Abstract
Background: Epidemiologic studies have reported that the geographical distribution of the prevalence of allelic variants of serine protein inhibitor-A1 (SERPINA1) and severe cases of COVID-19 were similar. Methods: A multicenter, cross-sectional, observational study to evaluate the frequency of alpha-1 antitrypsin deficiency (AATD) in patients with COVID-19 and whether it was associated with having suffered severe COVID-19. Results: 2022 patients who had laboratory-confirmed SARS-CoV-2 infection. Mutations associated with AATD were more frequent in severe COVID versus non-severe (23% vs. 18.8%, p = 0.022). The frequency of Pi*Z was 37.8/1000 in severe COVID versus 17.5/1000 in non-severe, p = 0.001. Having an A1AT level below 116 was more frequent in severe COVID versus non-severe (29.5% vs. 23.1, p = 0.003). Factors associated with a higher likelihood of severe COVID-19 were being male, older, smoking, age-associated comorbidities, and having an A1AT level below 116 mg/dL [OR 1.398, p = 0.003], and a variant of the SERPINA1 gene that could affect A1AT protein [OR 1.294, p = 0.022]. Conclusions: These observations suggest that patients with AATD should be considered at a higher risk of developing severe COVID-19. Further studies are needed on the role of A1AT in the prognosis of SARS-CoV-2 infection and its possible therapeutic role.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in a pandemic, with more than 6 million deaths [1]. There are remarkably different infection and mortality rates for SARS-CoV-2 between different countries [2]. Moreover, there are remarkably interindividual differences in the clinical severity of coronavirus disease 2019 (COVID-19) that cannot be completely explained by environmental factors, comorbidities, and age-related fragility [3]. On the basis of these observations and the susceptibility of hosts, it could be argued that genetic differences among populations, ethnicities, and individuals may contribute to the different epidemiological and clinical manifestations of COVID-19 [4]. Recent studies have investigated genetic susceptibility to SARS-CoV-2 and reported that approximately 20% of life-threatening COVID-19 cases are associated with genetic errors and gene loci, most of which are involved in immune signaling pathways [5].
In-hospital COVID-19 patient, studies have described a proinflammatory syndrome with a disproportionately high rate of progression to acute respiratory distress syndrome [2]. Recent data indicates that the COVID-19 cytokinemia is distinct in critical care presentations, showing marked differences in the balance between proinflammatory and anti-inflammatory cytokines and a blunted alpha-1 antitrypsin (A1AT) acute phase response. Cytokine ratios, such as high IL-6:A1AT levels, are related to worse prognosis in COVID-19 patients [2].
Alpha-1 antitrypsin deficiency (AATD) is the most common inherited disorder in adults; it is often under-diagnosed [6] and characterized by reduced plasma levels or the abnormal functioning of A1AT, a human blood serine protease inhibitor, which is encoded by the serine protein inhibitor-A1 (SERPINA1) gene. Recent studies confirmed a correlation between the COVID-19 pandemic and the prevalence of AATD in the same geographical areas [7].
A1AT is a tissue protector, as well as an antiviral and anti-inflammatory molecule. Indeed, A1AT has several biological functions that may antagonize SARS-CoV-2 infection and pathophysiologic processes resulting in cellular entry. Recent studies have demonstrated that A1AT is an inhibitor of SARS-CoV-2 infection and two of the most important proteases in the pathophysiology of COVID-19: transmembrane serine protease 2 and the disintegrin and metalloproteinase 17, as was well as an inhibitor of inflammatory molecules, such as IL-8, TNF-α, and neutrophil elastase [8,9]. Other potential A1AT protective mechanisms of action are the inhibitory effect on thrombin and delayed thrombus formation [10] and decreased oxidative stress, inflammation, and cell wall deterioration [11].
Therefore, we focused on the possible role of AATD as a risk factor for severe COVID-19 progression. A poor prognosis for COVID-19 patients may be related to A1AT levels. In our study, we examined the presence of genetic mutations associated with AATD and A1AT levels in patients who had suffered a SARS-CoV-2 infection in order to assess whether AATD was associated with having suffered severe COVID-19.
2. Materials and Methods
COVID-AATD is a multicenter, cross-sectional, observational study conducted from 1 May 2021 to 1 September 2022. The sample population was adults who had laboratory confirmed SARS-CoV-2 infection and were treated by a pneumology department. Participants were enrolled consecutively at 9 centers in the inpatient ward or in follow-up consultation after discharge. There were no exclusion criteria, except for patients’ or families’ explicit refusal to participate. The study was performed according to the Declaration of Helsinki and its amendments. All patients gave written informed consent. The study was approved by the Research Ethics Committee at Hospital Clínico San Carlos, Madrid, Spain (internal code 20/809-E). The personal data of the patients was kept under strict confidentiality in compliance with the provisions of Spanish Organic Law 3/2018, of December 5, on the Protection of Personal Data and Guarantee of Digital Rights (LOPDGDD) and its development regulations, and in accordance with the provisions of Regulation (EU) 2016/679 of the European Parliament and of the Council of 27 April 2016, regarding the protection of natural persons with regard to the processing of personal data and the free circulation of these data.
