Computational Analysis Predicts Correlations among Amino Acids in SARS-CoV-2 Proteomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieving Amino Acid Sequences
2.2. Data Pre-Processing
2.3. Analysis
2.3.1. Determining Amino Acid Frequencies
2.3.2. Correlation Analyses
2.3.3. Test for Normality
2.4. Validating Correlation Analysis
3. Results and Discussion
- B.1.1.7 (Alpha)
- B.1.351 (Beta)
- P.1 (Gamma)
- B.1.617.2 (Delta) and B.1.617.1 (Kappa)
- B.1.525 (Eta)
- B.1.526 (Iota)
- C.37 (Lambda)
- BA.1 and BA.2 (both Omicron)
- BA.4 (Omicron)
- BA.5 (Omicron)
- B.1.177
- B.1.160.
3.1. The Range of Amino Acid Residues in the SARS-CoV-2 Samples
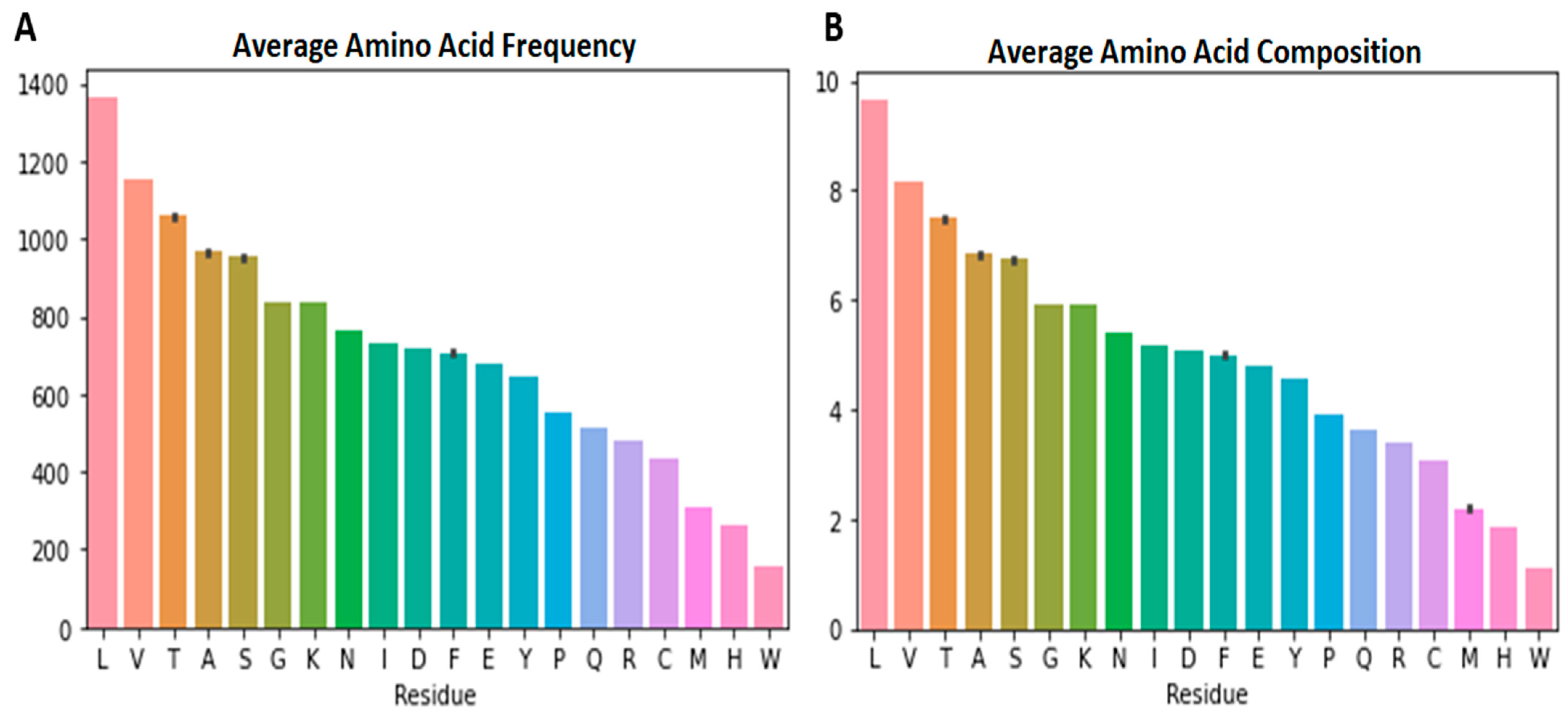
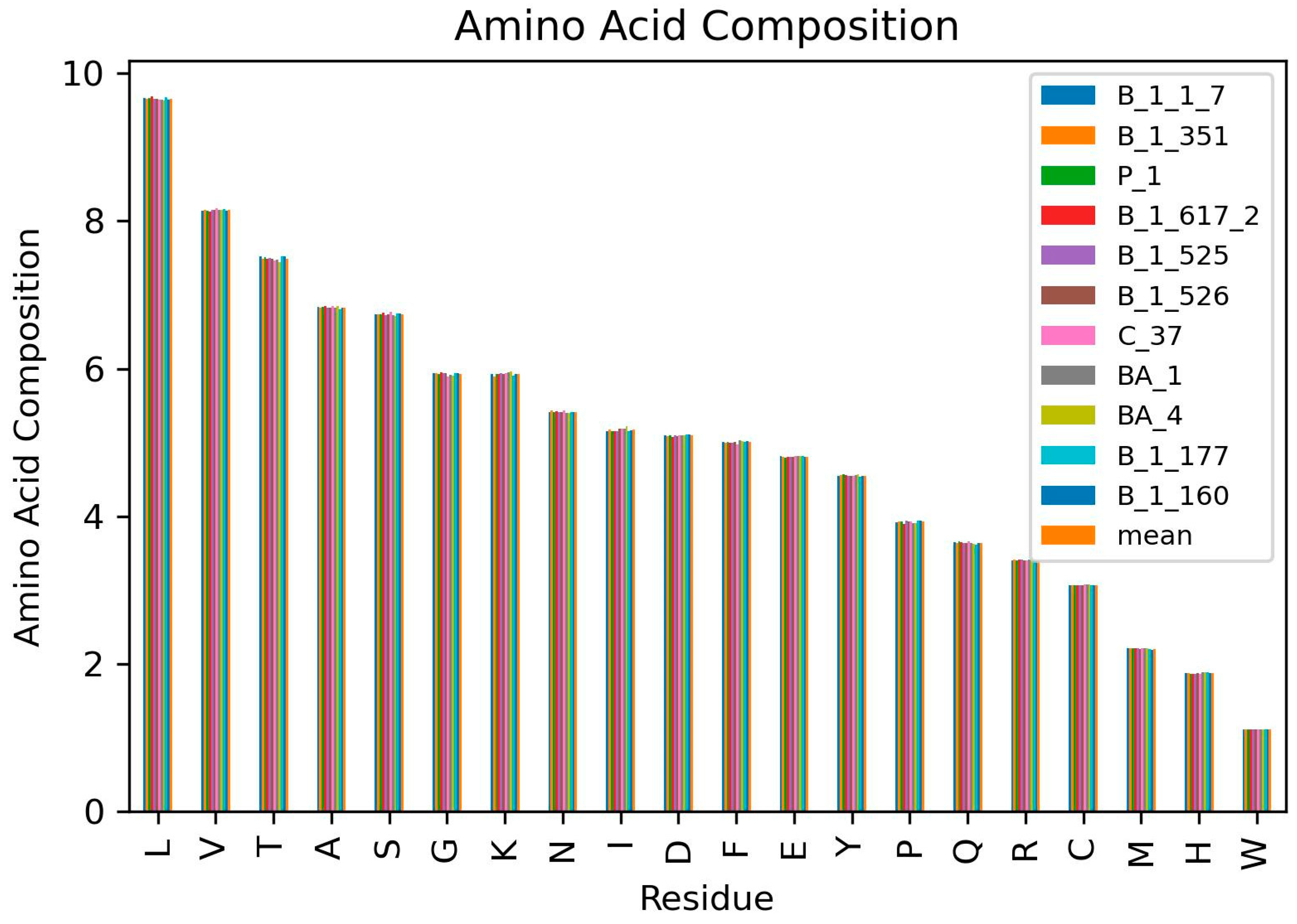
3.2. Amino Acid Frequencies per Variant
3.3. Highest and Least Represented Amino Acids in the SARS-CoV-2 Proteomes
3.4. Tryptophan Less Likely to Mutate (Intolerant to Mutation)
3.5. Lysine and Glycine Counts and Ranking
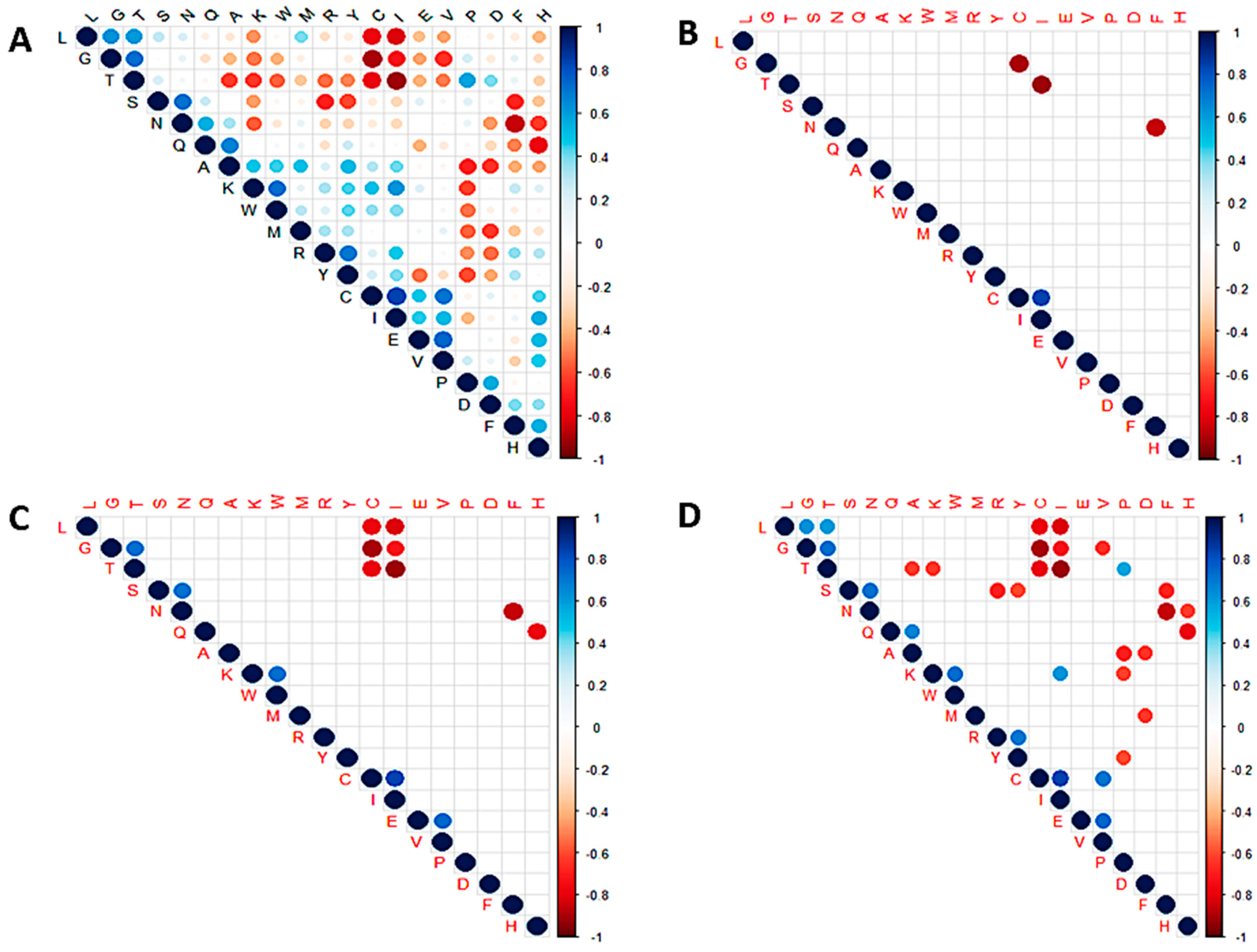
3.6. Correlation Analysis
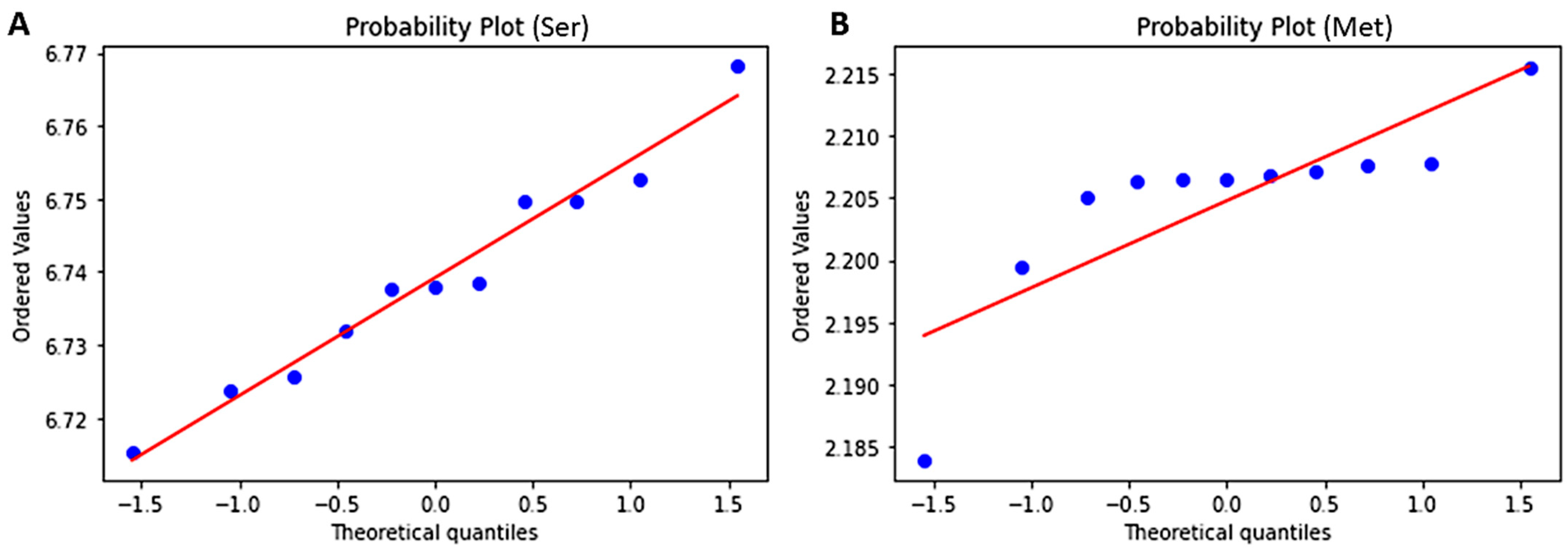
3.7. Test for Normality Using Shapiro and Predicting Probability Using Z-Score
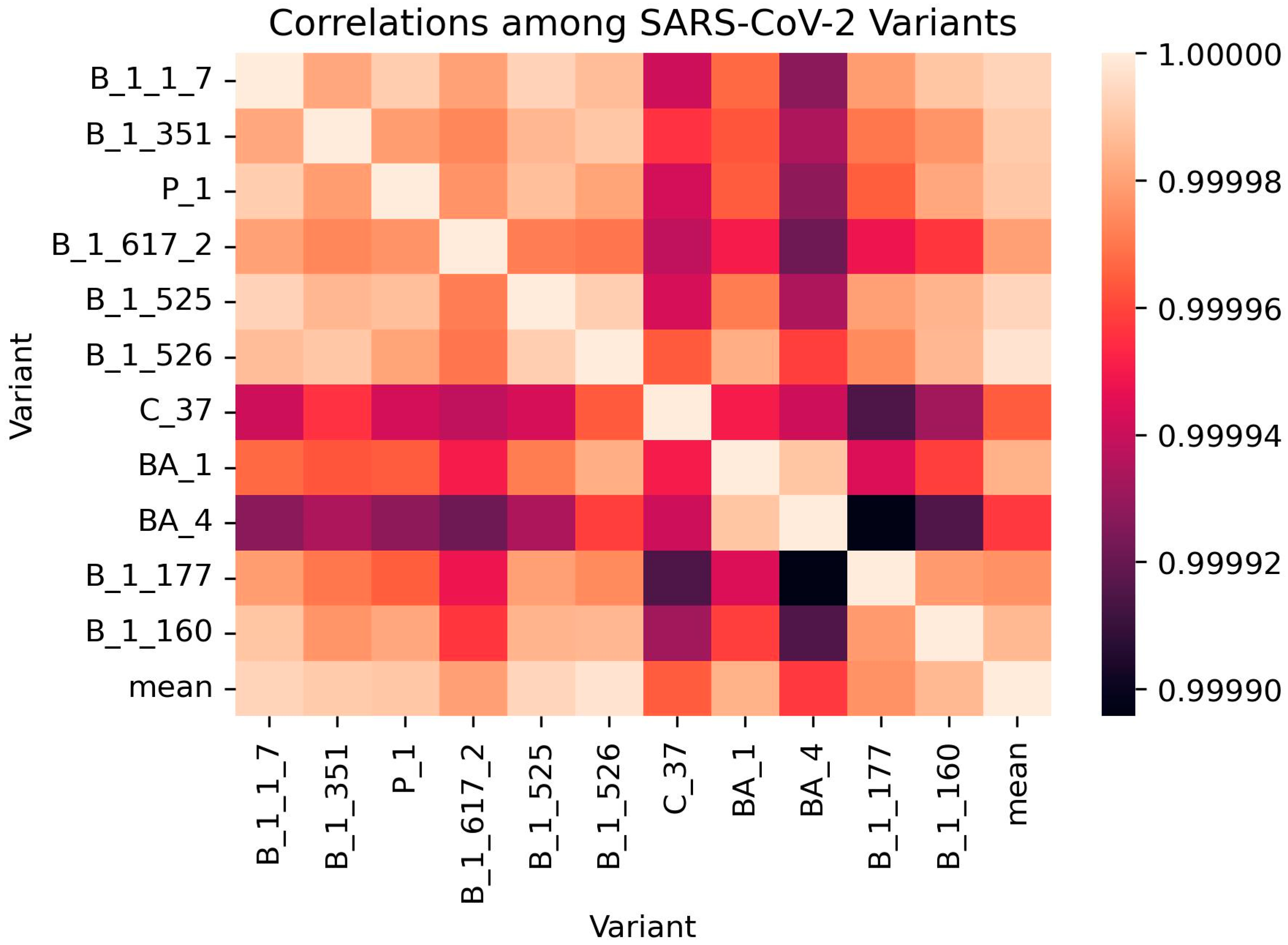
3.8. Validation of Correlation Analysis
