Peptides That Block RAS-p21 Protein-Induced Cell Transformation
Abstract
1. Background
1.1. The Ras-Gene-Encoded p21 Protein
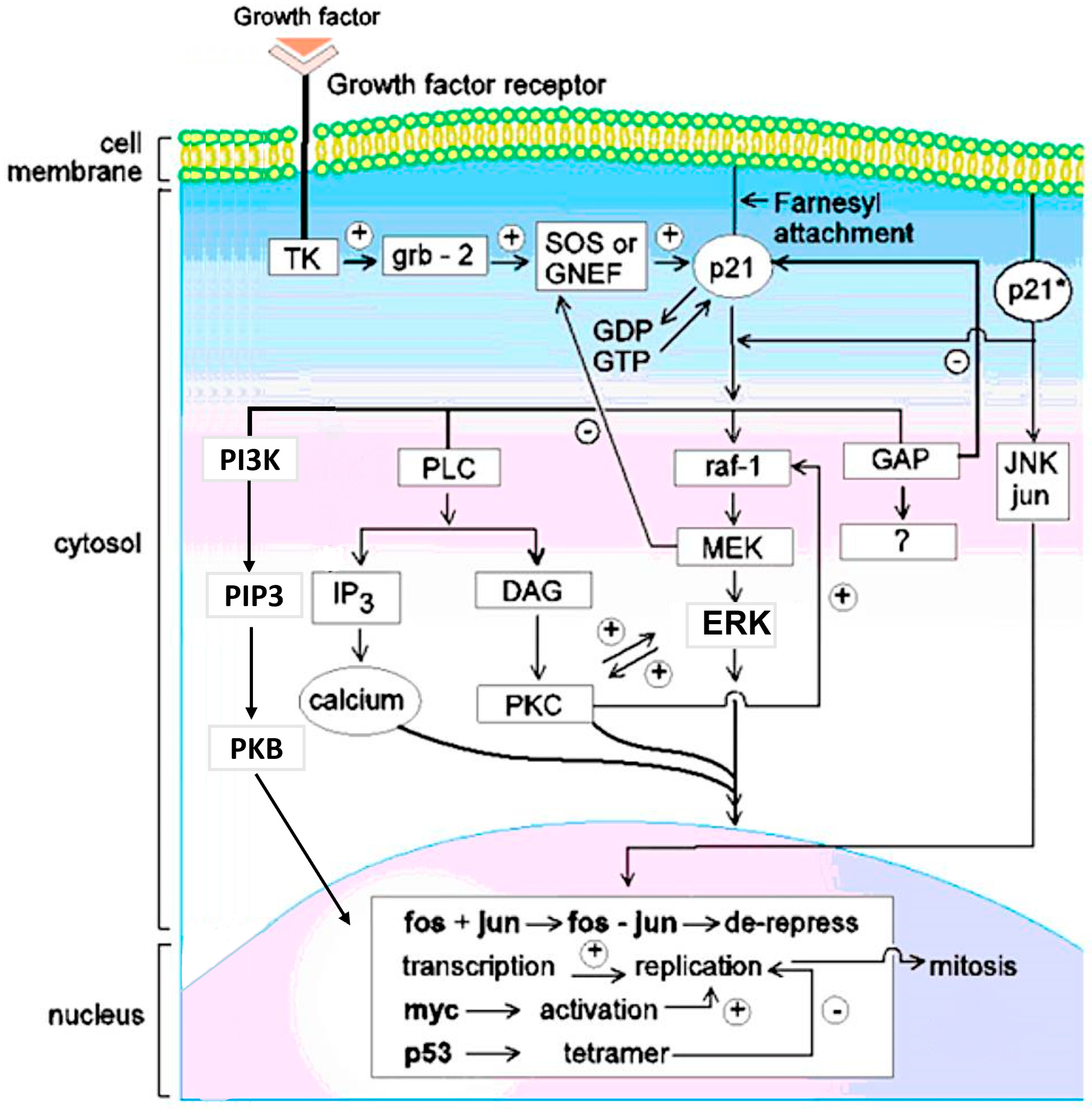
1.2. Mitogenic Signal Transduction Pathways Induced by RAS-p21
1.3. Structure of RAS-p21
1.4. Effects of GTP Binding and Oncogenic Amino Acid Substitutions on the Structure and Function of RAS-p21
1.5. Peptides and Small Molecules Are Promising Agents That Can Selectively Destroy Oncogenic RAS-p21-Induced Cancers
2. Anti-Oncogenic RAS-p21 Peptides from RAS-p21 Itself
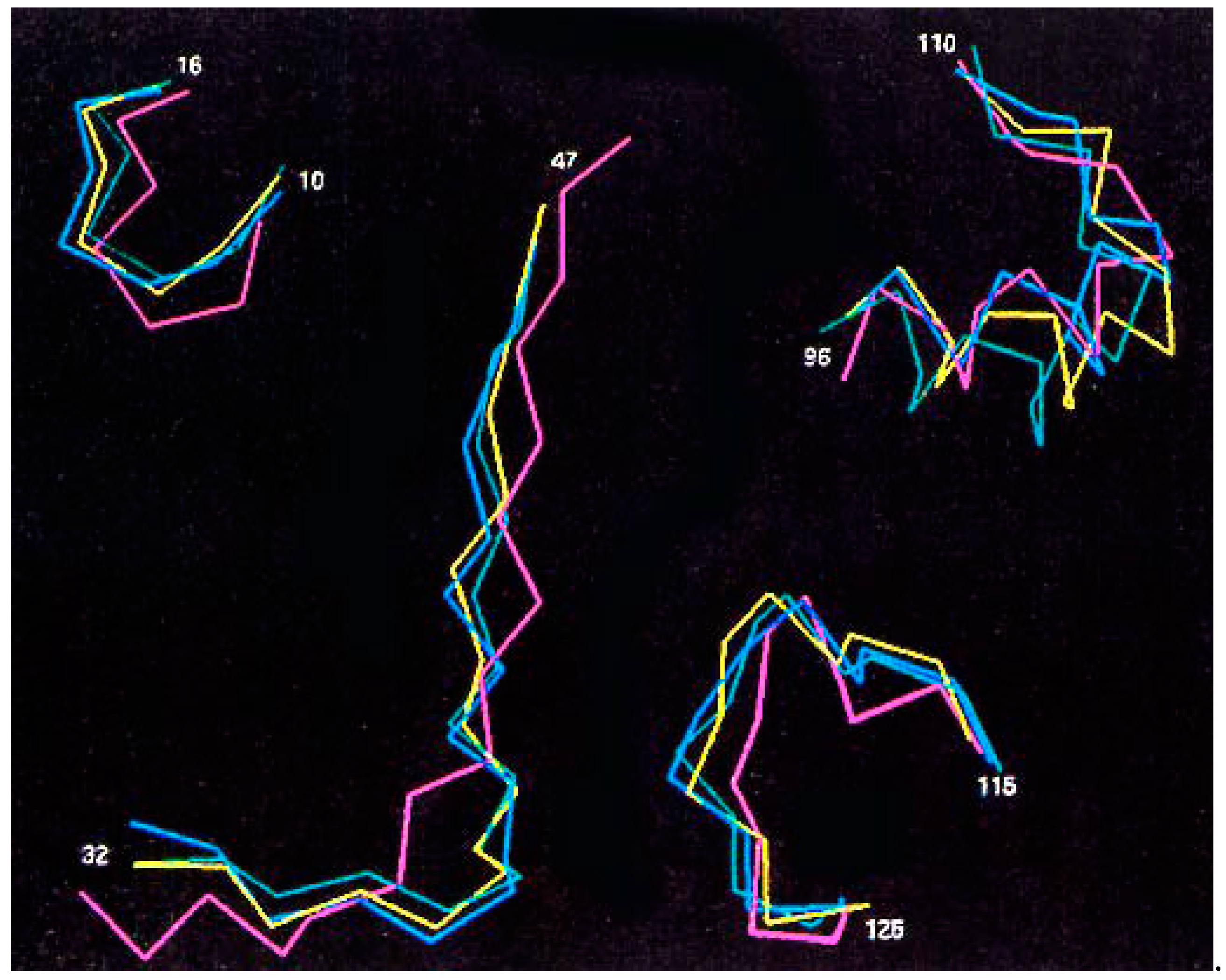
2.1. Three RAS-p21 Peptides Selectively Block Oncogenic RAS-p21
2.1.1. Site of Action of PNC-7 (RAS-p21 Residues 35–47)
Alternate Targets of RAF
2.1.2. Site of Action of PNC-1 (GNKCDLAARTVE) and PNC-2 (YREQIKRVKDSDDVP)
2.2. Effects of PNC-2 and PNC-7 in Cancer and Normal Cells
2.3. Peptides from Proteins That Interact Directly with RAS-p21 or Selectively Block Targets on the Oncogenic RAS-p21 Pathway
2.4. Use of Overlapping Peptides from RAS-p21 to Target Ras-RAF Interactions
3. Anti-RAS-p21 Peptides from Expression Libraries
3.1. Cyclic Peptides Freeze Oncogenic G12D-RAS-p21 in an Inactive GTP-Bound State
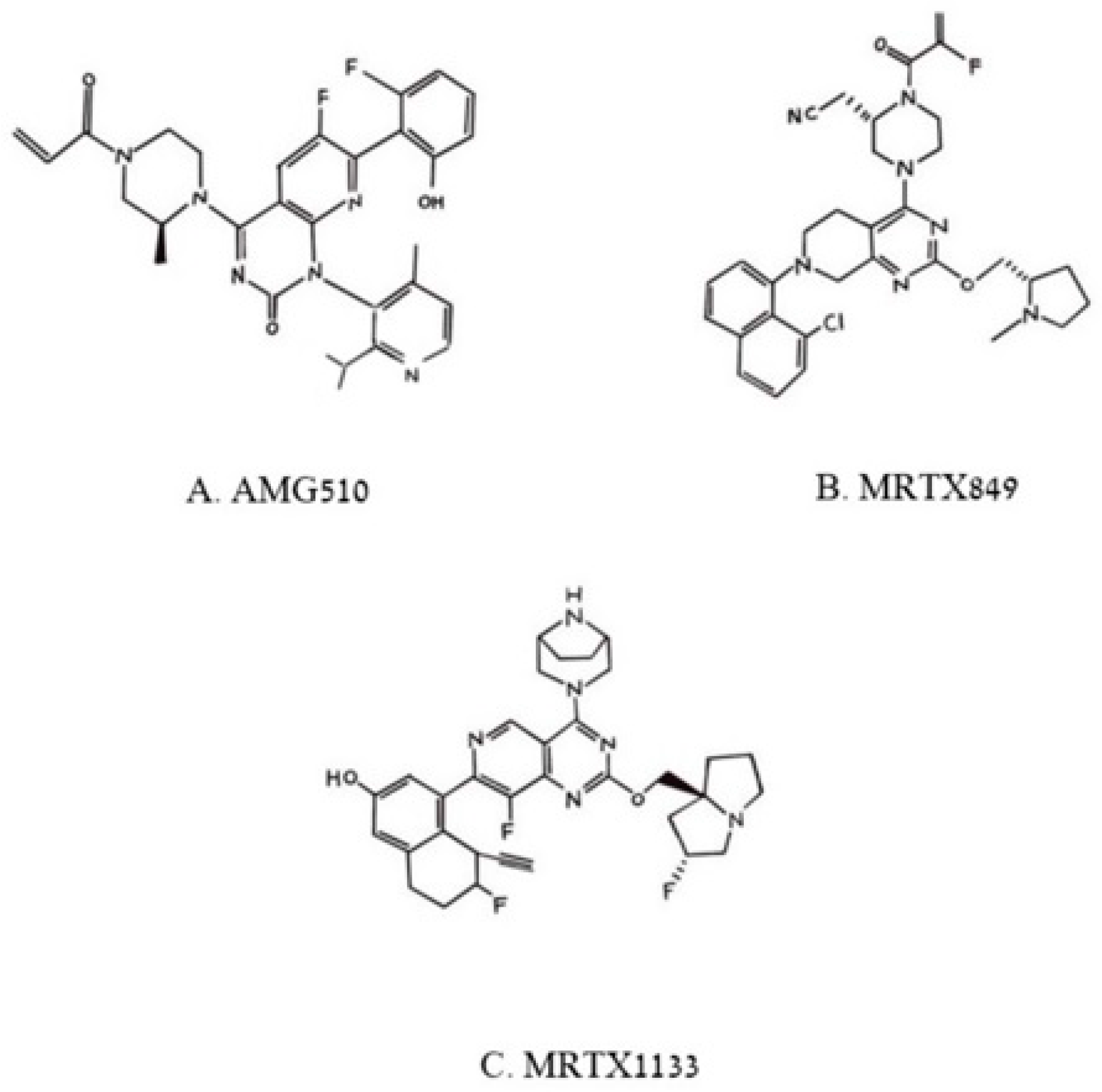
3.2. Small Molecules Freeze Oncogenic RAS-p21 in an Inactive GDP Bound State
3.3. Peptides and Small Molecules from Libraries That Bind to Multiple Forms of RAS-p21-Gpp-NHp
4. Use of Cancer Cell-Penetrating Peptides Linked to an Anti-Ras Antibody to Inactivate Oncogenic RAS-p21
5. Assessment of Peptide Therapy for Ras-Induced Tumors