A retrospective review was performed through the analysis of electronic medical records where SARS-CoV-2 infection clinical data were collected. Patients were defined as suffering severe COVID-19 if they had been treated with high-flow nasal cannula (HFNC) oxygenation, non-invasive ventilation therapy, or were admitted to the intensive care unit at any stage of the disease according to the WHO Clinical Progression Scale [12], or if they died as a result of COVID-19.
The data collected during the only visit were concurrent. The information collected was clinical data (demographic data, smoking status, comorbidities). Allele-specific genotyping testing was carried out in all patients using the Progenika A1AT Genotyping Test. The test allows the identification of the 14 most frequent deficiency variants of the SERPINA1 gene: PI*S, PI*Z, PI*I, PI*Mprocida, PI*Mmalton, PI*Siiyama, PI*Q0granite falls, PI*Q0west, PI*Q0bellingham, PI*F, PI*Plowell, PI*Q0mattawa, PI*Q0clayton, and PI*Mheerlen. SERPINA1 gene sequencing was performed in the cases where none of the 14 mutations were found and the A1AT serum level was <60 mg/dL. The test is CE marked and United States Food and Drug Administration approved. The test is intended for use with genomic DNA extracted from human whole blood samples collected in K3-ethylenediaminetetraacetic acid (EDTA) tubes or as dried blood spots (DBS), or from human buccal swab samples [13]. The biological samples related to the study were numbered with a code to guarantee the confidentiality of the sample and the associated clinical data. There were no data in the database that could be used to identify patients. The patients signed a written informed consent authorizing the genetic study to be carried out according to Spanish legislation.
In the clinical stability phase, serum A1AT levels were analyzed using nephelometric and C-reactive protein (CRP) in plasma as a potential confounder by the immunonephelometry method. Although the lower limit of normal A1AT by nephelometry is 90 mg/dL, the use of a higher than normal cut-off value was established as a threshold value to study the possible presence of a deficient allele. The variability of A1AT levels has been described for different AATD genotypes and how it may be influenced by increased systemic inflammation [14].
Statistical Analysis
Descriptive statistics are reported as mean (standard deviation [SD]) or median (interquartile range [IQR]). Differences between the non-severe and severe COVID-19 groups were analyzed for statistical significance using the chi-square or Fisher’s exact test for categorical variables and the two-sample t-test or Wilcoxon rank sum test for continuous variables, as applicable. Adjustment variables (patient characteristics, genotyping test, and serum A1AT levels) with a p-value < 0.05 in the univariate analysis were included in the simple logistic regression analysis. Statistical significance was assumed as p < 0.05. All analyses were performed with Stata software version 17 (Stata Corp LLC, College Station, TX, USA). The study size was determined by the number of patients referred to the follow-up clinic in a pneumology department during the enrolment period.
3. Results
3.1. Characteristics of the Study Population
In total, 2022 patients were included in the analysis. Table 1 describes the sociodemographic and clinical characteristics of the enrolled patients. An amount of 43.2% had severe COVID-19 infection and six (0.3%) deaths occurred. The mean (SD) age of the overall COVID-19 cohort was 60.3 ± 14 years; 59.9% were men and 45.6% of patients were current or former smokers. Comorbidities were common in the study population.

Table 1.
Characteristics of the study population.
A1AT serum levels were available in 1691 (83.6%) cases, with a mean value of 132.1 (28.8) mg/dL. There were 390 (19.9%) carrying frequent mutations (S or Z), and 14 (0.7%) carrying rare alleles. In total, 67 samples were not processed due to the poor quality of the sample or due to errors recording the identification code on the web. The prevalence of the frequent allele combinations in this selected population was as follows: MS 16.3%, MZ 2.1%, SS 1.1%, SZ 0.3%, and ZZ 0.2%. Considered globally, 2.5% were Z carriers and 17.6% S carriers.
3.2. Characteristics According to the Presence of Genetic Mutations Associated with AATD
Patients with variants of the SERPINA1 gene that could affect A1AT protein activity or expression were older than patients without mutations (mean [SD] age: 61.8 [14.1] versus 60 [13.9] years; p = 0.021) and current smokers were more prevalent (7.9% versus 5.3%; p = 0.004). There were no differences in respiratory or non-respiratory comorbidities, Table 2. The frequency of severe COVID was also higher in patients positive for A1AT genotyping testing (48.8% vs. 42.4%; p = 0.022). A1AT serum levels were significantly lower in patients with mutations associated with AATD (106.3 [24] versus 138.8 [25.8]; p < 0.001).