3.9. Limitations of the Study
3.10. Potential Implications of the Study and Future Perspective
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chai, T.; Tian, M.; Yang, X.; Qiu, Z.; Lin, X.; Chen, L. Association of Circulating Cathepsin B Levels With Blood Pressure and Aortic Dilation. Front. Cardiovasc. Med. 2022, 9, 762468. [Google Scholar] [CrossRef] [PubMed]
- Sternke, M.; Tripp, K.W.; Barrick, D. Consensus sequence design as a general strategy to create hyperstable, biologically active proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 11275–11284. [Google Scholar] [CrossRef] [PubMed]
- Lermyte, F. Roles, Characteristics, and Analysis of Intrinsically Disordered Proteins: A Minireview. Life 2020, 10, 320. [Google Scholar] [CrossRef]
- Owen, I.; Shewmaker, F. The Role of Post-Translational Modifications in the Phase Transitions of Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2019, 20, 5501. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Dhanda, S.K. Bird Eye View of Protein Subcellular Localization Prediction. Life 2020, 10, 347. [Google Scholar] [CrossRef]
- Arapoglou, D.; Labropoulos, A.; Varzakas, T. Enzymes Applied in Food Technology. In Advances in Food Biochemistry; CRC Press: Boca Raton, FL, USA, 2009; pp. 101–129. ISBN 9781420007695. [Google Scholar]