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pincus, M.R.; Bluth, M.; Brandt-Rauf, P.W.; Bowne, W.; LaDoulis, C. Oncoproteins and Early Tumor Detection, Chapter 77. In Henry’s Clinical Diagnosis and Management by Laboratory Methods, 24th ed.; McPherson, R.A., Pincus, M.R., Eds.; Elsevier: New York, NY, USA; Philadelphia, PA, USA, 2021; pp. 1525–1541. [Google Scholar]
- Barbacid, M. ras Genes. Ann. Rev. Biochem. 1987, 56, 779–827. [Google Scholar] [CrossRef] [PubMed]
- Stacey, D.W.; Kung, H.-F. Transformation of NIH 3T3 Cells by Microinjection of Ha-ras p21 Protein. Nature 1984, 310, 508–511. [Google Scholar] [CrossRef]
- Birchmeier, C.; Broeck, D.; Wigler, M. RAS Proteins Can Induce Meiosis in Xenopus Oocytes. Cell 1985, 43, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.K.; Kung, H.-F. Insulin Induction of Xenopus laevis Oocyte Maturation Is Inhibited by Monoclonal Antibody Against p21 ras Proteins. Mol. Cell Biol. 1987, 7, 1285–1288. [Google Scholar]
- Lowenstein, E.J.; Daly, R.J.; Batzer, A.G.; Li, W.; Margolis, B.; Lammers, R.; Ullrich, A.; Skolnik, E.Y.; Bar-Sagi, D.; Schlessinger, J. The SH2 and SH3 Domain-Containing Protein GRB2 Links Receptor Tyrosine Kinases to ras Signaling. Cell 1992, 70, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Boriak-Sjodin, P.A.; Margarit, S.M.; Bar-Sagi, D.; Kuriyan, J. The Structural Basis of the Activation of ras by SOS. Nature 1998, 394, 337–343. [Google Scholar] [CrossRef]
- Scheffzek, K.; Ahmadian, M.R.; Kabsch, W.; Weismuller, L.; Lautwein-Schmitz, F.; Wittinghofer, A. The Ras-RasGAP Complex: Structural Basis for GTPase Activation and Its Loss in Oncogenic ras Mutants. Science 1997, 227, 333–338. [Google Scholar] [CrossRef]
- Smith, M.R.; A-Lun, Y.; Kim, L.; Rhee, S.G.; Kung, H.-F. Inhibition of Serum- and Ras-Stimulated DNA Synthesis by Antibodies to Phospholipase C. Science 1990, 247, 1074–1077. [Google Scholar] [CrossRef]