Table 2.
Characteristics of enrolled patients according A1AT genotyping test.
3.3. Characteristics According to A1AT Levels
There were 440 (26%) patients with A1AT serum levels below 116 mg/dL, Table 3. The frequency of severe COVID was higher in patients with A1AT serum levels below 116 mg/dL compared with those above or equal to 116 mg/dL (51.9% versus 43.9%, p = 0.003), Figure 1.

Table 3.
Characteristics of patients, according A1AT level (≥116 mg/dL versus <116 mg/dL).
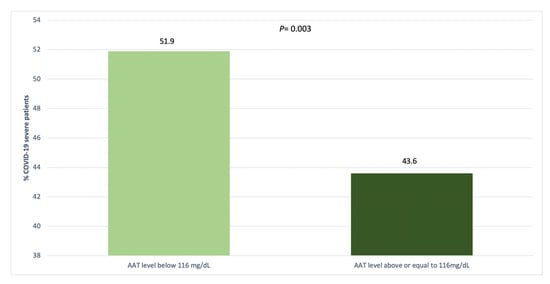
Figure 1.
Distribution of COVID-19 severity by serum AAT levels.
3.4. Characteristics According to COVID-19 Severity
There were 872 (43.2) patients defined as suffering severe COVID-19. Cases with severe COVID were older than patients with non-severe COVID (mean [SD] age: 62.8 [12.8] versus 58.9 [14.7] years; p < 0.001) and being male was more frequent (67.4% versus 54.3%, p < 0.001), Table 4. Having A1AT levels below 116 was more frequent in cases with severe COVID versus non-severe COVID (29.5% versus 23.1, p = 0.003). Cases carrying mutations associated with AATD were more frequent in severe COVID versus non-severe COVID (23% versus 18.8%, p = 0.022), Figure 2.

Table 4.
Characteristics of patients, according to severity of COVID-19.
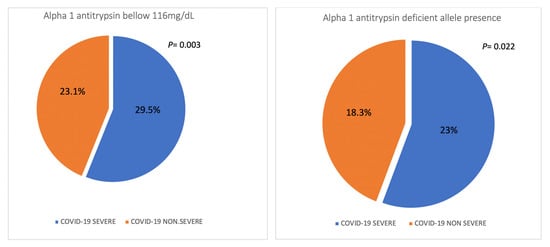
Figure 2.
Distribution of the severity of COVID-19 in population with AAT levels < 116 mg/dL and in population with deficiency-related mutations.
3.5. Factors Related to COVID-19 Severity
In the simple logistic regression analysis, the factors associated with a greater likelihood of having suffered severe COVID-19 were being older, being a male, being a former smoker, having cardiovascular comorbidities and a history of cancer, having A1AT levels below 116 mg/dL [OR 1.398 (CI95%: 1.124–1.739), p = 0.003], and having a mutation associated with AATD [OR 1.294 (CI95%: 1.038–1.612), p = 0.022], Table 5.

Table 5.
Clinical associations with a severe COVID-19 disease.