- Saidijam, M.; Azizpour, S.; Patching, S.G. Comprehensive analysis of the numbers, lengths and amino acid compositions of transmembrane helices in prokaryotic, eukaryotic and viral integral membrane proteins of high-resolution structure. J. Biomol. Struct. Dyn. 2018, 36, 443–464. [Google Scholar] [CrossRef]
- Bogatyreva, N.S.; Finkelstein, A.V.; Galzitskaya, O.V. Trend of Ino Acid Composition of Proteins of Different Taxa. J. Bioinform. Comput. Biol. 2006, 4, 597–608. [Google Scholar] [CrossRef]
- Lobry, J.R. Influence of genomic G + C content on average amino-acid composition of proteins from 59 bacterial species. Gene 1997, 205, 309–316. [Google Scholar] [CrossRef]
- Bharanidharan, D.; Ramya Bhargavi, G.; Uthanumallian, K.; Gautham, N. Correlations between nucleotide frequencies and amino acid composition in 115 bacterial species. Biochem. Biophys. Res. Commun. 2004, 315, 1097–1103. [Google Scholar] [CrossRef]
- Wang, G.-Z.; Lercher, M. Biased amino acid composition in warm-blooded animals. Nat. Preced. 2009. [Google Scholar] [CrossRef]
- Cabello-Yeves, P.J.; Rodriguez-Valera, F. Marine-freshwater prokaryotic transitions require extensive changes in the predicted proteome. Microbiome 2019, 7, 117. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Savageau, M.A.; Alves, R. Relative Amino Acid Composition Signatures of Organisms and Environments. PLoS ONE 2013, 8, e77319. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Wang, J. Predicting enzyme class with Rough Sets. In Proceedings of the 2016 9th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Datong, China, 15–17 October 2016; pp. 1832–1836. [Google Scholar]
- Bao, W.; Chen, Y.; Wang, D. Prediction of protein structure classes with flexible neural tree. Biomed. Mater. Eng. 2014, 24, 3797–3806. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, Y.; Gromiha, M.M. Application of amino acid occurrence for discriminating different folding types of globular proteins. BMC Bioinform. 2007, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- Ofran, Y.; Margalit, H. Proteins of the same fold and unrelated sequences have similar amino acid composition. Proteins Struct. Funct. Bioinform. 2006, 64, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Davari, M.D.; Li, W. Recent Advances in the Prediction of Protein Structural Classes: Feature Descriptors and Machine Learning Algorithms. Crystals 2021, 11, 324. [Google Scholar] [CrossRef]
