Abstract
NAFLD is the most common cause of chronic liver disease worldwide. The miRNAs and lncRNAs are important endogenous ncRNAs families that can regulate molecular mechanisms. The aim of this study was to analyze the miRNA and lncRNA expression profiles in serum samples of NAFLD patients with different types of hepatosteatosis compared to healthy controls by the qPCR method. A total of180 NAFLD patients and 60 healthy controls were included. miRCURY LNA miRNA miRNome PCR human panel I + II kit and LncProfiler qPCR Array Kit were used to detect miRNA and lncRNA expression, respectively. DIANA miRPath and DIANA-lncBase web servers were used for interaction analysis. As a result, 75 miRNA and 24 lncRNA expression changes were determined. For miRNAs and lncRNAs, 30 and 5 were downregulated and 45 and 19 were upregulated, respectively. hsa-miR-21 was upregulated 2-fold whereas miR-197 was downregulated 0.25-fold. Among lncRNAs, NEAT1 was upregulated 2.9-fold while lncRNA MEG3 was downregulated 0.41-fold. A weak correlation was found between hsa-miR-122 and lncRNA MALAT1. As a conclusion, it is clear that lncRNA–miRNA interaction is involved in the molecular mechanisms of the emergence of NAFLD. The lncRNAs MEG3 and PTENP1 interacted with hsa-miR-21. It was thought that this interaction should be investigated as a biomarker for the development of NAFLD.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most prevalent form of chronic liver disease globally, and the leading cause of chronic liver damage. It is a significant public health issue that is frequently overlooked [1,2]. NAFLD is a health condition in which imaging methods identify a fatty liver. Still, secondary factors such as substance abuse and alcoholism are not identified [3]. NAFLD is also connected with type 2 diabetes, metabolic syndrome, and obesity. Non-alcoholic steatohepatitis (NASH), a form of NAFLD, can also proceed to liver cirrhosis and hepatocellular cancer, resulting in severe complications that may necessitate liver transplantation [4]. While microRNAs (miRNAs) are known as short (20–24 nucleotides in length) endogenous non-coding RNAs (ncRNAs), long non-coding RNAs (lncRNAs) are defined as a family of endogenous ncRNAs with a length greater than 200 nt [5]. In recent years, breakthroughs in molecular methods have led to a vast increase in understanding regarding the impact of ncRNAs on human cells. It is known that miRNAs and lncRNAs are key RNA families that may influence biological processes, such as gene expression, but have little protein-coding activity. LncRNAs regulate gene expression during the transcriptional and post-transcriptional phases, whereas miRNAs target mRNA and regulate the gene expressions of agents that induce its degradation or inhibition [6]. Although it has been shown that the development of NAFLD is dependent on genetic, environmental, and metabolic variables, the pathogenesis process remains unclear. Additionally, how a simple fat deposition turns into a life-threatening, complex process is not yet understood. Although ncRNAs such as miRNA and lncRNA have been shown to have a role in the pathogenesis of NAFLD, it was predicted that the common interaction of these ncRNAs would help to explain the situation [7]. In the development of NAFLD, miRNAs are thought to be involved in metabolic and inflammatory processes and play an important role in the progression of the disease towards more severe stages. Furthermore, studies suggest that miRNAs influence the pathophysiology of NAFLD by directing the regulators that govern lipid metabolism, oxidative stress, and inflammation in the liver [8]. This study aimed to analyze the expression profile of 753 miRNAs and 90 lncRNAs in serum samples of NAFLD patients with different types of hepatosteatosis compared to healthy controls using the qPCR method, as well as the relationship between detected miRNA-LncRNAs and their association with regulatory pathways.