4. Discussion
This multicenter observational study investigates the association between AATD and the severity of COVID-19 in patients with a SARS-CoV-2 infection that were treated by pneumology departments in Spain. This analysis demonstrates that, having mutations, variants of the SERPINA1 gene that could affect A1AT protein activity or expression and that having decreased A1AT levels was significantly associated with a higher likelihood of suffering from a severe COVID-19 case. This is consistent with data that suggested that AATD might explain the high COVID-19 mortality in countries with a high AATD prevalence. During the COVID-19 pandemic, several epidemiologic studies have reported that the geographical distributions of the prevalence of SERPINA1 allelic variants and severe cases of COVID-19 were similar, although confounding factors should be considered in these analyses, such as the different control measures established by governments, SARS-CoV-2 vaccination, socioeconomic status, and population health [3,7,15]. Other observational studies in patients with AATD also found a higher frequency of SARS-CoV-2 infection and a higher risk for symptomatic SARS-CoV-2 infection in patients with severe AATD with lung disease [16,17]. Recently, an EARCO ERS Clinical Research Collaboration analysis that investigated the impact of COVID-19 on patients with severe AATD (PiZZ, PiSZ, or rare variants with an equivalent serum A1AT level < 60 mg/dL) [18] showed that while a poor outcome was more frequent in PiZZ compared with PiSZ, this did not reach statistical significance; non-respiratory comorbidities were more strongly associated with a poor outcome than genotype, baseline FEV1, or oxygen saturation. However, it should be noted that in this cohort of patients with AATD, although 88% were diagnosed with COVID-19 with a positive PCR, only 31% required hospitalization. In addition, an analysis of a community-based cohort with > 500,000 participants that assessed the association between AATD and COVID-19 in the United Kingdom Biobank showed that the most common and mild AATD genotypes were not associated with increased SARS-CoV-2 infection rates or increased SARS-CoV-2 fatalities, although it must be noted that there were very few cases of severe AATD in this study [19]. In our population of patients with SARS-CoV-2 pneumonia, the frequency of Pi*S was 176/1000 and Pi*Z 25/1000. These figures are high in relation to the estimated prevalence in Spain [20], with a mean SZ prevalence of 278/1000, Pi*Z 17/1000, and Pi*S 104/1000. This higher frequency of mutations related to severe impairment (ZZ, SZ) found in our cohort could support our hypothesis that a poor prognosis for COVID-19 patients may be related to the presence of genetic mutations associated with AATD. A large proportion of patients in our cohort required supportive therapies and intensive care for COVID-19, which could be explained by the fact that patients with more severe COVID-19 are usually referred to the pneumology follow-up clinic because they are at higher risk of developing complications [21]. Indeed, our data showed that cases carrying mutations associated with AATD were more frequent in severe COVID versus non-severe COVID (23% vs. 18.8%, p = 0.022). The frequency of Pi*Z was 37.8/1000 in severe COVID versus 17.5/1000 in non-severe COVID, p = 0.001. The presence of genetic mutations associated with AATD was found to be a predictor factor associated with a higher likelihood of suffering a severe COVID-19 case [OR 1.294 (CI95%: 1.038–1.612), p = 0.022], which was consistent with studies that confirmed a correlation between the frequency of Pi*Z and Pi*S alleles and mortality rates due to COVID-19 [7]. Furthermore, recent studies have investigated genetic susceptibility to SARS-CoV-2 and reported that approximately 20% of life-threatening COVID-19 cases were associated with genetic errors and gene loci, most of which are involved in two immune signaling pathways [5,22]. Thus, we could hypothesize that upon exposure to the same virus, while some individuals show asymptomatic or mild illness, plausibly due to effective immune reactions, severe COVID-19 patients may reflect dysfunctional immune reactions that lead to increased lung injury.
Regarding information on risk factors for the development of SARS-CoV-2 pneumonia and a severe disease course, our study supports many of the findings from previous reports indicating that the epidemiology of COVID-19 shows a diverse pattern across people who are different in age, sex, ethnicity, and particularly among those with pre-existing medical conditions [3,23,24,25]. In our cohort, being male, being older, having a history of smoking, and having age-associated comorbidities significantly contributes to the severity of acute COVID-19. However, it should be noted that in our analysis, patients with the presence of genetic mutations associated with AATD or A1AT levels below 116 mg/dL do not have a higher prevalence of hypertension, diabetes, heart disease, chronic kidney disease, or chronic obstructive pulmonary disease. However, there are other potential factors, such as the dominant COVID strain at the time of infection, as this is not an assessment that is performed in daily clinical practice, or vaccination status against SARS-CoV-2; however, vaccination coverage in adults in Spain was very high with more than 85% are vaccinated.
Several studies have focused on the possibility of shared pathogenic pathways between AATD and SARS-CoV-2 infection. Indeed, A1AT has several biological functions that may antagonize SARS-CoV-2 infection and pathophysiological processes. Alpha-1-antitrypsin is a tissue protector with antiviral and anti-inflammatory properties [26,27]. The main function of A1AT is inactivating proteolytic enzymes [28], which are released in pulmonary tissue. Furthermore, a protective role for A1AT has been described for several viral infections. A1AT levels may be relevant to the development of viral diseases as rhinovirus infection, human immunodeficiency virus, hepatitis B and C, and complications [29,30,31].