- Roy, S.; Martinez, D.; Platero, H.; Lane, T.; Werner-Washburne, M. Exploiting Amino Acid Composition for Predicting Protein-Protein Interactions. PLoS ONE 2009, 4, e7813. [Google Scholar] [CrossRef]
- Higa, R.H.; Tozzi, C.L. A simple and efficient method for predicting protein-protein interaction sites. Genet. Mol. Res. 2008, 7, 898–909. [Google Scholar] [CrossRef]
- Lee, S.; Lee, B.; Kim, D. Prediction of protein secondary structure content using amino acid composition and evolutionary information. Proteins Struct. Funct. Bioinform. 2005, 62, 1107–1114. [Google Scholar] [CrossRef]
- Majumdar, P.; Niyogi, S. SARS-CoV-2 mutations: The biological trackway towards viral fitness. Epidemiol. Infect. 2021, 149, e110. [Google Scholar] [CrossRef]
- Malik, J.A.; Ahmed, S.; Mir, A.; Shinde, M.; Bender, O.; Alshammari, F.; Ansari, M.; Anwar, S. The SARS-CoV-2 mutations versus vaccine effectiveness: New opportunities to new challenges. J. Infect. Public Health 2022, 15, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Cosar, B.; Karagulleoglu, Z.Y.; Unal, S.; Ince, A.T.; Uncuoglu, D.B.; Tuncer, G.; Kilinc, B.R.; Ozkan, Y.E.; Ozkoc, H.C.; Demir, I.N.; et al. SARS-CoV-2 Mutations and their Viral Variants. Cytokine Growth Factor Rev. 2022, 63, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, D.; Giorgi, F.M. