurofibromatosis type I. Dis. Model. Mech. 2016, 9, 759–767. [Google Scholar] [PubMed]
- Kiser, K.; Webb-Jones, K.D.; Bowne, S.J.; Sullivan, S.L.; Daiger, S.P.; Birch, D.G. Time Course of Disease Progression of PRPF31-mediated Retinitis Pigmentosa. Am. J. Ophthalmol. 2019, 200, 76–84. [Google Scholar] [CrossRef]
- Nogueira, G.; Fernandes, R.; García-Moreno, J.F.; Romão, L. Nonsense-mediated RNA decay and its bipolar function in cancer. Mol. Cancer. 2021, 20, 72. [Google Scholar] [CrossRef]
- Frischmeyer, P.A.; Dietz, H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999, 8, 1893–1900. [Google Scholar] [CrossRef]
- Mendell, J.R.; Buzin, C.H.; Feng, J.; Yan, J.; Serrano, C.; Sangani, D.S.; Wall, C.; Prior, T.W.; Sommer, S.S. Diagnosis of Duchenne dystrophy by enhanced detection of small mutations. Neurology 2001, 57, 645–650. [Google Scholar] [CrossRef]
- Pulak, R.; Anderson, P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993, 7, 1885–1897. [Google Scholar] [CrossRef] [PubMed]
- Leeds, P.; Wood, J.M.; Lee, B.S.; Culbertson, M.R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 2165–2177. [Google Scholar]
- Popp, M.W.; Maquat, L.E. Nonsense-mediated mRNA Decay and Cancer. Curr. Opin. Genet. Dev. 2018, 48, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Boehm, V.; Gehring, N.H. Exon Junction Complexes: Supervising the Gene Expression Assembly Line. Trends Genet. 2016, 11, 724–735. [Google Scholar] [CrossRef]
- Bono, F.; Ebert, J.; Lorentzen, E.; Conti, E. The Crystal Structure of the Exon Junction Complex Reveals How It Maintains a Stable Grip on mRNA. Cell 2006, 126, 713–725. [Google Scholar] [CrossRef]
- Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000, 24, 6860–6869. [Google Scholar] [CrossRef]
- Gardner, L.B. Nonsense mediated RNA decay regulation by cellular stress; implications for tumorigenesis NIH Public Access. Mol. Cancer Res. 2010, 8, 295–308. [Google Scholar] [CrossRef]
- Ishigaki, Y.; Li, X.; Serin, G.; Maquat, L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 2001, 106, 607–617. [Google Scholar] [CrossRef]
- Dostie, J.; Dreyfuss, G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 2002, 13, 1060–1067. [Google Scholar] [CrossRef]
- Lykke-Andersen, J.; Shu, M.D.; Steitz, J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 2000, 103, 1121–1131. [Google Scholar] [CrossRef]
- Gehring, N.H.; Neu-Yilik, G.; Schell, T.; Hentze, M.W.; Kulozik, A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003, 11, 939–949. [Google Scholar] [CrossRef]
- Cheng, Z.; Saito, K.; Pisarev, A.V.; Wada, M.; Pisareva, V.P.; Pestova, T.V.; Gajda, M.; Round, A.; Kong, C.; Lim, M. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009, 23, 1106–1118. [Google Scholar] [CrossRef]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef]
- Schoenberg, D.R.; Maquat, L.E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012, 13, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Isken, O.; Kim, Y.K.; Hosoda, N.; Mayeur, G.L.; Hershey, J.W.B.; Maquat, L.E. Upf1 Phosphorylation Triggers Translational Repression during Nonsense-Mediated mRNA Decay. Cell 2008, 133, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Maquat, L.E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 2016, 129, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Muhlrad, D.; Parker, R. Premature translational termination triggers mRNA decapping. Nature 1994, 370, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Pawlicka, K.; Kalathiya, U.; Alfaro, J. Nonsense-Mediated mRNA Decay: Pathologies and the Potential for Novel Therapeutics. Cancers 2020, 12, 765. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Jensen, T.H. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015, 16, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Kashima, I.; Fauser, M.; Saulière, J.; Izaurralde, E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 2008, 14, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Melero, R.; Hug, N.; López-Perrote, A.; Yamashita, A.; Cáceres, J.F.; Llorca, O. The RNA helicase DHX34 functions as a scaffold for SMG1-mediated UPF1 phosphorylation. Nat. Commun. 2016, 7, 10585. [Google Scholar] [CrossRef] [PubMed]
- Hug, N.; Caceres, J.F. The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep. 2014, 8, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Hanson, G.; Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell. Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef]
- Hia, F.; Takeuchi, O. The effects of codon bias and optimality on mRNA and protein regulation. Cell. Mol. Life Sci. 2021, 78, 1909–1928. [Google Scholar]
- Yu, C.H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 2015, 59, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 2020, 182, 404–416.e14. [Google Scholar] [CrossRef]