The relationship between a worse prognosis of SARS-CoV-2 infection and lower levels of AAT could be explained by the possible protective role of A1AT against COVID-19. A1AT reduces transmembrane serine protease 2 activity [27], protection against acute lung injury [32], and strong anti-inflammatory properties [10]. In relation to the potential A1AT protective mechanisms of action, our analysis showed that having A1AT levels below 116 was more frequent in cases with severe COVID versus non-severe COVID (29.5% versus 23.1, p = 0.003), and the presence of A1AT levels below 116mg/dL was identified as a predictor factor associated with a higher likelihood of suffering a severe COVID-19 case, which was consistent with studies that demonstrated the COVID-19 cytokinemia is distinct from that of other types of pneumonia. In these studies, the production and sialylation of A1AT are increased in COVID-19, but this anti-inflammatory response is overwhelmed in severe illness, with the IL-6: A1AT ratio being markedly higher in patients requiring ICU admission. In critically unwell patients with COVID-19, increases in IL-6: A1AT predicted a prolonged ICU stay and mortality, whereas improvement in IL-6:A1AT was associated with clinical resolution [2]. In this regard, supplementation of the acute A1AT response with exogenous A1AT may merit consideration, as it has been shown to modulate the production and activity of the key proinflammatory cytokines described in [28,33] while preserving the production of IL-10 [34]. Indeed, it has recently been reported that abrupt cessation of A1AT augmentation therapy for patients with AATD resulted in marked increases in levels of these specific proinflammatory cytokines, a loss of IL-10, and subsequent progression to respiratory failure [35].
Our study has several limitations. First, COVID-AATD is a cross-sectional, observational study in patients treated by pneumology departments and this may carry some bias. Patients with more severe COVID-19 are controlled mainly by pulmonologists, which may result in overestimates since it is more likely the most unwell patients are selected. On the other hand, most cases were included at the follow-up consultation after discharge, and very few were during hospitalization for COVID-19, which results in underestimated fatal COVID cases, despite the fact that the sample size is quite large and stratified by COVID infection severity. Second, the multiplex system studies the 14 most frequent mutations that include more than 99% of the deficient variants observed in the world. Therefore, the identification of Pi*M is achieved through exclusion, since the absence of any of these 14 alleles suggests with more than 99% probability that it is an M. However, when none of the 14 mutations were found and the A1AT serum level was <60 mg/dL, SERPINA1 gene sequencing was performed, which did not occur in our study. Third, although the lower limit of normal A1AT by nephelometry is 90 mg/dL, we established an above-normal cut-off value in our analysis on the basis that deficient mutations can be detected above this level and may also be influenced by increased systemic inflammation [36]. Consequently, the use of an above-normal cut-off value could be argued as a threshold value to screen for the possible presence of a deficient allele. Fourth, in the logistic regression analysis, other variables are not considered such as vaccines and specific therapies that could impact association estimates.
5. Conclusions
Our study identifies the presence of mutations associated with A1AT and that have A1AT levels below 116 as predictors associated with an increased likelihood of severe COVID-19. These observations suggest that patients with AATD should be considered at a higher risk of developing severe COVID-19. These findings highlight the need for further studies on the role of the A1AT in the pathogenesis and prognosis of SARS-CoV-2 infection and a potential therapeutic role.
Author Contributions
Conceptualization, M.C.R. and J.L.R.H.; Methodology, M.C.R. and J.L.R.H.; M.C.R. and J.L.R.H. wrote the manuscript. Investigation and writing—review and editing, M.C.R., J.L.R.H., G.V.C., M.E.G.C., M.M., L.L.-A., B.M.J.-R., R.A.R., R.M.M., M.T.-D. and J.M.H.-P.; A.M.H.-N. did the statistical analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This study was promoted by the Madrid Society of Pneumology and Thoracic Surgery (Neumomadrid). We thank Grifols for its financial support to carry out the study. The financing entities did not participate in the design of the study, data collection, analysis, publication, or preparation of this manuscript. The participation of the Vall d’Hebron University Hospital in this study has been funded by a research grant from the Fundació Catalana de Pneumologia (FUCAP) 2021.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Research Ethics Committee at Hospital Clínico San Carlos, Madrid, Spain (internal code 20/809-E).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
Lucía Gómez Martín-Caro, Jorge García Aragón, Íñigo García Peñuelas (Hospital Clínico San Carlos, Madrid), Alexa Núñez, Gerard Orriols, Georgina Farago (Hospital Universitari Vall d’Hebron, Barcelona), Teresa Peña Miguel, Rosario González Campo, Elsa Merino García (Complejo Asistencial Universitario de Burgos), Marta Núñez-Fernández y Cristina Ramos-Hernández (Hospital Álvaro Cunqueiro, Vigo), Ester Sánchez Guerra, Jin-Seok Yo (Hospital Clinic, Barcelona), Ana Bustamante Ruiz (Hospital de Torrelavega, Santander), Marta Arroyo Cózar, Pilar Alba (Hospital Infanta Cristina, Madrid), Olga Rajas (Hospital de La Princesa, Madrid), for their participation in the collection of data.