2. Materials and Methods
2.1. Study Design and NAFLD Diagnosis
In this observational study, 180 NAFLD patients and 60 healthy controls were included randomly. The collection and use of data and samples of NAFLD patients and healthy controls were approved by the ethics committee of the Istanbul MedicalPark Private Hospital (Approval no: 2021-1-7; Date: 26 April 2021). All participants provided their permission with full knowledge of the procedure. All subjects were diagnosed and graded for NAFLD utilizing abdominal ultrasonography exams based on the liver’s brightness and the presence of diffuse echogenicity in the liver parenchyma. Patients with NAFLD do not consume alcohol or specific medications that induce fatty liver. All subjects were negative for hepatitis conditions, including autoimmune hepatitis, HBV, HCV, and HDV. The NAFLD grade was recorded as none (0), mild (1), moderate (2), or severe (3) [9]. Inclusion criteria were participants who were aged >18 years, who used abdominal USG to evaluate hepatic steatosis grading (0 to 3), and who did not consume alcohol. Exclusion criteria were diagnosis with different hepatitis conditions, consumption of alcohol, positive results for HBsAg or Anti-HCV, presence of a cancer diagnosis, other chronic liver diseases or chronic infectious, a history of autoimmune disease or metabolic disease, and use of hepatotoxic medication.
2.2. Sample Collection
Blood samples were collected from every participant. Samples were immediately delivered to the laboratory in a chiller, followed by 10 min of centrifugation at 3000 rpm. In sterile microcentrifuge tubes, serum samples were separated and aliquoted. These aliquoted samples were kept at −80 degrees Celsius until RNA extraction.
2.3. miRNA Gene Expression Profilling Using qPCR Array
miRNeasy Serum/Plasma RNA isolation kit (Qiagen, Hilden, Germany) was used for miRNA isolation from the 200 µL of aliquoted serum samples. The DNase digestion protocol in the miRNeasy Serum/Plasma RNA isolation kit (Qiagen, Hilden, Germany) directive was applied during RNA isolations to avoid genomic DNA contamination. The miRCURY LNA RT Kit (Qiagen, Hilden, Germany) was used according to the manufacturer’s instructions to synthesize cDNA from 25 pg/L of extracted miRNAs. miRNA gene expression profiling was performed with the miR-CURY LNA miRNA miRNome PCR human panel I + II kit (YAHS-312Y) (Qiagen, Hilden, Germany). A pair of 384-well qPCR plates with predesigned PCR primer sets for specific miRNAs were included in the kit. The qPCR reaction was conducted using a LightCycler 480 (Roche Diagnostics, Mannheim, Germany) and a miRCURY LNA SYBR Green PCR Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. qPCR conditions were 95 °C for 2 min for denaturation, followed by 40 cycles at 95 °C for 1 min and 56 °C for 1 min [10]. The SNORD38B, SNORD49A, and U6 genes were used as housekeeping genes. Livak and Schmittgen’s 2−CT (delta-delta Ct) method was utilized to detect specific miRNA gene expression levels through relative quantification. Relative ratio values were presented. A result of 0 to 1 was evaluated as downregulated, and >1 was evaluated as upregulated [11].
2.4. LncRNA Expression Profiling Using qPCR Array
RNA was extracted from 200 µL of aliquoted serum samples using the miRNeasy Serum/Plasma RNA isolation kit (Qiagen, Hilden, Germany). Then, 25 pg/µL of RNA was used for cDNA synthesis, which was performed using the RT2 First Strand Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The LncProfiler qPCR Array Kit (System Biosciences (SBI), Palo Alto, CA, USA) was used to conduct LncRNA profiling analysis. The kit was provided with one 96-well ready-to-use qPCR plate containing predesigned PCR primer sets for specific lncRNAs. The qPCR reaction was performed on a LightCycler 480 (Roche Diagnostics, Mannheim, Germany) instrument using a LightCycler 480 SYBR Green Master Kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions. The 18S rRNA, RNU43, GAPDH, LAMIN A/C, and U6 genes were used as housekeeping genes. Relative quantification analysis was used to detect specific lncRNA profiling and then analyzed by Livak and Schmittgen’s 2−ΔΔCT (delta-delta Ct) method. Relative ratio values were presented. A result of 0 to 1 was evaluated as downregulated and >1 was evaluated as upregulated [12].