- Ikeuchi, K.; Tesina, P.; Matsuo, Y.; Sugiyama, T.; Cheng, J.; Saeki, Y.; Tanaka, K.; Becker, T.; Beckmann, R.; Inada, T. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 2019, 38, e100276. [Google Scholar] [CrossRef]
- Tesina, P.; Lessen, L.N.; Buschauer, R.; Cheng, J.; Wu, C.C.; Berninghausen, O.; Buskirk, A.R.; Becker, T.; Beckmann, R.; Green, R. Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J. 2020, 39, e103365. [Google Scholar] [CrossRef]
- Diament, A.; Feldman, A.; Schochet, E.; Kupiec, M.; Arava, Y.; Tuller, T. The extent of ribosome queuing in budding yeast. PLoS Comput. Biol. 2018, 14, e1005951. [Google Scholar] [CrossRef]
- Matsuo, Y.; Ikeuchi, K.; Saeki, Y.; Iwasaki, S.; Schmidt, C.; Udagawa, T.; Sato, F.; Tsuchiya, H.; Becker, T.; Tanaka, K.; et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Shichino, Y.; Schneider-Poetsch, T.; Mito, M.; Hashimoto, S.; Udagawa, T.; Kohno, K.; Yoshida, M.; Mishima, Y.; Inada, T.; et al. Genome-wide Survey of Ribosome Collision. Cell Rep. 2020, 31, 107610. [Google Scholar] [CrossRef]
- Meydan, S.; Guydosh, N.R. Disome and Trisome Profiling Reveal Genome-wide Targets of Ribosome Quality Control. Mol. Cell. 2020, 79, 588–602.e6. [Google Scholar] [CrossRef]
- Ouranidis, A.; Vavilis, T.; Mandala, E.; Davidopoulou, C.; Stamoula, E.; Markopoulou, C.K.; Karagianni, A.; Kachrimanis, K. mRNA Therapeutic Modalities Design, Formulation and Manufacturing under Pharma 4.0 Principles. Biomedicines 2021, 10, 50. [Google Scholar] [CrossRef]
- Damase, T.R.; Sukhovershin, R.; Boada, C.; Taraballi, F.; Pettigrew, R.I.; Cooke, J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 628137. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Andresen, J.L.; Manan, R.S.; Langer, R. Nucleic Acid Delivery for Therapeutic Applications. Adv. Drug Deliv. Rev. 2021, 178, 113834. [Google Scholar] [CrossRef] [PubMed]
- Wayment-Steele, H.K.; Kim, D.S.; Choe, C.A.; Nicol, J.J.; Wellington-Oguri, R.; Watkins, A.M.; Parra Sperberg, R.A.; Huang, P.S.; Participants, E.; Das, R. Theoretical Basis for Stabilizing Messenger RNA through Secondary Structure Design. Nucleic Acids Res. 2021, 49, 10604–10617. [Google Scholar] [CrossRef]
- Lee, H.L.; Dougherty, J.P. Pharmaceutical therapies to recode nonsense mutations in inherited diseases. Pharmacol. Ther. 2012, 136, 227–266. [Google Scholar] [CrossRef]
- Lombardi, S.; Testa, M.F.; Pinotti, M.; Branchini, A. Molecular Insights into Determinants of Translational Readthrough and Implications for Nonsense Suppression Approaches. Int. J. Mol. Sci. 2020, 21, 9449. [Google Scholar] [CrossRef]
- Bonetti, B.; Fu, L.; Moon, J.; Bedwell, D.M. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995, 251, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.T.; Shirts, B.H.; Petros, L.M.; Flanigan, K.M.; Gesteland, R.F.; Atkins, J.F. Sequence specificity of aminoglycoside-induced stop codon readthrough: Potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 2000, 48, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Jones, M.K.; Hill, L.S.; Atkinson, J.; Martin, R. Context effects on misreading and suppression at UAG codons in human cells. Mol. Cell Biol. 1995, 15, 6593–6600. [Google Scholar] [CrossRef] [PubMed]
- Manuvakhova, M.; Keeling, K.; Bedwell, D.M. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA 2000, 6, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- McCaughan, K.K.; Brown, C.M.; Dalphin, M.E.; Berry, M.J.; Tate, W.P. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. USA 1995, 92, 5431–5435. [Google Scholar] [CrossRef]
- Martins-Dias, P.; Romão, L. Nonsense suppression therapies in human genetic diseases. Cell Mol. Life Sci. 2021, 78, 4677–4701. [Google Scholar] [CrossRef]
- Schlautmann, L.P.; Gehring, N.H. A Day in the Life of the Exon Junction Complex. Biomolecules 2020, 10, 866. [Google Scholar] [CrossRef] [PubMed]
- Hug, N.; Longman, D.; Cáceres, J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016, 44, 1483–1495. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Lin, Y.; Yan, X.X.; Ye, J.Z.; Li, Y.Q.; Ye, H.H. Enhanced RBM8A expression in human hepatocellular carcinoma. Int. J. Clin. Exp. Med. 2017, 10, 598–607. [Google Scholar]
- Pastor, F.; Kolonias, D.; Giangrande, P.H.; Gilboa, E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 2010, 465, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Stupack, D.G.; Wilkinson, M.F. Nonsense-mediated RNA decay: An emerging modulator of malignancy. Nat. Rev. Cancer 2022, 22, 437–451. [Google Scholar] [CrossRef]
- Liang, R.; Lin, Y.; Ye, J.Z.; Yan, X.X.; Liu, Z.H.; Li, Y.Q.; Luo, X.L.; Ye, H.H. High expression of RBM8A predicts poor patient prognosis and promotes tumor progression in hepatocellular carcinoma. Oncol. Rep. 2017, 37, 2167–2176. [Google Scholar] [CrossRef] [PubMed]