Conflicts of Interest
J.L.R.H. has received speaker fees from Bial, Boehringer Ingelheim, CSL Behring, GlaxoSmithKline, Grifols, and Zambon, and consulting fees from Bial. G.V.C. has received speaker fees from GlaxoSmithKline, and Grifols. Marc Miravitlles has received speaker fees from AstraZeneca, Boehringer Ingelheim, Chiesi, Cipla, Menarini, Rovi, Bial, Kamada, Takeda, Sandoz, Zambon, CSL Behring, Specialty Therapeutics, Janssen, Grifols, and Novartis, consulting fees from AstraZeneca, Atriva Therapeutics, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Bial, Gebro Pharma, CSL Behring, Inhibrx, Laboratorios Esteve, Ferrer, Menarini, Mereo Biopharma, Verona Pharma, Spin Therapeutics, ONO Pharma, pH Pharma, Palobiofarma SL, Takeda, Novartis, Sanofi, and Grifols and research grants from Grifols. L.L.A. has received speaker fees from AstraZeneca, Boehringer Ingelheim, Chiesi, CSL Bering, GlaxoSmithKline, Gebro Pharma, Grifols, Novartis, and Pfizer, and consulting feels from CSL Bering. M.C.R. has received speaker fees from AstraZeneca, Bial, Chiesi, CSL Behring, GlaxoSmithKline, Menarini, and Grifols, and consulting fees from GlaxoSmithKline, and Bial. The other authors declare that they have no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
AATD = alpha-1 antitrypsin deficiency; A1AT = alpha-1 antitrypsin; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; COVID-19 = coronavirus disease 2019; IPA: Pack-years; COPD: chronic obstructive pulmonary disease, ILD: diffuse interstitial lung disease, AOS: sleep apnea syndrome, ICU: intensive care unit, UCRI: intermediate respiratory care unit; HFNC = high-flow nasal cannula; NIV/CPAP: non-invasive ventilation/continuous positive airway pressure; CRP: C-reactive protein. IQR = interquartile range.
References
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 28 September 2022).
- McElvaney, O.J.; McEvoy, N.L.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Choileáin, O.N.; Clarke, J.; O’Connor, E.; Hogan, G.; et al. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care Med. 2020, 202, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Yamamoto, R.; Ariumi, Y.; Mizokami, M.; Shimotohno, K.; Yoshikura, H. Does Genetic Predisposition Contribute to the Exacerbation of COVID-19 Symptoms in Individuals with Comorbidities and Explain the Huge Mortality Disparity between the East and the West? Int. J. Mol. Sci. 2021, 22, 5000. [Google Scholar] [CrossRef] [PubMed]
- van der Made, C.I.; Simons, A.; Schuurs-Hoeijmakers, J.; van den Heuvel, G.; Mantere, T.; Kersten, S.; van Deuren, R.C.; Steehouwer, M.; van Reijmersdal, S.V.; Jaeger, M.; et al. Presence of Genetic Variants Among Young Men with Severe COVID-19. Jama 2020, 324, 663–673. [Google Scholar] [CrossRef]
- McCoy, K.; Peterson, A.; Tian, Y.; Sang, Y. Immunogenetic Association Underlying Severe COVID-19. Vaccines 2020, 8, 700. [Google Scholar] [CrossRef] [PubMed]
- Calle Rubio, M.; Soriano, J.B.; López-Campos, J.L.; Soler-Cataluña, J.J.; Alcázar Navarrete, B.; Rodríguez González-Moro, J.M.; Miravitlles, M.; Barrecheguren, M.; Ferrer, M.E.F.; Hermosa, J.L.R.; et al. Testing for alpha-1 antitrypsin in COPD in outpatient respiratory clinics in Spain: A multilevel, cross-sectional analysis of the EPOCONSUL study. PLoS ONE 2018, 13, e0198777. [Google Scholar] [CrossRef]
- Shapira, G.; Shomron, N.; Gurwitz, D. Ethnic differences in alpha-1 antitrypsin deficiency allele frequencies may partially explain national differences in COVID-19 fatality rates. FASEB J. 2020, 34, 14160–14165. [Google Scholar] [CrossRef]