2.5. miRNA and Pathway Interaction Analysis by DIANA-miRPath Web Server
DIANA miRPath v3.0, a web server (https://dianalab.e-ce.uth.gr/html/mirpathv3/index.php?r=mirpath, accessed on 15 December 2022) was used for the analysis between the target miRNA and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. This was an in silico miRNA target prediction algorithm [13].
LncRNA and miRNA connection analysis was conducted by the DIANA-lncBase web server.
DIANA-lncBase v3.0, a web server (https://diana.e-ce.uth.gr/lncbasev3/home, accessed on 15 December 2022) was used to conduct connection analysis between the miRNAs and lncRNAs.
2.6. Statistical Analysis
IBM SPSS version 20.0 software (SPSS Inc., Chicago, IL, USA) was used to perform statistical analysis. Data are presented as mean ± standard deviation. The Kruskal–Wallis test, Mann–Whitney U test and Spearman correlation analysis were used. p values below 0.05 (p < 0.05) were considered statistically significant. For Spearman’s analysis, which was used for correlation and values (rs), rs < 0.25 was evaluated as not statistically correlated, rs = 0.25–0.5 was evaluated as a weak correlation, rs = 0.5–0.75 was evaluated as a moderate correlation, rs = 0.76–0.85 was evaluated as a strong correlation, and rs > 0.85 was evaluated as a very strong correlation [13].
3. Results
Table 1 provides demographic information for the 180 NAFLD patients and 60 healthy controls included in our study. The mean age of the NAFLD patients in our study was 39.09 ± 11.91 years, while the mean age of the healthy controls was 38.82 ± 8.5 years (Table 1).

Table 1.
Distribution of demographic data in NAFLD patients and healthy controls.
NAFLD patients were graded according to ultrasound examinations, and their demographic data are presented in Table 2.

Table 2.
Distribution of demographic data of NAFLD patient groups, separated by grade.
A total of 75 of 753 miRNA alterations in NAFLD patients were found to be significant compared to healthy controls (p < 0.05) by qPCR. Of these 75, 30 miRNAs were downregulated and 45 were upregulated compared to healthy controls. When lncRNA expression was examined, it was determined that 24 of 90 lncRNAs were altered significantly (p < 0.05); 19 of 24 lncRNAs were upregulated and 5 were downregulated (Figure 1).

Figure 1.
Heatmap of statistically significant miRNA and lncRNA expression profiles detected by qPCR in NAFLD patients compared to the healthy controls.
Among the upregulated miRNAs, hsa-miR-21 showed the highest increase, at 2.3 ± 0.22 fold, followed by hsa-miR-222, hsa-miR-122, hsa-miR-10b, hsa-miR-221, hsa-miR-155, hsa-miR-33a, hsa-miR-223, hsa-miR-378a, and hsa-miR-193a, respectively. Among the downregulated miRNAs, hsa-miR-197 showed the greatest decrease, at 0.24 ± 0.07 fold, followed by hsa-miR-129, hsa-miR-99a, hsa-miR-422a, hsa-miR-27a, hsa-miR-139, hsa-miR-29a, hsa-miR-146b, hsa-miR-99b, and hsa-miR-296.
Among lncRNAs, only NEAT1 (family) (2.87 ± 0.23), MALAT1 (2.51 ± 0.31), MEG3 (family) (0.41 ± 0.12), and PTENP1 (0.48 ± 0.05) were detected in all samples. While NEAT1 (family) (2.87 ± 0.23) and MALAT1 (2.51 ± 0.31) were detected as upregulated, MEG3 (family) (0.41 ± 0.12) and PTENP1 (0.48 ± 0.05) were found to be downregulated. HOTAIR, EgoA, and Nespas, which were found to be upregulated in NAFLD patients, were found to be expressed in 114 (63.34%), 97 (53.88%), and 91 (50.56%) samples, respectively. MEG9, which was found to be downregulated in NAFLD patients, was found to be expressed in 93 (51.67%) samples. In healthy controls, HOTAIR, EgoA, Nespas, and MEG9 were detected in 34 (56.67%), 24 (40%), 28 (46.67%), and 38 (63.34%) samples, respectively.