- Chung, D.L.; Brandt-Rauf, P.W.; Weinstein, I.B.; Nishimura, S.; Yamaizumi, Z.; Murphy, R.B.; Pincus, M.R. Evidence That the ras Oncogene-Encoded p21 Protein Induces Oocyte Maturation via Activation of Protein Kinase C. Proc. Natl. Acad. Sci. USA 1992, 89, 1993–1996. [Google Scholar] [CrossRef]
- Adler, V.; Pincus, M.R.; Brandt-Rauf, P.W.; Ronai, Z. Complexes of RAS-p21 with jun-N-Terminal Kinase and c-jun Proteins. Proc. Natl. Acad. Sci. USA 1995, 92, 10585–10589. [Google Scholar] [CrossRef]
- Pai, E.F.; Krengel, U.; Petsko, G.A.; Goody, R.S.; Kabsch, W.; Wittinghofer, A. Refined Crystal Structure of the Triphosphate Conformation of H-ras p21 at 1.35 A Resolution: Implications for the Mechanism of GTP Hydrolysis. EMBO J. 1990, 9, 2351–2359. [Google Scholar] [CrossRef]
- Krengel, U.; Schlichting, I.; Scherer, A.; Schumann, R.; Frech, M.; John, J.; Kabsch, W.; Pai, E.F.; Wittinghofer, A. Three-Dimensional Structures of H-RAS-p21 Mutants:Molecular Basis for Their Inability to Function As Signal Switch Molecules. Cell 1990, 62, 539–548. [Google Scholar] [CrossRef]
- Clanton, D.J.; Lu, Y.; Blair, D.G.; Shih, T.Y. Structural Significance of the GTP-binding Domain of ras p21 Studied by Site-directed Mutagenesis. Mol. Cell Biol. 1987, 7, 3092–3097. [Google Scholar]
- Pincus, M.R. Development of New Anti-Cancer Peptides from Conformational Energy Analysis of the Oncogenic RAS-p21 Protein and Its Complexes with Target Proteins. Front. Biosci. 2004, 9, 3486–3509. [Google Scholar] [CrossRef]
- Weiner, S.J.; Kollman, P.A.; Case, D.A.; Singh, V.C.; Ghio, C.; Alagona, G.; Profeta, S.; Weiner, P.K. A New Force Field for Molecular Simulations of Nucleic Acids and Proteins. J. Amer. Chem. Soc. 1986, 106, 765–784. [Google Scholar] [CrossRef]