- Bruno, I.G.; Karam, R.; Huang, L.L.; Bhardwaj, A.; Lou, C.H.; Shum, E.Y.; Song, H.W.; Corbett, M.A.; Gifford, W.D.; Gecz, J.; et al. Identification of a MicroRNA that Activates Gene Expression by Repressing Nonsense-Mediated RNA Decay. Mol. Cell. 2011, 42, 500–510. [Google Scholar] [CrossRef]
- Jia, J.S.; Werkmeister, E.; Gonzalez-Hilarion, S.; Leroy, C.; Gruenert, D.C.; Lafont, F.; Tulasne, D.; Lejeune, F. Premature termination codon readthrough in human cells occurs in novel cytoplasmic foci and requires UPF proteins. J. Cell Sci. 2017, 130, 3009–3022. [Google Scholar]
- Fatscher, T.; Boehm, V.; Weiche, B.; Gehring, N.H. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA 2014, 20, 1579–1592. [Google Scholar] [CrossRef]
- Toma, K.G.; Rebbapragada, I.; Durand, S.; Lykke-Andersen, J. Identification of elements in human long 3 UTRs that inhibit nonsense-mediated decay. RNA 2015, 21, 887–897. [Google Scholar] [CrossRef]
- Friesen, W.J.; Johnson, B.; Sierra, J.; Zhuo, J.; Vazirani, P.; Xue, X.; Tomizawa, Y.; Baiazitov, R.; Morrill, C.; Ren, H.; et al. The minor gentamicin complex component, X2, is a potent premature stop codon readthrough molecule with therapeutic potential. PLoS ONE 2018, 13, e0206158. [Google Scholar] [CrossRef]
- Leubitz, A.; Frydman-Marom, A.; Sharpe, N.; van Duzer, J.; Campbell, K.C.M.; Vanhoutte, F. Safety, Tolerability, and Pharmacokinetics of Single Ascending Doses of ELX-02, a Potential Treatment for Genetic Disorders Caused by Nonsense Mutations, in Healthy Volunteers. Clin. Pharmacol. Drug Dev. 2019, 8, 984–994. [Google Scholar] [CrossRef]
- Forge, A.; Schacht, J. Aminoglycoside antibiotics. Audiol. Neurootol. 2000, 5, 3–22. [Google Scholar] [CrossRef]
- Obrecht, D.; Bernardini, F.; Dale, G.; Dembowsky, K. Emerging new therapeutics against key gram-negative pathogens. Annu. Rep. Med. 2011, 46, 245–262. [Google Scholar]
- Prokhorova, I.; Altman, R.B.; Djumagulov, M.; Shrestha, J.P.; Urzhumtsev, A.; Ferguson, A.; Chang, C.T.; Yusupov, M.; Blanchard, S.C.; Yusupova, G. Aminoglycoside interactions and impacts on the eukaryotic ribosome. Proc. Natl. Acad. Sci. USA 2017, 114, E10899–E10908. [Google Scholar] [CrossRef]
- Keeling, K.M.; Xue, X.; Gunn, G.; Bedwell, D.M. Therapeutics based on stop codon readthrough. Annu. Rev. Genom. Hum. Genet. 2014, 15, 371–394. [Google Scholar] [CrossRef]
- Fan-Minogue, H.; Bedwell, D.M. Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity. RNA 2008, 14, 148–157. [Google Scholar] [CrossRef]
- François, B.; Russell, R.J.M.; Murray, J.B.; Aboul-ela, F.; Masquida, B.; Vicens, Q.; Westhof, E. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: Role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005, 33, 5677–5699. [Google Scholar] [CrossRef]
- Jin, Y.; Watkins, D.; Degtyareva, N.N.; Green, K.D.; Spano, M.N.; Garneau-Tsodikova, S.; Arya, D.P. Arginine-linked neomycin B dimers: Synthesis, rRNA binding, and resistance enzyme activity. Med. Chem. Comm. 2016, 7, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Arya, D.P.; Coffee, R.L. DNA triple helix stabilization by aminoglycoside antibiotics. Bioorg. Med. Chem. Lett. 2000, 10, 1897–1899. [Google Scholar] [CrossRef]
- Arya, D.P.; Coffee, R.L.; Charles, I. Neomycin-induced hybrid triplex formation. J. Am. Chem. Soc. 2001, 123, 11093–11094. [Google Scholar] [CrossRef] [PubMed]
- Wesgate, R.; Evangelista, C.; Atkinson, R.; Shepard, A.; Adegoke, O.; Maillard, J. Y Understanding the risk of emerging bacterial resistance to over the counter antibiotics in topical sore throat medicines. J. Appl. Microbiol. 2020, 129, 916–925. [Google Scholar]
- Burke, J.F.; Mogg, A.E. Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin. Nucleic Acids Res. 1985, 13, 6265–6272. [Google Scholar] [CrossRef]
- Bidou, L.; Bugaud, O.; Belakhov, V.; Baasov, T.; Namy, O. Characterization of new-generation aminoglycoside promoting premature termination codon readthrough in cancer cells. RNA Biol. 2017, 14, 378–388. [Google Scholar]
- Recht, M.I.; Douthwaite, S.; Puglisi, J.D. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999, 18, 3133–3138. [Google Scholar] [CrossRef]
- Lynch, S.R.; Puglisi, J.D. Structural origins of aminoglycoside specificity for prokaryotic ribosomes. J. Mol. Biol. 2001, 306, 1037–1058. [Google Scholar] [CrossRef] [PubMed]
- Floquet, C.; Hatin, I.; Rousset, J.P.; Bidou, L. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet. 2018, 8, e1002608. [Google Scholar] [CrossRef] [PubMed]
- Bedwell, D.M.; Kaenjak, A.; Benos, D.J.; Bebok, Z.; Bubien, J.K.; Hong, J.; Tousson, A.; Clancy, J.P.; Sorscher, E.J. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat. Med. 1997, 3, 1280–1284. [Google Scholar] [CrossRef]