According to in silico analysis conducted with the DIANA miRPath v3.0 online software, miRNAs that were found to be upregulated at the highest rates and downregulated at the lowest rates in our study had the greatest statistical impact on the “Fatty acid biosynthesis” pathway. Following this analysis, it was discovered that the pathways including the greatest number of genes were the “cancer pathways.” Moreover, among the miRNAs included in this research, the “Hippo signaling pathway” was shown to include the greatest amount of miRNAs (Table 3).

Table 3.
Interaction of upregulated and downregulated miRNAs with pathways after in silico analysis with DIANA miRPath v3.0 online software.
Figure 2 depicts the cluster analysis and heatmap of the relationship with the pathways of miRNAs that were upregulated at the greatest rate and downregulated at the lowest rate in our study, as determined by in silico analysis with the DIANA miRPath v3.0 online web server.
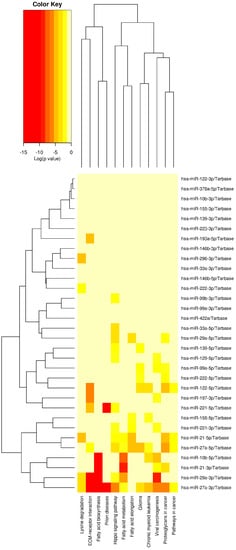
Figure 2.
Cluster analysis and the heatmap of the relationship with the pathways of miRNAs.
Figure 3 displays the heatmap linked with gene ontology (GO) analysis of miRNAs that were upregulated at the greatest relative rate and downregulated at the lowest relative rate in our study. According to our analysis, the associated miRNAs affected 6910 and 6536 genes involved in cellular components (GO:0005575) and biological processes (GO:0008150), respectively.
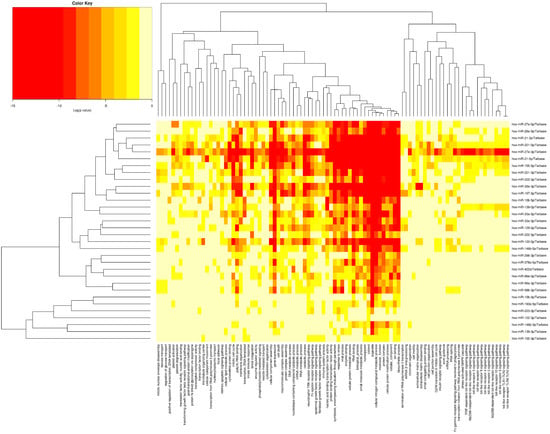
Figure 3.
The heatmap associated with gene ontology (GO) analysis of miRNAs.
The connection between the miRNA and lncRNAs which were found to be upregulated at the highest rate and downregulated at the lowest rate in our study, was examined by in silico analysis performed with DIANA lncBase v3.0 online software. This analysis found a correlation between lncRNAs with the MEG3 family, PTENP1, and miRNAs detected with HAR1B. The associated miRNAs are presented in Table 4.

Table 4.
Distribution of miRNAs associated with lncRNA with DIANA lncBase v3.0 online software.
When the demographic data of the patients and their correlation with miRNA and lncRNAs detected in the circulation were investigated, a weak positive correlation was found between hsa-miR-122-5p and BMI among miRNAs (rs: 0.351; p < 0.05). There was no significant relationship between other parameters and miRNAs. Moreover, among the lncRNAs, a weak positive correlation was found with MALAT1 between FBS (rs: 0.403; p < 0.05), insulin (rs: 0.384; p < 0.05), HOMA-IR (rs: 0.298; p < 0.05), and HbA1c levels (rs: 0.306; p < 0.05).
4. Discussion
Currently, NAFLD is the most common chronic liver condition worldwide. Although it is known that NAFLD affects several organs and regulatory pathways outside of the liver, the underlying molecular processes of the illness remain obscure [14]. In this investigation, we focused on identifying the miRNAs and lncRNAs, two different families of non-coding RNAs (ncRNAs), in the serum samples of NAFLD patients, demonstrating their existence and rates in circulation and evaluating their interrelationships.