- Dauber-Osguthorpe, P.; Roberts, V.A.; Osguthorpe, D.J.; Wolff, J.; Genest, M.; Hagler, A.T. Structure and Energetics of Ligand Binding to Proteins: E. coli Dihydrofolate Reductase Trimethoprim, a Drug-Receptor System. Proteins Struct. Funct. Genet. 1988, 4, 31–47. [Google Scholar] [CrossRef]
- Ripol, D.; Scheraga, H.A. On the multiple-minima problem in the conformational analysis of polypeptides. II. An Electrostatically-Driven Monte Carlo method. Tests on poly-(L-Alanine). Biopolymers 1988, 27, 1283–1303. [Google Scholar] [CrossRef]
- Arnautova, Y.A.; Jagielska, A.; Scheraga, H.A. A New Force Field (ECEPP-05) for Peptides, Proteins, and Organic Molecules. J. Phys. Chem B 2006, 110, 5025–5044. [Google Scholar] [CrossRef]
- Liwo, A.; Gibson, K.D.; Scheraga, H.A.; Brandt-Rauf, P.W.; Monaco, R.; Pincus, M.R. Comparison of the Low Energy Conformations of an Oncogenic and a Non-Oncogenic p21 Protein, Neither of Which Binds GDP or GTP. J. Protein Chem. 1994, 13, 237–251. [Google Scholar] [CrossRef]
- Ranginwale, M.; Smith, S.; Flom, J.; Chie, L.; Kanovsky, M.; Chung, D.; Friedman, F.K.; Robinson, R.C.; Brandt-Rauf, P.W.; Yamaizumi, Z.; et al. Differences in Patterns of Activation of MAP Kinases Induced by Oncogenic RAS-p21 and Insulin in Oocytes. Exp. Cell Res. 2001, 269, 162–169. [Google Scholar] [CrossRef]
- Qu, Y.; Adler, V.; Chu, T.; Platica, O.; Michl, J.; Pestka, S.; Izotova, L.; Boutjdir, M.; Pincus, M.R. Two Dual Specificity Kinases Are Preferentially Induced by Wild-Type Rather than by Oncogenic RAS-p21 in Xenopus Oocytes. Front. Biosci. 2006, 11, 2420–2427. [Google Scholar] [CrossRef]