- Bar-Nun, S.; Beckmann, J.S. G-418, an elongation inhibitor of 80 S ribosomes. Biochim. Biophys. Acta Gene. Struct. Expr. 1983, 741, 123–127. [Google Scholar] [CrossRef]
- Sharma, J.; Keeling, K.M.; Rowe, S.M. Pharmacological approaches for targeting cystic fibrosis nonsense mutations. Eur. J. Med. Chem. 2020, 200, 112436. [Google Scholar] [CrossRef] [PubMed]
- Nudelman, I.; Glikin, D.; Smolkin, B.; Hainrichson, M.; Belakhov, V.; Baasov, T. Repairing faulty genes by aminoglycosides: Development of new derivatives of geneticin (G418) with enhanced suppression of diseases-causing nonsense mutations. Bioorg. Med. Chem. 2010, 18, 3735–3746. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Abbott, J.; Klaskala, L.; Zhao, G.; Birket, S.E.; Rowe, S.M. A Novel G542X CFTR Rat Model of Cystic Fibrosis Is Sensitive to Nonsense Mediated Decay. Front. Physiol. 2020, 11, 611294. [Google Scholar] [CrossRef]
- de Poel, E.; Spelier, S.; Suen, S.W.F.; Kruisselbrink, E.; Graeber, S.Y.; Mall, M.A.; Weersink, E.J.M.; van der Eerden, M.M.; Koppelman, G.H.; van der Ent, C.K.; et al. Functional Restoration of CFTR Nonsense Mutations in Intestinal Organoids. J. Cyst. Fibros. 2022, 2, 246–253. [Google Scholar] [CrossRef]
- Beauchamp, D.; Laurent, G.; Maldague, P.; Abid, S.; Kishore, B.K.; Tulkens, P.M. Protection against gentamicin-induced early renal alterations (phospholipidosis and increased DNA synthesis) by coadministration of poly-L-aspartic acid. J. Pharmacol. Exp. Ther. 1990, 255, 858–866. [Google Scholar]
- Ben Ismail, T.H.; Ali, B.H.; Bashir, A.A. Influence of iron, deferoxamine and ascorbic acid on gentamicin-induced nephrotoxicity in rats. Gen. Pharmacol. 1994, 25, 1249–1252. [Google Scholar] [CrossRef]
- Du, M.; Keeling, K.M.; Fan, L.; Liu, X.; Bedwell, D.M. Poly-L-aspartic acid enhances and prolongs gentamicin-mediated suppression of the CFTR-G542X mutation in a cystic fibrosis mouse model. J. Biol. Chem. 2009, 284, 6885–6892. [Google Scholar] [CrossRef]
- Sharma, J.; Du, M.; Wong, E.; Mutyam, V.; Li, Y.; Chen, J.; Wangen, J.; Thrasher, K.; Fu, L.; Peng, N.; et al. A small molecule that induces translational readthrough of CFTR nonsense mutations by eRF1 depletion. Nat. Commun. 2021, 12, 4358. [Google Scholar] [CrossRef]
- Venturini, A.; Borrelli, A.; Musante, I.; Scudieri, P.; Capurro, V.; Renda, M.; Pedemonte, N.; Galietta, L.J.V. Comprehensive Analysis of Combinatorial Pharmacological Treatments to Correct Nonsense Mutations in the CFTR Gene. Int. J. Mol. Sci. 2021, 22, 11972. [Google Scholar] [CrossRef]
- Kuschal, C.; Khan, S.G.; Enk, B.; DiGiovanna, J.J.; Kraemer, K.H. Readthrough of stop codons by use of aminoglycosides in cells from xeroderma pigmentosum group C patients. Exp. Dermatol. 2015, 24, 296–297. [Google Scholar] [CrossRef] [PubMed]
- Zilberberg, A.; Lahav, L.; Rosin-Arbesfeld, R. Restoration of APC gene function in colorectal cancer cells by aminoglycoside- and macrolide-induced read-through of premature termination codons. Gut 2010, 59, 496–507. [Google Scholar] [CrossRef]
- Keeling, K.M.; Bedwell, D.M. Clinically relevant aminoglycosides can suppress disease-associated premature stop mutations in the IDUA and P53 cDNAs in a mammalian translation system. J. Mol. Med. 2002, 80, 367–376. [Google Scholar] [CrossRef]
- Floquet, C.; Deforges, J.; Rousset, J.P.; Bidou, L. Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 2011, 39, 3350–3362. [Google Scholar] [CrossRef]
- Du, L.; Damoiseaux, R.; Nahas, S.; Gao, K.; Hu, H.; Pollard, J.M.; Goldstine, J.; Jung, M.E.; Henning, S.M.; Bertoni, C.; et al. Non-aminoglycoside compounds induce readthrough of nonsense mutations. J. Exp. Med. 2009, 206, 2285–2297. [Google Scholar] [CrossRef]
- Novotny, G.W.; Jakobsen, L.; Andersen, N.M.; Poehlsgaard, J.; Douthwaite, S. Ketolide antimicrobial activity persists after disruption of interactions with domain II of 23S rRNA. Antimicrob. Agents Chemother. 2004, 48, 3677–3683. [Google Scholar] [CrossRef]
- GLOBOCAN 2020: New Global Cancer Data|UICC. Available online: https://www.uicc.org/news/globocan-2020-new-globalcancer-data (accessed on 27 July 2022).
- Kaur, J.; Gulati, M.; Jha, N.K.; Disouza, J.; Patravale, V.; Dua, K.; Singh, S.K. Recent advances in developing polymeric micelles for treating cancer: Breakthroughs and bottlenecks in their clinical translation. Drug Discov. Today 2022, 27, 1495–1512. [Google Scholar] [CrossRef]
- Ioele, G.; Chieffallo, M.; Occhiuzzi, M.A.; De Luca, M.; Garofalo, A.; Ragno, G.; Grande, F. Anticancer Drugs: Recent Strategies to Improve Stability Profile, Pharmacokinetic and Pharmacodynamic Properties. Molecules 2022, 27, 5436. [Google Scholar] [CrossRef]