Numerous studies have been conducted on miRNAs that may have an effect on the molecular mechanism of NAFLD, and the reviews or meta-analyses that resulted from these studies reported that the expression levels of numerous miRNAs, such as miR-122, miR-34, miR-33, miR-21, miR-192, miR-375, miR-146b, miR-221, and miR-222, varied. Several studies suggest that miR-122, miR-33, miR-34a, and miR-21 regulate liver metabolism, and it has been claimed that they can be employed as invasive biomarkers [14,15]. Similarly, it has been reported that circular miRNAs can be used as biomarkers in NAFLD as well as in NASH, as these miRNAs also differentiate throughout the development to NASH [16]. Our research yielded comparable results to those of earlier studies. Specifically, the fact that certain miRNA levels varied with different grades of the disease suggested that they may be used as biomarkers if the cut-off values were determined for larger cohort groups.
The pathways of fatty acid biosynthesis and fatty acid metabolism have been linked to the development of NAFLD. It has been reported that this pathway can be used for therapeutic purposes [17]. According to our study, both upregulated and downregulated miRNAs had a significant impact on these pathways. Therefore, it has been hypothesized that silencing and stimulating these circulating miRNAs to affect these fatty acid-related pathways might serve as a therapeutic option. In addition, our research revealed that “cancer pathways” and the “Hippo signaling pathway” were significantly affected. Similar to our results, Wu et al. showed in their research that “pathways in cancer” are affected by the progression of the disease using bioinformatic techniques [18]. By regulating IRS2 expression, Jeong et al. found that the Hippo signaling system’s communication with the AKT signaling pathway protected against NAFLD in a study on a mouse model. Moreover, they reported that disruption of this pathway may contribute to the advancement of liver cancer [19]. In our study, 11 miRNAs affecting the “Hippo signaling pathway” were detected, and these were among the miRNAs that showed significant changes, supporting the study by Jeong et al. The gene ontology analysis of the miRNAs obtained in this study revealed that these miRNAs regulate genes related to cellular components and biological processes. Moreover, Jia et al. found that cellular component and biological process-related gene expressions in the microarray data of NAFLD patients are essential for understanding the molecular processes underlying NAFLD [20]. This result also confirms the conclusions which we reached in our study on miRNAs.
In mouse models, Zou et al. demonstrated that lncRNA MEG3 (maternally expressed gene 3) upregulates the sirtuin 6 (SIRT6) and enhancer of zeste homolog 2 (EZH2) genes, which regulate fat accumulation, inflammation, and the development of NAFLD. In addition, they stated that MEG3 should be increased for NAFLD control [21]. Zamani et al. found that curcumin increased MEG3 while decreasing its expression in hepatocellular carcinomas. Additionally, they reported that MEG3 exerted this effect through miRNAs [22]. Furthermore, lncRNAs such as nuclear paraspeckle assembly transcript 1 (NEAT1), hox transcript antisense RNA (HOTAIR), metastasis-associated lung adeno-carcinoma transcript 1 (MALAT1), and HOXA distal transcript antisense RNA (HOTTIP) are upregulated and play a role in the progression of liver fibrosis and hepatocellular carcinoma [23]. Similarly to previous studies, our analysis found that NEAT1, MALAT1, and MEG3 were upregulated in all samples, whereas MEG3 was downregulated. Although not detected in all samples, upregulation of HOTAIR was also observed. Nevertheless, there are contradictory data addressing the downregulation of lncRNA NEAT1 and its contribution to the correction of NAFLD in rats via the mTOR/S6K1 signaling pathway [24]. In addition, studies indicate that lncRNA PTENP1 suppresses the oncogenic PI3K/AKT pathway in hepatocellular carcinomas, and can be used to treat hepatocellular carcinomas via miRNAs such as miR-17, miR-19b, and miR-20 [25]. Similarly, lncRNA PTENP1, which is believed to be involved in a complicated autophagy process in the liver, was downregulated in all NAFLD patients in our study [26].