- Adler, V.; Pincus, M.R.; Polatskaya, A.; Montano, X.; Friedman, F.K.; Ronai, Z. Activation of c-jun NH2 Kinase by UV Irradiation Is Dependent on p21ras. J. Biol. Chem. 1996, 271, 23304–23309. [Google Scholar] [CrossRef]
- Sarafraz-Yazdi, E.; Mumin, S.; Cheung, D.; Fridman, D.; Lin, B.; Wong, L.; Rosal, R.; Rudolph, R.; Frenkel, M.; Thadi, A.; et al. PNC-27, a Chimeric p53-Penetratin Peptide Binds to HDM-2 in a p53 Peptide-like Structure, Induces Selective Membrane-Pore Formation and Leads to Cancer Cell Lysis. Biomedicines 2022, 10, 945. [Google Scholar] [CrossRef]
- Adler, V.; Bowne, W.B.; Kamran, I.; Michl, J.; Friedman, F.K.; Chiu, E.; Zenilman, M.; Pincus, M.R. Two Peptides Derived from RAS-p21 Induce Either Phenotypic Reversion or Tumor Cell Necrosis of ras-Transformed Human Cancer Cells. Cancer Chemother. Pharmacol. 2007, 62, 491–498. [Google Scholar] [CrossRef]
- Kanovsky, M.; Michl, J.; Botzolaki, G.; Morin, J.; Kovac, C.; Chunh, D.; Friedman, F.K.; Pincus, M.R. Peptides, Designed from Molecular Modeling Studies of the RAS-p21 Protein, Induce Phenotypic Reversion of a Pancreatic Carcinoma Cell Line But Have No Effect on Normal Pancreatic Acinar Cell Growth. Cancer Chemother. Pharmacol. 2003, 52, 202–208. [Google Scholar] [CrossRef]
- Adler, V.; Yin, Z.; Fuchs, S.; Benezra, M.; Rosario, L.; Tew, K.; Pincus, M.R.; Sardana, M.; Henderson, C.; Wolf, C.R.; et al. GSTπ-a Regulator of JNK Signaling. EMBO J. 1999, 18, 1321–1334. [Google Scholar] [CrossRef]