- Bidou, L.; Allamand, V.; Rousset, J.P.; Namy, O. Sense from nonsense: Therapies for premature stop codon diseases. Trends Mol. Med. 2012, 18, 679–688. [Google Scholar] [CrossRef]
- Nudelman, I.; Rebibo-Sabbah, A.; Cherniavsky, M.; Belakhov, V.; Hainrichson, M.; Chen, F.; Schacht, J.; Pilch, D.S.; Ben-Yosef, T.; Baasov, T. Development of novel aminoglycoside (NB54) with reduced toxicity and enhanced suppression of disease-causing premature stop mutations. J. Med. Chem. 2009, 52, 2836–2845. [Google Scholar] [CrossRef]
- Nudelman, I.; Rebibo-Sabbah, A.; Shallom-Shezifi, D.; Hainrichson, M.; Stahl, I.; Ben-Yosef, T.; Baasov, T. Redesign of aminoglycosides for treatment of human genetic diseases caused by premature stop mutations. Bioorg. Med. Chem. Lett. 2006, 16, 6310–6315. [Google Scholar] [CrossRef]
- Goldmann, T.; Rebibo-Sabbah, A.; Overlack, N.; Nudelman, I.; Belakhov, V.; Baasov, T.; Ben-Yosef, T.; Wolfrum, U.; Nagel-Wolfrum, K. Beneficial read-through of a USH1C nonsense mutation by designed aminoglycoside NB30 in the retina. Physiol. Pharmacol. 2010, 51, 6671–6680. [Google Scholar]
- Goldmann, T.; Overlack, N.; Möller, F.; Belakhov, V.; Wyk, M.V.; Baasov, T.; Wolfrum, U.; Nagel-Wolfrum, K. A comparative evaluation of NB30, NB54 and PTC124 in translational read-through efficacy for treatment of an USH1C nonsense mutation. EMBO Mol. Med. 2012, 4, 1186–1199. [Google Scholar] [CrossRef]
- Mattis, V.B.; Ebert, A.D.; Fosso, M.Y.; Chang, C.W.; Lorson, C.L. Delivery of a read-through inducing compound, TC007, lessens the severity of a spinal muscular atrophy animal model. Hum. Mol. Genet. 2009, 18, 3906–3913. [Google Scholar] [CrossRef]
- Tambuyzer, E.; Vandendriessche, B.; Austin, C.P.; Brooks, P.J.; Larsson, K.; Miller Needleman, K.I.; Valentine, J.; Davies, K.; Groft, S.C.; Preti, R.; et al. Therapies for Rare Diseases: Therapeutic Modalities, Progress and Challenges Ahead. Nat. Rev. Drug Discov. 2020, 19, 93–111. [Google Scholar] [CrossRef]
- Schilff, M.; Sargsyan, Y.; Hofhuis, J.; Thoms, S. Stop Codon Context-Specific Induction of Translational Readthrough. Biomolecules 2021, 7, 1006. [Google Scholar] [CrossRef]
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A Meta-Analysis of Nonsense Mutations Causing Human Genetic Disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar] [CrossRef]
- Omachi, K.; Kai, H.; Roberge, M.; Miner, J.H. NanoLuc reporters identify COL4A5 nonsense mutations susceptible to drug-induced stop codon readthrough. iScience 2022, 3, 103891. [Google Scholar] [CrossRef]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.; Hwang, S.; et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef]
- Hirawat, S.; Welch, E.M.; Elfring, G.L.; Northcutt, V.; Paushkin, S.; Hwang, S.; Leonard, E.; Almstead, N.; Ju, W.; Peltz, S.; et al. Safety, tolerability, and pharmacokinetics of PTC124, a nonaminoglycoside nonsense mutation suppressor, following single- and multiple-dose administration to healthy male and female adult volunteers. J. Clin. Pharmacol. 2007, 47, 430–444. [Google Scholar] [CrossRef]
- Allamand, V.; Pascale, G. Drug-induced readthrough of premature stop codons leads to the stabilization of laminin alpha2 chain mRNA in CMD myotubes. J. Gene Med. 2008, 10, 217–224. [Google Scholar] [CrossRef]
- Roy, B.; Friesen, W.J.; Tomizawa, Y.; Jacobson, A. Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proc. Natl. Acad. Sci. USA 2016, 113, 12508–12513. [Google Scholar] [CrossRef]
- Tutone, M.; Pibiri, I.; Lentini, L.; Pace, A. Deciphering the nonsense readthrough mechanism of action of Ataluren: An insilico compared study. ACS Med. Chem. Lett. 2019, 10, 522–527. [Google Scholar] [CrossRef]
- Ng, M.Y.; Li, H.; Ghelfi, M.D.; Goldman, Y.E.; Cooperman, B.S. Ataluren and aminoglycosides stimulate read-through of nonsense codons by orthogonal mechanisms. Proc. Natl. Acad. Sci. USA 2021, 118, e2020599118. [Google Scholar] [CrossRef]
- Bolze, F.; Mocek, S.; Zimmermann, A.; Klingenspor, M. Aminoglycosides, but not PTC124 (Ataluren), rescue nonsense mutations in the leptin receptor and in luciferase reporter genes. Sci. Rep. 2018, 7, 1020. [Google Scholar] [CrossRef]
- Torriano, S.; Erkilic, N.; Baux, D.; Cereso, N.; De Luca, V.; Meunier, I.; Moosajee, M.; Roux, A.F.; Hamel, C.P.; Kalatzis, V. The effect of PTC124 on choroideremia fibroblasts and iPSC-derived RPE raises considerations for therapy. Sci. Rep. 2018, 8, 8234. [Google Scholar] [CrossRef]
- Krall, M.; Htun, S.; Slavotinek, A. Use of PTC124 for nonsense suppression therapy targeting BMP4 nonsense variants in vitro and the bmp4st72 allele in zebrafish. PLoS ONE 2019, 14, e0212121. [Google Scholar] [CrossRef]