- Wettstein, L.; Weil, T.; Conzelmann, C.; Müller, J.A.; Groß, R.; Hirschenberger, M.; Seidel, A.; Klute, S.; Zech, F.; Bozzo, C.P.; et al. Alpha-1 antitrypsin inhibits TMPRSS2 protease activity and SARS-CoV-2 infection. Nat. Commun. 2021, 12, 1726. [Google Scholar] [CrossRef]
- Azouz, N.P.; Klingler, A.M.; Callahan, V.; Akhrymuk, I.V.; Elez, K.; Raich, L.; Henry, B.M.; Benoit, J.L.; Benoit, S.W.; Noé, F.; et al. Alpha 1 Antitrypsin is an Inhibitor of the SARS-CoV-2-Priming Protease TMPRSS2. Pathog. Immun. 2021, 6, 55–74. [Google Scholar] [CrossRef]
- Wang, J.Z.; Zhang, R.Y.; Bai, J. An anti-oxidative therapy for ameliorating cardiac injuries of critically ill COVID-19-infected patients. Int. J. Cardiol. 2020, 312, 137–138. [Google Scholar] [CrossRef]
- Petrache, I.; Fijalkowska, I.; Medler, T.R.; Skirball, J.; Cruz, P.; Zhen, L.; Petrache, H.I.; Flotte, T.R.; Tuder, R.M. Alpha-1 antitrypsin inhibits caspase-3 activity, preventing lung endothelial cell apoptosis. Am. J. Pathol. 2006, 169, 1155–1166. [Google Scholar] [CrossRef]
- WHO Working Group on the Clinical Characterisation; Management of COVID-19 infection. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 2020, 20, e192–e197. [Google Scholar] [CrossRef]
- López-Campos, J.L.; Casas-Maldonado, F.; Torres-Duran, M.; Medina-Gonzálvez, A.; Rodriguez-Fidalgo, M.L.; Carrascosa, I.; Calle, M.; Osaba, L.; Rapun, N.; Drobnic, E.; et al. Results of a diagnostic procedure based on multiplex technology on dried blood spots and buccal swabs for subjects with suspected alpha1 antitrypsin deficiency. Arch. Bronconeumol. 2021, 57, 42–50. [Google Scholar] [CrossRef]
- Janciauskiene, S.; DeLuca, D.S.; Barrecheguren, M.; Welte, T.; Miravitlles, M. Serum Levels of Alpha1-antitrypsin and their relationship with COPD in the General Spanish Population. Arch. Bronconeumol. 2020, 56, 76–83. [Google Scholar] [CrossRef]
- Yoshikura, H. Epidemiological correlation between COVID-19 epidemic and prevalence of α-1 antitrypsin deficiency in the world. Glob. Health Med. 2021, 3, 73–81. [Google Scholar] [CrossRef]
- Ferrarotti, I.; Ottaviani, S.; Balderacchi, A.M.; Barzon, V.; De Silvestri, A.; Piloni, D.; Mariani, F.; Corsico, A. COVID-19 infection in severe Alpha 1-antitrypsin deficiency: Looking for a rationale. Respir. Med. 2021, 183, 106440. [Google Scholar] [CrossRef]
- Faria, N.; Inês Costa, M.; Gomes, J.; Sucena, M. Alpha-1 antitrypsin deficiency severity and the risk of COVID-19: A Portuguese cohort. Respir. Med. 2021, 181, 106387. [Google Scholar] [CrossRef]
- Parr, D.G.; Chorostowska-Wynimko, J.; Corsico, A.; Esquinas, C.; McElvaney, G.N.; Sark, A.D.; Sucena, M.; Tanash, H.; Turner, A.M.; Miravitlles, M. Impact of COVID-19 in Patients with Severe Alpha-1 Antitrypsin Deficiency: The IMCA1 Study of the EARCO Clinical Research Collaboration. Arch. Bronconeumol. 2022, 58, 840–842, Epub ahead of print. [Google Scholar] [CrossRef]
- Schneider, C.V.; Strnad, P. SARS-CoV-2 infection in alpha1-antitrypsin deficiency. Respir. Med. 2021, 184, 106466. [Google Scholar] [CrossRef]
- Blanco, I.; Bueno, P.; Diego, I.; Pérez-Holanda, S.; Lara, B.; Casas-Maldonado, F.; Esquinas, C.; Miravitlles, M. Alpha-1 antitrypsin Pi*SZ genotype: Estimated prevalence and number of SZ subjects worldwide. Int. J. Chronic Obstr. Pulm. Dis. 2017, 12, 1683–1694. [Google Scholar] [CrossRef]
- Vargas Centanaro, G.; Calle Rubio, M.; Álvarez-Sala Walther, J.L.; Martinez-Sagasti, F.; Albuja Hidalgo, A.; Herranz Hernández, R.; Rodríguez Hermosa, J.L. Long-term Outcomes and Recovery of Patients who Survived COVID-19: LUNG INJURY COVID-19 Study. Open Forum Infect. Dis. 2022, 9, ofac098. [Google Scholar] [CrossRef]