- Tian, L.; Zhang, X.; Nemati, F.; Vallerand, D.; Decaudin, D.; Brossas, J.Y.; Zini, J.M.; Sylvain Choquet, S.; Scoazec, M.F.; Feillant, M.; et al. Identification of ras/raf Binding Site and Design of Interfering Peptide with Potential Clinical Application. Integr. Mol. Med. 2016, 3, 1–9. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, R.; Hu, Q.; Peacock, H.; Peacock, D.M.; Dai, S.; Shokat, K.M.; Suga, H. GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf. ACS Cent. Sci. 2020, 6, 1753–1761. [Google Scholar] [CrossRef]
- Spoerner, M.; Herrmann, C.; Vetter, I.R.; Kalbitzer, H.R.; Wittinghofer, A. Dynamic Properties of the Ras Switch I Region and Its Importance for Binding To Effectors. Proc. Natl. Acad. Sci. USA 2001, 98, 4944–4949. [Google Scholar] [CrossRef]
- Patricelli, M.P.; Janes, M.R.; Li, L.-S.; Hansen, R.; Peters, U.; Kessler, L.V.; Chen, Y.; Kucharski, J.M.; Feng, J.; Ely, T.; et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 2016, 6, 316–329. [Google Scholar] [CrossRef]
- Janes, M.R.; Zhang, J.; Li, L.-S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babber, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589. [Google Scholar] [CrossRef]
- Wang, X.; Allen, S.; Blake, J.F.; Bowcut, V.; Briere, D.M.; Calinisan, A.; Dahlke, J.R.; Fell, J.B.; Fischer, J.P.; Gunn, R.J.; et al. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRASG12D Inhibitor. J. Med. Chem. 2022, 65, 3123–3133. [Google Scholar] [CrossRef]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaidal, K.; Holt, T.; Knudson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) Inhibitor AMG 510 Drives Anti-Tumour Immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Clinicaltrials.gov A Phase 1,2 Study Evaluating the Safety, Tolerability, PK and Efficacy of AMG510 in Subjects with Solid Tumors with a Specific KRAS Mutation. Available online: https://clinicaltrials.gov/ct2/show/NCT03600883(2018) (accessed on 1 December 2022).