- Harmer, S.C.; Mohal, J.S.; Kemp, D.; Tinker, A. Readthrough of long-QT syndrome type 1 nonsense mutations rescues function but alters the biophysical properties of the channel. Biochem. J. 2012, 443, 635–642. [Google Scholar] [CrossRef]
- Kosmidis, G.; Veerman, C.C.; Casini, S.; Verkerk, A.O.; Pas, S.V.D.; Bellin, M.; Wilde, A.A.M.; Mummery, C.L.; Bezzina, C.R. SCN5A, Readthrough-promoting drugs gentamicin and PTC124 fail to rescue Nav1.5 function of human-induced pluripotent stem cell-derived cardiomyocytes carrying nonsense mutations in the sodium channel gene. Circ. Arrhythmia Electrophysiol. 2016, 9, e004227. [Google Scholar] [CrossRef]
- Hamada, M.; Takeuchi, T.; Kondo, S.; Ikeda, Y.; Naganawa, H.; Maeda, K.; Okami, Y.; Umezawa, H. A new antibiotic, negamycin. J. Antibiot. 1970, 23, 170–171. [Google Scholar] [CrossRef]
- Polikanov, Y.S.; Szal, T.; Jiang, F.; Gupta, P.; Matsuda, R.; Shiozuka, M.; Steitz, T.A.; Vázquez-Laslop, N.; Mankin, A.S. Negamycin interferes with decoding and translocation by simultaneous interaction with rRNA and tRNA. Mol. Cell. 2016, 56, 541–550. [Google Scholar] [CrossRef]
- Schroeder, S.J.; Blaha, G.; Moore, P.B. Negamycin binds to the wall of the nascent chain exit tunnel of the 50S ribosomal subunit. Antimicrob. Agents Chemother. 2007, 51, 4462–4465. [Google Scholar] [CrossRef]
- Taguchi, A.; Nishiguchi, S.; Shiozuka, M. Negamycin analogue with readthrough-promoting activity as a potential drug candidate for Duchenne muscular dystrophy. ACS Med. Chem. Lett. 2012, 3, 2736–2740. [Google Scholar] [CrossRef]
- Hamad, K.; Omura, N.; Taguchi, A.; Baradaran-Heravi, A.; Kotake, M.; Arai, M.; Takayama, K.; Taniguchi, A.; Roberge, M.; Hayashi, Y. New negamycin-based potent readthrough derivative effective against TGA-type nonsense mutations. ACS Med. Chem. Lett. 2019, 10, 1450–1456. [Google Scholar] [CrossRef]
- Baltz, R.H.; Seno, E.T. Genetics of Streptomyces fradiae and tylosin biosynthesis. Annu. Rev. Microbiol. 1988, 42, 547–574. [Google Scholar] [CrossRef]
- Garza-Ramos, G.; Xiong, L.; Zhong, P.; Mankin, A. Binding site of macrolide antibiotics on the ribosome: New resistance mutation identifies a specific interaction of ketolides with rRNA. J. Bacteriol. 2001, 183, 6898–6907. [Google Scholar] [CrossRef]
- Du, L.; Jung, M.E.; Damoiseaux, R.; Completo, G.; Fike, F.; Ku, J.M.; Nahas, S.; Piao, C.; Hu, H.; Gatti, R.A. A new series of small molecular weight compounds induce read through of all three types of nonsense mutations in the ATM gene. Mol. Ther. 2013, 21, 1653–1660. [Google Scholar] [CrossRef]
- Kayali, R.; Ku, J.M.; Bertoni, C. Read-through compound 13 restores dystrophin expression and improves muscle function in the mdx mouse model for Duchenne muscular dystrophy. Hum. Mol. Genet. 2012, 21, 4007–4020. [Google Scholar] [CrossRef]
- Tutone, M.; Pibiri, I.; Perriera, R.; Campofelice, A.; Culletta, G.; Melfi, R.; Pace, A.; Almerico, A.M.; Lentini, L. Pharmacophore-based design of new chemical scaffolds as translational readthrough-inducing drugs (TRIDs). ACS Med. Chem. Lett. 2020, 11, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Moosajee, M.; Tracey-White, D.; Smart, M.; Weetall, M.; Torriano, S.; Kalatzis, V.; da Cruz, L.; Coffey, P.; Webster, A.R.; Welch, E. Functional rescue of REP1 following treatment with PTC124 and novel derivative PTC-414 in human choroideremia fibroblasts and the nonsense-mediated zebrafish model. Hum. Mol. Genet. 2016, 25, 3416–3431. [Google Scholar] [CrossRef] [PubMed]
- Friesen, W.J.; Trotta, C.R.; Tomizawa, Y.; Zhuo, J.; Welch, E.M. The nucleoside analog clitocine is a potent and efficacious readthrough agent. RNA 2017, 23, 567–577. [Google Scholar] [CrossRef]
- Trzaska, C.; Amand, S.; Bailly, C.; Leroy, C.; Marchand, V. 2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations. Nat. Commun. 2020, 11, 1509. [Google Scholar] [CrossRef]
- Gonzalez-Hilarion, S.; Beghyn, T.; Jia, J.; Debreuck, N.; Berte, G.; Mamchaoui, K.; Mouly, V.; Gruenert, D.C.; Déprez, B.; Lejeune, F. Rescue of nonsense mutations by amlexanox in human cells. Orphanet J. Rare Dis. 2012, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, V.S.; Jiang, Q.; Prisco, M.; Gruber, C.; South, A.P. Amlexanox enhances premature termination codon read-through in COL7A1 and expression of full length type VII collagen: Potential therapy for recessive dystrophic epidermolysis bullosa. J. Investig. Dermatol. 2017, 137, 1842–1849. [Google Scholar] [CrossRef]
- Dabrowski, M.; Bukowy-Bieryllo, Z.; Jackson, C.L.; Zietkiewicz, E. Properties of Non-Aminoglycoside Compounds Used to Stimulate Translational Readthrough of PTC Mutations in Primary Ciliary Dyskinesia. Int. J. Mol. Sci. 2021, 22, 4923. [Google Scholar] [CrossRef]