Matboli et al. [6] reported that miRNAs (mir-650 and miR-1205) and lncRNAs (RPARP-AS1 and SRD5A3-AS1) interact in the pathogenesis of NAFLD. In our study, in silico analysis showed that there may be more miRNA–lncRNA interactions in addition to this one.
While there have been studies demonstrating that circulating miRNAs or lncRNAs were correlated with many parameters in metabolic diseases such as NAFLD [27,28], there was also research demonstrating that they were not correlated with these parameters [29]. In our investigation, only circulating hsa-miR-122-5p and lncRNA MALAT1 levels were observed to have a weak connection with certain parameters.
Our study’s limitations included its single-center design and the absence of biopsies and histopathologies in grade evaluations of NAFLD patients. Ultrasound examinations were used in our study instead of an invasive procedure such as a biopsy, which was considered sufficient for evaluating the data obtained from patients. RNA-Seq methods, which can discover novel miRNAs and lncRNAs, were not utilized in our study, which was another limitation. We used commercial qPCR profiler kits that were comparatively less expensive than RNA-Seq, and our approach supplied comprehensive data without the requirement for qPCR data validation, which is necessary in methods such as RNA-Seq and micro-array.
5. Conclusions
In this study, in vitro and in silico miRNA and lncRNA interactions in NAFLD patients were investigated. Compared to healthy controls, in NAFLD patients, miR-21 was upregulated by about 2-fold and LncRNA NEAT1 by 2.9-fold, whereas miR-197 and lncRNA MEG3 were downregulated by 0.25- and 0.41-fold, respectively. In addition, while lncRNA MEG3 and PTENP1 interacted with distinct miRNAs, miRPath analysis demonstrated that the fatty acid biosynthesis pathway was affected. It is evident that the interaction between lncRNAs and miRNAs is involved in the molecular pathways underlying the development of NAFLD. In light of our results, we recommend that thorough multicenter clinical research on the silencing and stimulating miRNAs and lncRNAs discovered in this interaction should be designed.
Author Contributions
Conceptualization, M.G.E., O.U. and M.D.; design, M.G.E. and M.D.; data collection and/or processing, M.G.E., O.U. and M.D.; analysis and/or interpretation, M.G.E., O.U. and M.D.; literature review: M.D.; writing manuscript, M.G.E., O.U. and M.D.; critical review, M.G.E., O.U. and M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the local ethics committee of the Istanbul MedicalPark Private Hospital (Approval no: 2021-1-7; Date: 26 April 2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
No new data were created.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| NAFLD | non-alcoholic fatty liver disease |
| NASH | non-alcoholic steatohepatitis |
| miRNAs | microRNAs |
| ncRNAs | non-coding RNAs |
| lncRNAs | long non-coding RNAs |
| qPCR | quantitative polymerase chain reaction |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MEG3 | maternally expressed gene 3 |
| SIRT6 | sirtuin 6 |
| EZH2 | enhancer of zeste homolog 2 |
| NEAT1 | nuclear paraspeckle assembly transcript 1 |
| HOTAIR | hox transcript antisense RNA |
| MALAT1 | metastasis-associated lung adenocarcinoma transcript 1 |
| HOTTIP | HOXA distal transcript antisense RNA |