- Zhu, C.; Guan, X.; Zhang, X.; Luan, X.; Song, Z.; Cheng, X.; Zhang, W.; Qin, J.-J. Targeting KRAS Mutant Cancers: From Druggable Therapy To Drug Resistance. Mol. Cancer 2022, 21, 159–177. [Google Scholar] [CrossRef]
- Poorebrahim, M.; Abazari, M.F.; Moradi, L.; Shahbazi, B.; Mahmoudi, R.; Kalhor, H.; Askari, H.; Teimoori-Toolabi, L. Multi-Targeting of K-Ras Domains and Mutations by Peptide and Small Molecule Inhibitors. PLOS Comput. Biol. 2022, 18, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.J.; Sung, B.H.; Shin, J.R.; Lee, Y.W.; Kim, D.J.; Yang, K.S.; Kim, S.C. A Cancer Specific Cell-Penetrating Peptide, BR2, for the Efficient Delivery of an scFv into CancerCells. PLoS ONE 2013, 8, e66084. [Google Scholar] [CrossRef]
- Timar, J.; Kashofer, K. Molecular Epidemiology and Diagnostics of KRAS Mutations in Human Cancer. Cancer Metastasis Rev. 2020, 39, 1029–1038. [Google Scholar] [CrossRef] [PubMed]




| Source of Peptide or Small Molecule | Amino Acid Sequence or Small Molecule | Site of Action | Effect on Cancer Cells | Effect on Normal Cells |
|---|---|---|---|---|
| RAS-p21,35–47 (PNC-7) 1 | TIEDSYRKQVVID and TIEDSYRKQVVID-Leader 2 | G12V-H-RAS-p21 binding to RAF | Leader form induces phenotypic reversion of rat TUC-3 pancreatic cancer cells and HT1080 human fibrosarcoma cells; causes 100% cell death of MIA-PaCa-2 human pancreatic cancer cells. | Leader form has no effect on rat BMRPA1 pancreatic acinar cells and human keritinocytes. |
| RAS-p21,96–110 (PNC-2) 1 | YREQIKRVKDSDDVP and YREQIKRVKDSDDVP-Leader 2 | G12V-H-RAS-p21 binding to JNK, SOS | Leader form induces phenotypic reversion of rat TUC-3 pancreatic cancer cells and HT1080 human fibrosarcoma cells; causes 100% cell death of MIA-PaCa-2 human pancreatic cancer cells. | Leader form has no effect on rat BMRPA1 pancreatic acinar cells and human keritinocytes. |
| RAS-p21,115–126 (PNC-1) 1 | GNKCDLAARTVE and GNKCDLAARTVE-Leader 2 | G12V-H-RAS-p21 binding to JNK, SOS | Leader form induces phenotypic reversion of rat TUC-3 pancreatic cancer cells and HT1080 human fibrosarcoma cells; causes 100% cell death of MIA-PaCa-2 human pancreatic cancer cells. | Leader form has no effect on rat BMRPA1 pancreatic acinar cells and human keritinocytes. |
| RBD of RAF 97–110 1 | AVFRLLHEHKGKKA | G12V-H-RAS-p21 binding to RAF and MEK | NT 3 | NT |
| SOS 994–1004 1 | LNPMGNSMEKE | G12V-H-RAS-p21 binding to SOS | NT | NT |
| GST-π 34–50 1 | TIDTWMQGLLKPT CLYG | G12V-H-RAS-p21 binding to JNK/jun | NT | NT |
| GST-π 169–182 1 | CLDNFPLLSAYVAR | G12V-H-RAS-p21 binding to JNK/jun | NT | NT |
| CPP-RAS-p21 169–188 (Mut3DPTSh1) | VKK-KIKAEIKI-KMSKDGKKKKKKSRTRCTVM CPP (Cell-Penetrating Peptide) is the italicized sequence | K-RAS-p21 binding to RAF | Apoptosis in MDA-MB-231 and HBCx17 human breast cancer, SW626 human ovarian cancer and SW480 human colon cancer. Lower viability of H1650, HBEC human NSCCL. Minimal effect on HCT-116 and HT-29 human colon cancer cell | NT |