- Linde, L.; Boelz, S.; Nissim-Rafinia, M.; Oren, Y.; Wilschanski, M.; Yaacov, Y.; Virgilis, D.; Neu-Yilik, G.; Kulozik, A.; Kerem, E.; et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Investig. 2007, 117, 683–692. [Google Scholar] [CrossRef]
- Xi, B.; Guan, F.; Lawrence, D.S. Enhanced production of functional proteins from defective genes. J. Am. Chem. Soc. 2004, 126, 5660–5661. [Google Scholar] [CrossRef]
- Barton-Davis, E.R.; Cordier, L.; Shoturma, D.I.; Leland, S.E.; Sweeney, H.L. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J. Clin. Investig. 1999, 104, 375–381. [Google Scholar] [CrossRef]
- Kishore, B.K.; Ibrahim, S.; Lambricht, P.; Laurent, G.; Maldague, P.; Tulkens, P.M. Comparative assessment of poly-L-aspartic and poly-L-glutamic acids as protectants against gentamicin-induced renal lysosomal phospholipidosis, phospholipiduria and cell proliferation in rats. J. Pharmacol. Exp. Ther. 1992, 262, 424–432. [Google Scholar]
- Buck, N.E.; Wood, L.; Hu, R.; Peters, H.L. Stop codon read-through of a methylmalonic aciduria mutation. Mol. Genet. Metab. 2009, 97, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Abi-Hachem, R.N.; Zine, A.; Van De Water, T.R. The injured cochlea as a target for inflammatory processes, initiation of cell death pathways and application of related otoprotectives strategies. Recent Pat. CNS Drug Discov. 2010, 5, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Moestrup, S.K.; Cui, S.; Vorum, H.; Bregengård, C.; Bjørn, S.E.; Norris, K.; Gliemann, J.; Christensen, E.I. Evidence that epithelial glycoprotein 330/megalin mediates uptake of polybasic drugs. J. Clin. Investig. 1995, 96, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Reasor, M.J.; Kacew, S. Drug-Induced Phospholipidosis: Are There Functional Consequences? Exp. Biol. Med. 2001, 226, 825–830. [Google Scholar] [CrossRef]
- Xie, J.; Talaska, A.E.; Schacht, J. New developments in aminoglycoside therapy and ototoxicity. Hear Res. 2011, 281, 28–37. [Google Scholar] [CrossRef]
- Shulman, E.; Belakhov, V.; Wei, G.; Kendall, A.; Meyron-Holtz, E.G.; Ben-Shachar, D.; Schacht, J.; Baasov, T. Designer aminoglycosides that selectively inhibit cytoplasmic rather than mitochondrial ribosomes show decreased ototoxicity: A strategy for the treatment of genetic diseases. J. Biol. Chem. 2014, 289, 2318–2330. [Google Scholar] [CrossRef]
- Myrdal, S.E.; Johnson, K.C.; Steyger, P.S. Cytoplasmic and intra-nuclear binding of gentamicin does not require endocytosis. Hear Res. 2005, 204, 156–169. [Google Scholar] [CrossRef]
- Miller, J.N.; Pearce, D.A. Nonsense-mediated decay in genetic disease: Friend or foe? Mutat. Res. Rev. Mutat. Res. 2014, 762, 52–64. [Google Scholar] [CrossRef]
- Kuzmiak, H.A.; Maquat, L.E. Applying nonsense-mediated mRNA decay research to the clinic: Progress and challenges. Trends Mol. Med. 2006, 12, 306–316. [Google Scholar] [CrossRef]
- Hwang, H.J.; Park, Y.; Kim, Y.K. UPF1: From mRNA Surveillance to Protein Quality Control. Biomedicines 2021, 9, 995. [Google Scholar] [CrossRef]
- Dem Bokhari, A.; Jonchere, V.; Lagrange, A.; Bertrand, R.; Svrcek, M.; Marisa, L.; Collura, A. Oncogenesis Targeting nonsense-mediated mRNA decay in colorectal cancers with microsatellite instability. Oncogenesis 2018, 7, 70. [Google Scholar] [CrossRef]
- Duval, A.; Hamelin, R. Mutations at Coding Repeat Sequences in Mismatch Repair-deficient Human Cancers: Toward a New Concept of Target Genes for Instability. Cancer Res. 2002, 62, 2447–2454. [Google Scholar]
- Durand, S.; Cougot, N.; Mahuteau-Betzer, F.; Nguyen, C.H.; Grierson, D.S.; Bertrand, E.; Tazi, J.; Lejeune, F. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J. Cell Biol. 2007, 178, 1145–1160. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Low, W.K.; Xu, J.; Gehring, N.H.; Dietz, H.C.; Romo, D.; Liu, J.O. Inhibition of nonsense-mediated mRNA decay by the natural product pateamine A through eukaryotic initiation factor 4AIII. J. Biol. Chem. 2009, 284, 23613–23621. [Google Scholar] [CrossRef]
- Martin, L.; Grigoryan, A.; Wang, D.; Wang, J.; Breda, L.; Rivella, S.; Cardozo, T.; Gardner, L.B. Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 2014, 74, 3104–3113. [Google Scholar] [CrossRef]
- Feng, D.; Su, R.C.; Zou, L.; Triggs-Raine, B.; Huang, S.; Xie, J. Increase of a group of PTC (+) transcripts by curcumin through inhibition of the NMD pathway. Biochim. Biophys. Acta 2015, 1849, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Gopalsamy, A.; Bennett, E.M.; Shi, M.; Zhang, W.G.; Bard, J.; Yu, K. Identification of pyrimidine derivatives as hSMG-1 inhibitors. Bioorg. Med. Chem. Lett. 2012, 22, 6636–6641. [Google Scholar] [CrossRef] [PubMed]