| PTENP1 | phosphatase and tensin homolog pseudogene 1 |
References
- Lujan, P.V.; Esmel, E.V.; Meseguer, E.S. Overview of Non-Alcoholic Fatty Liver Disease (NAFLD) and the Role of Sugary Food Consumption and Other Dietary Components in Its Development. Nutrients 2021, 13, 1442. [Google Scholar] [CrossRef] [PubMed]
- Alsaif, F.; Al-Hamoudi, W.; Alotaiby, M.; Alsadoon, A.; Almayouf, M.; Almadany, H.; Abuhaimed, J.; Ghufran, N.; Merajuddin, A.; Khan, I.A. Molecular Screening via Sanger Sequencing of the Genetic Variants in Non-Alcoholic Fatty Liver Disease Subjects in the Saudi Population: A Hospital-Based Study. Metabolites 2022, 12, 1240. [Google Scholar] [CrossRef] [PubMed]
- Kupčová, V.; Fedelešová, M.; Bulas, J.; Kozmonová, P.; Turecký, L. Overview of the Pathogenesis, Genetic, and Non-Invasive Clinical, Biochemical, and Scoring Methods in the Assessment of NAFLD. Int. J. Environ. Res. Public Health 2019, 16, 3570. [Google Scholar] [CrossRef] [PubMed]
- Sheka, A.C.; Adeyi, O.; Thompson, J.; Hameed, B.; Crawford, P.A.; Ikramuddin, S. Nonalcoholic Steatohepatitis: A Review. JAMA 2020, 323, 1175–1183. [Google Scholar] [CrossRef]
- Meng, X.; Li, A.; Yu, B.; Li, S. Interplay between miRNAs and lncRNAs: Mode of action and biological roles in plant development and stress adaptation. Comput. Struct. Biotechnol. J. 2021, 19, 2567–2574. [Google Scholar] [CrossRef]
- Lanzillotti, C.; De Mattei, M.; Mazziotta, C.; Taraballi, F.; Rotondo, J.C.; Tognon, M.; Martini, F. Long Non-Coding RNAs and MicroRNAs Interplay in Osteogenic Differentiation of Mesenchymal Stem Cells. Front. Cell Dev. Biol. 2021, 9, 646032. [Google Scholar] [CrossRef]
- Matboli, M.; Gadallah, S.; Rashed, W.; Hasanin, A.; Essawy, N.; Ghanem, H.; Eissa, S. mRNA-miRNA-lncRNA Regulatory Network in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2021, 22, 6770. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Yang, Y.-L.; Wang, P.-W.; Wang, F.-S.; Huang, Y.-H. The Emerging Role of MicroRNAs in NAFLD: Highlight of MicroRNA-29a in Modulating Oxidative Stress, Inflammation, and Beyond. Cells 2020, 9, 1041. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Kwon, O.-S.; Her, K.H. The grade of nonalcoholic fatty liver disease is an independent risk factor for gallstone disease. Medicine 2019, 98, e16018. [Google Scholar] [CrossRef]
- Fayyad-Kazan, M.; Makki, R.; Skafi, N.; El Homsi, M.; Hamade, A.; El Majzoub, R.; Hamade, E.; Fayyad-Kazan, H.; Badran, B. Circulating miRNAs: Potential diagnostic role for coronavirus disease 2019 (COVID-19). Infect. Genet. Evol. 2021, 94, 105020. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]
- Demirci, M.; Taner, Z.; Keskin, F.E.; Ozyazar, M.; Kiraz, N.; Kocazeybek, B.S.; Tokman, H.B. Similar bacterial signatures in the gut microbiota of type 1 and type 2 diabetes patients and its association with G protein-coupled receptor 41 and 43 gene expression. J. Diabetes Metab. Disord. 2022, 21, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, O.; Errafii, K.; Al-Akl, N.S.; Arredouani, A. Noncoding RNAs in Nonalcoholic Fatty Liver Disease: Potential Diagnosis and Prognosis Biomarkers. Dis. Markers 2020, 2020, 8822859. [Google Scholar] [CrossRef] [PubMed]
- Gjorgjieva, M.; Sobolewski, C.; Dolicka, D.; de Sousa, M.C.; Foti, M. miRNAs and NAFLD: From pathophysiology to therapy. Gut 2019, 68, 2065–2079. [Google Scholar] [CrossRef]