| High diversity cDNA Library | KD2 Cyclic Peptide | Binds to crevice in G12D-K-RAS-p21 bound to GppNHp blocking its activation | Peptide blocks binding specifically of G12D-K-RAS-p21 to RAF in vitro, but peptide does not enter cells. | Peptide blocks binding specifically of G12D-K-RAS-p21 to RAF in vitro, but peptide does not enter cells. |
| Polyaromatic molecule library | AMG510 | Binds to crevice in G12C-K-RAS-p21-GDP maintaining it in the inactive state | Induces cell death in CT26, MiaPaCa-2 human pancreatic cancer cells, NCI-H358 human NSCCL, CT26 murine colorectal ca in culture and in mice. FDA approved for treatment of human cancers with G12C-K-RAS-p21-induced tumors. Treatment of 4 patients with NSCCL treatment of four patients with NSCCL resulted in 34 and 67% remission in two of the patients and stable disease in the other two patients. | |
| Polyaromatic molecule library | MRTX849 | Binds to crevice in G12D-K-RAS-p21-GDP maintaining it in the inactive state | MRTX 849 Cell Lines: cytotoxic to human NSCCL, H2030, H358, 1573, 1792 cells. In vivo: Eradicates Colorectal-CR6243 Lung-Calu-1 (KRAS-G12C) Pancreas-MIA PaCa-2 (KRASG12C Lung-LU65 Lung-H1373. FDA approved for human cancers with G12C-K-RAS-p21-induced tumors. FDA approved for treatment of human cancers with G12C-K-RAS-p21-induced tumors. | |
| Cancer PPD Data Base, docking programs including PRODIGY, SwarmDock and FlexPepDock. 1st black sequence is Retro peptide, blue sequence is linker, and last black sequence is LfcinB peptide. Red R (Arg) residues were substituted to facilitate cell penetration. | LGRIVSAVKKIVRFLGGGGGS-FKRRRWQWRRKK | Docks to known K-RAS-p21 site exposed in wt-, G12V-,G12D-, G12C-,G13D-Q61H-K-RAS-p21. | Reduces viability (by about 6-%) of MiaPaCa-2 and AsPC-2 human pancreatic cancer cells. | NT |
| Construct peptide using BR2 as cancer cell CPP linked to the variable heavy chain domain of the anti-ras MAb, Y13 259. This is linked in turn to a linker sequence that is linked to the variable light chain that is attached to the hexaHis sequence. | BR2-VH-(GGGGS)3-VL-H6 BR2 = RAGLQFPVGRLLRRLLR. | Enters cancer cells selectively due to BR2; the scFv is directed against ras-21 and blocks its mitogenic signaling. | Induced 60 percent apoptosis of HCT-116 human colon cancer cells. | NT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pincus, M.R.; Lin, B.; Patel, P.; Gabutan, E.; Zohar, N.; Bowne, W.B. Peptides That Block RAS-p21 Protein-Induced Cell Transformation. Biomedicines 2023, 11, 471. https://doi.org/10.3390/biomedicines11020471
Pincus MR, Lin B, Patel P, Gabutan E, Zohar N, Bowne WB. Peptides That Block RAS-p21 Protein-Induced Cell Transformation. Biomedicines. 2023; 11(2):471. https://doi.org/10.3390/biomedicines11020471
Chicago/Turabian StylePincus, Matthew R., Bo Lin, Purvi Patel, Elmer Gabutan, Nitzan Zohar, and Wilbur B. Bowne. 2023. "Peptides That Block RAS-p21 Protein-Induced Cell Transformation" Biomedicines 11, no. 2: 471. https://doi.org/10.3390/biomedicines11020471
APA StylePincus, M. R., Lin, B., Patel, P., Gabutan, E., Zohar, N., & Bowne, W. B. (2023). Peptides That Block RAS-p21 Protein-Induced Cell Transformation. Biomedicines, 11(2), 471. https://doi.org/10.3390/biomedicines11020471