- López-Rodríguez, R.; Del Pozo-Valero, M.; Corton, M.; Minguez, P.; Ruiz-Hornillos, J.; Pérez-Tomás, M.E.; Barreda-Sánchez, M.; Mancebo, E.; Villaverde, C.; Núñez-Moreno, G.; et al. Presence of rare potential pathogenic variants in subjects under 65 years old with very severe or fatal COVID-19. Sci. Rep. 2022, 12, 10369. [Google Scholar] [CrossRef] [PubMed]
- von der Thüsen, J.; van der Eerden, M. Histopathology and genetic susceptibility in COVID-19 pneumonia. Eur. J. Clin. Investig. 2020, 50, e13259. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.J.; Liang, W.H.; Zhao, Y.; Liang, H.R.; Chen, Z.S.; Li, Y.M.; Liu, X.Q.; Chen, R.C.; Tang, C.L.; Wang, T.; et al. China Medical Treatment Expert Group for COVID-19. Comorbidity and its impact on 1590 patients with COVID-19 in China: A nationwide analysis. Eur. Respir. J. 2020, 55, 2000547. [Google Scholar] [CrossRef] [PubMed]
- Gassen, N.C.; Niemeyer, D.; Muth, D.; Corman, V.M.; Martinelli, S.; Gassen, A.; Hafner, K.; Papies, J.; Mösbauer, K.; Zellner, A.; et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019, 10, 5770. [Google Scholar] [CrossRef]
- Oguntuyo, K.Y.; Stevens, C.S.; Siddiquey, M.N.; Schilke, R.M.; Woolard, M.D.; Zhang, H.; Acklin, J.A.; Ikegame, S.; Huang, C.-T.; Lim, J.K.; et al. In plain sight: The role of alpha-1-antitrypsin in COVID-19 pathogenesis and therapeutics. bioRxiv 2020, Preprint. [Google Scholar] [CrossRef]
- Bergin, D.A.; Reeves, E.P.; Hurley, K.; Wolfe, R.; Jameel, R.; Fitzgerald, S.; McElvaney, N.G. The circulating proteinase inhibitor α-1 antitrypsin regulates neutrophil degranulation and autoimmunity. Sci. Transl. Med. 2014, 6, 217ra1. [Google Scholar] [CrossRef]
- Berman, R.; Jiang, D.; Wu, Q.; Chu, H.W. alpha1-Antitrypsin reduces rinovirus infection in primary human airway epithelial cells exposed to cigarette smoke. Int. J. Chronic Obstr. Pulm. Dis. 2016, 11, 1279. [Google Scholar] [CrossRef]
- Stephenson, S.E.; Wilson, C.L.; Crothers, K.; Attia, E.F.; Wongtrakool, C.; Petrache, I.; Schnapp, L.M. Impact of HIV infection on α1-antitrypsin in the lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 314, L583–L592. [Google Scholar] [CrossRef]
- Hashemi, M.; Alavian, S.M.; Ghavami, S.; de Serres, F.J.; Salehi, M.; Doroudi, T.; Fard, A.H.M.; Mehrabifar, H.; Milani, B.; Shahri, S.J.S. High prevalence of Alpha 1 antitrypsin phenotypes in viral hepatitis B infected patients in Iran. Hepatol. Res. 2005, 33, 292–297. [Google Scholar] [CrossRef]
- Ishii, T.; Doi, K.; Okamoto, K.; Imamura, M.; Dohi, M.; Yamamoto, K.; Fujita, T.; Noiri, E. Neutrophil elastase contributes to acute lung injury induced by bilateral nephrectomy. Am. J. Pathol. 2010, 177, 1665–1673. [Google Scholar] [CrossRef]
- McCarthy, C.; Dunlea, D.M.; Saldova, R.; Henry, M.; Meleady, P.; McElvaney, O.J.; Marsh, B.; Rudd, P.M.; Reeves, E.P.; McElvaney, N.G. Glycosylation Repurposes Alpha-1 Antitrypsin for Resolution of Community-acquired Pneumonia. Am. J. Respir. Crit. Care Med. 2018, 197, 1346–1349. [Google Scholar] [CrossRef]
- Janciauskiene, S.M.; Nita, I.M.; Stevens, T. Alpha1-antitrypsin, old dog, new tricks. Alpha1-antitrypsin exerts in vitro anti-inflammatory activity in human monocytes by elevating cAMP. J. Biol. Chem. 2007, 282, 8573–8582. [Google Scholar] [CrossRef]
- McElvaney, O.J.; Carroll, T.P.; Franciosi, A.N.; Sweeney, J.; Hobbs, B.D.; Kowlessar, V.; Gunaratnam, C.; Reeves, E.P.; McElvaney, N.G. Consequences of Abrupt Cessation of Alpha1-Antitrypsin Replacement Therapy. N. Engl. J. Med. 2020, 382, 1478–1480. [Google Scholar] [CrossRef]
- Sanders, C.L.; Ponte, A.; Kueppers, F. The Effects of Inflammation on Alpha 1 Antitrypsin Levels in a National Screening Cohort. COPD 2018, 15, 10–16. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).