- Kim, T.H.; Lee, Y.; Lee, Y.-S.; Gim, J.-A.; Ko, E.; Yim, S.Y.; Jung, Y.K.; Kang, S.; Kim, M.Y.; Kim, H.; et al. Circulating miRNA is a useful diagnostic biomarker for nonalcoholic steatohepatitis in nonalcoholic fatty liver disease. Sci. Rep. 2021, 11, 14639. [Google Scholar] [CrossRef]
- Batchuluun, B.; Pinkosky, S.L.; Steinberg, G.R. Lipogenesis inhibitors: Therapeutic opportunities and challenges. Nat. Rev. Drug Discov. 2022, 21, 283–305. [Google Scholar] [CrossRef]
- Wu, C.; Zhou, Y.; Wang, M.; Dai, G.; Liu, X.; Lai, L.; Tang, S. Bioinformatics Analysis Explores Potential Hub Genes in Nonalcoholic Fatty Liver Disease. Front. Genet. 2021, 12, 772487. [Google Scholar] [CrossRef]
- Jeong, S.-H.; Kim, H.-B.; Kim, M.-C.; Lee, J.-M.; Lee, J.H.; Kim, J.-H.; Kim, J.W.; Park, W.-Y.; Kim, S.-Y.; Kim, J.B.; et al. Hippo-mediated suppression of IRS2/AKT signaling prevents hepatic steatosis and liver cancer. J. Clin. Investig. 2018, 128, 1010–1025. [Google Scholar] [CrossRef]
- Jia, X.; Zhai, T. Integrated Analysis of Multiple Microarray Studies to Identify Novel Gene Signatures in Non-Alcoholic Fatty Liver Disease. Front. Endocrinol. 2019, 10, 599. [Google Scholar] [CrossRef]
- Zou, D.; Liu, L.; Zeng, Y.; Wang, H.; Dai, D.; Xu, M. LncRNA MEG3 up-regulates SIRT6 by ubiquitinating EZH2 and alleviates nonalcoholic fatty liver disease. Cell Death Discov. 2022, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Zamani, M.; Sadeghizadeh, M.; Behmanesh, M.; Najafi, F. Dendrosomal curcumin increases expression of the long non-coding RNA gene MEG3 via up-regulation of epi-miRs in hepatocellular cancer. Phytomedicine 2015, 22, 961–967. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Yang, D.; Fan, X.; Zhang, M.; Li, Y.; Gu, X.; Yang, M. The Roles and Mechanisms of lncRNAs in Liver Fibrosis. Int. J. Mol. Sci. 2020, 21, 1482. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Down-regulation of lncRNA-NEAT1 alleviated the non-alcoholic fatty liver disease via mTOR/S6K1 signaling pathway. J. Cell. Biochem. 2018, 119, 1567–1574. [Google Scholar] [CrossRef]
- Chen, C.-L.; Tseng, Y.-W.; Wu, J.-C.; Chen, G.-Y.; Lin, K.-C.; Hwang, S.-M.; Hu, Y.-C. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials 2015, 44, 71–81. [Google Scholar] [CrossRef]
- Liu, L.; Liao, J.-Z.; He, X.-X.; Li, P.-Y. The role of autophagy in hepatocellular carcinoma: Friend or foe. Oncotarget 2017, 8, 57707–57722. [Google Scholar] [CrossRef]
- Ma, E.; Fu, Y.; Garvey, W.T. Relationship of Circulating miRNAs with Insulin Sensitivity and Associated Metabolic Risk Factors in Humans. Metab. Syndr. Relat. Disord. 2018, 16, 82–89. [Google Scholar] [CrossRef]
- Zhou, W.; Qiu, K. The correlation between lncRNA NEAT1 and serum hepcidin in the peripheral blood of non-alcoholic fatty liver disease patients. Am. J. Transl. Res. 2022, 14, 2593–2599. [Google Scholar]
- Paramasivam, P.; Meugnier, E.; Gokulakrishnan, K.; Ranjini, H.; Staimez, L.R.; Weber, M.B.; Narayan, K.M.V.; Vidal, H.; Tandon, N.; Prabhakaran, D.; et al. Blood-derived miRNA levels are not correlated with metabolic or anthropometric parameters in obese pre-diabetic subjects but with systemic inflammation. PLoS ONE 2022, 17, e0263479. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).