- Usuki, F.; Yamashita, A.; Higuchi, I.; Ohnishi, T.; Shiraishi, T.; Osame, M.; Ohno, S. Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrichs disease. Ann. Neurol. 2004, 55, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Gudikote, J.P.; Cascone, T.; Poteete, A.; Sitthideatphaiboon, P.; Wu, Q.; Morikawa, N.; Zhang, F.; Peng, S.; Tong, P.; Li, L.; et al. Inhibition of nonsense-mediated decay rescues p53β/γ isoform expression and activates the p53 pathway in MDM2-overexpressing and select p53-mutant cancers. J. Biol. Chem. 2021, 297, 101163. [Google Scholar] [CrossRef]
- Meraviglia-Crivelli, D.; Villanueva, H.; Menon, A.P.; Zheleva, A.; Moreno, B.; Villalba-Esparza, M.; Pastor, F. A pan-tumor-siRNA aptamer chimera to block nonsense-mediated mRNA decay inflames and suppresses tumor progression. Mol. Ther. Nucleic Acids 2022, 29, 413–425. [Google Scholar] [CrossRef]
- Nickless, A.; Jackson, E.; Marasa, J.; Nugent, P.; Mercer, R.W.; Piwnica-Worms, D.; You, Z. Intracellular calcium regulates nonsense-mediated mRNA decay. Nat. Med. 2014, 20, 961–966. [Google Scholar] [CrossRef]
- Bhuvanagiri, M.; Lewis, J.; Putzker, K.; Becker, J.P.; Leicht, S.; Krijgsveld, J.; Batra, R.; Turnwald, B.; Jovanovic, B.; Hauer, C. 5-azacytidine inhibits nonsense-mediated decay in a MYC-dependent fashion. EMBO Mol. Med. 2014, 6, 1593–1609. [Google Scholar] [CrossRef]
- Wang, D.; Wengrod, J.; Gardner, L.B. Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J. Biol. Chem. 2011, 286, 40038–40043. [Google Scholar] [CrossRef] [PubMed]
- Baradaran-Heravi, A.; Balgi, A.D.; Hosseini-Farahabadi, S.; Choi, K.; Has, C.; Roberge, M. Effect of small molecule eRF3 degraders on premature termination codon readthrough. Nucleic Acids Res. 2021, 49, 3692–3708. [Google Scholar] [CrossRef]
- Baradaran-Heravi, A.; Bauer, C.C.; Pickles, I.B.; Hosseini-Farahabadi, S.; Balgi, A.D.; Choi, K.; Linley, D.M.; Beech, D.J.; Roberge, M.; Bon, R.S. Nonselective TRPC channel inhibition and suppression of aminoglycoside-induced premature termination codon readthrough by the small molecule AC1903. J. Biol. Chem. 2022, 298, 101546. [Google Scholar] [CrossRef]
- Huang, L.; Low, A.; Damle, S.S.; Keenan, M.M.; Kuntz, S.; Murray, S.F.; Monia, B.P.; Guo, S. Antisense suppression of the nonsense mediated decay factor Upf3b as a potential treatment for diseases caused by nonsense mutations. Genome Biol. 2018, 19, 4. [Google Scholar] [CrossRef] [PubMed]
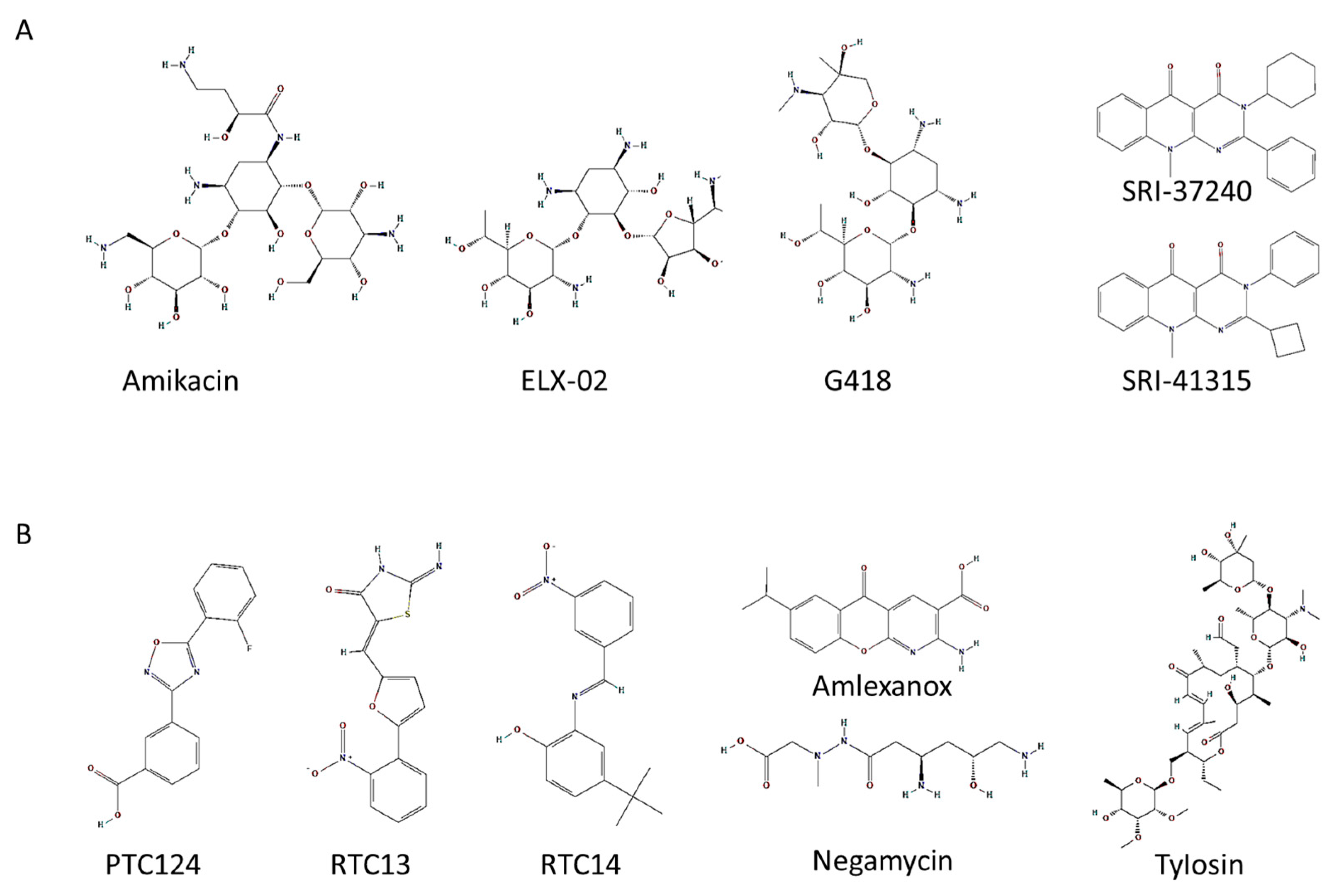
| Drugs | Gene | References |
|---|---|---|
| G418 | CFTR | [85] |
| Gentamicin | CFTR | [85] |
| ELX-02 | CFTR | [69,84] |
| NB-124 | CFTR | [87,89] |
| poly-L-aspartic acid with gentamicin | CFTR | [100] |
| SRI-37240 and its potent derivate SRI-41315 | CFTR | [101] |
| Elexacaftor | CFTR | [95] |
| Kanamycin, paromomycin and neomycin | xeroderma pigmentosum C (XPC) | [96] |
| Paromomycin | APC-mediated colon cancer | [97] |
| Amikacin | p53 gene | [98,99] |
| Drugs | Diseases/Gene | References |
|---|---|---|
| PTC124 | QT syndrome type 1 | [124] |
| Tylosin | APC gene | [97,101,132] |
| Negamycin | DMD patients | [129] |
| readthrough compound RTC13 and RTC14 | ATM gene | [93,133,134] |
| Clitocin | p53 protein | [137] |
| 2,6-diaminopurine (DAP) | TP53 gene in cancer cells | [138] |
| Amlexanox | COL7A1 gene in patients with recessive dystrophic epidermolysis bullosa (RDEB) | [140] |
| Drug | Gene Inhibition | References |
|---|---|---|
| NMD-1 | Stabilization of UPF1 protein during hyperphosphorylation | [156,157,158] |
| Pateamine A (Pat A) | Inhibited NMD pathway | [158,159] |
| Curcumin | Inhibited NMD pathway | [160,161] |
| Pyrimidine derivate | Inhibited SMG1 | [162] |
| Wortmanin and caffeine | Blocked NMD pathway | [161,163] |
| Digoxin and ouabain | Repressed NMD pathway | [163,166] |
| AC1903 | Inhibited multiple TRPC channels in renal cancer | [170] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Temaj, G.; Telkoparan-Akillilar, P.; Nuhii, N.; Chichiarelli, S.; Saha, S.; Saso, L. Recoding of Nonsense Mutation as a Pharmacological Strategy. Biomedicines 2023, 11, 659. https://doi.org/10.3390/biomedicines11030659
Temaj G, Telkoparan-Akillilar P, Nuhii N, Chichiarelli S, Saha S, Saso L. Recoding of Nonsense Mutation as a Pharmacological Strategy. Biomedicines. 2023; 11(3):659. https://doi.org/10.3390/biomedicines11030659
Chicago/Turabian StyleTemaj, Gazmend, Pelin Telkoparan-Akillilar, Nexhibe Nuhii, Silvia Chichiarelli, Sarmistha Saha, and Luciano Saso. 2023. "Recoding of Nonsense Mutation as a Pharmacological Strategy" Biomedicines 11, no. 3: 659. https://doi.org/10.3390/biomedicines11030659
APA StyleTemaj, G., Telkoparan-Akillilar, P., Nuhii, N., Chichiarelli, S., Saha, S., & Saso, L. (2023). Recoding of Nonsense Mutation as a Pharmacological Strategy. Biomedicines, 11(3), 659. https://doi.org/10.3390/biomedicines11030659