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front. Microbiol. 2020, 11, 1800. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.G.; Mahmud, M.M.; Nazir, K.H.M.N.H.; Ueda, K. PreS1 Mutations Alter the Large HBsAg Antigenicity of a Hepatitis B Virus Strain Isolated in Bangladesh. Int. J. Mol. Sci. 2020, 21, 546. [Google Scholar] [CrossRef]
- Zhou, W.; Xu, C.; Wang, P.; Anashkina, A.A.; Jiang, Q. Impact of mutations in SARS-COV-2 spike on viral infectivity and antigenicity. Brief. Bioinform. 2022, 23, bbab375. [Google Scholar] [CrossRef]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Jin, S.; Zhang, Y.; Sun, L.; Hu, X.; Zhao, M.; Li, F.; Wang, T.; Sun, W.; et al. PB1 S524G mutation of wild bird-origin H3N8 influenza A virus enhances virulence and fitness for transmission in mammals. Emerg. Microbes Infect. 2021, 10, 1038–1051. [Google Scholar] [CrossRef]
- Mair, C.M.; Ludwig, K.; Herrmann, A.; Sieben, C. Receptor binding and pH stability—How influenza A virus hemagglutinin affects host-specific virus infection. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1153–1168. [Google Scholar] [CrossRef]
- Ozono, S.; Zhang, Y.; Ode, H.; Sano, K.; Tan, T.S.; Imai, K.; Miyoshi, K.; Kishigami, S.; Ueno, T.; Iwatani, Y.; et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021, 12, 848. [Google Scholar] [CrossRef]
- Yan, D.; Wang, B.; Shi, Y.; Ni, X.; Wu, X.; Li, X.; Liu, X.; Wang, H.; Su, X.; Teng, Q.; et al. A Single Mutation at Position 120 in the Envelope Protein Attenuates Tembusu Virus in Ducks. Viruses 2022, 14, 447. [Google Scholar] [CrossRef]
- Li, J.; Du, P.; Yang, L.; Zhang, J.; Song, C.; Chen, D.; Song, Y.; Ding, N.; Hua, M.; Han, K.; et al. Two-step fitness selection for intra-host variations in SARS-CoV-2. Cell Rep. 2022, 38, 110205. [Google Scholar] [CrossRef] [PubMed]
- Dodds, W.J. Coronavirus SARS-CoV-2 (COVID-19) and Companion Animal Pets. J. Immunol. Allergy 2020, 1, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Laine, P.; Nihtilä, H.; Mustanoja, E.; Lyyski, A.; Ylinen, A.; Hurme, J.; Paulin, L.; Jokiranta, S.; Auvinen, P.; Meri, T. SARS-CoV-2 variant with mutations in N gene affecting detection by widely used PCR primers. J. Med. Virol. 2022, 94, 1227–1231. [Google Scholar] [CrossRef] [PubMed]
- Correia, V.; Abecasis, A.B.; Rebelo-de-Andrade, H. Molecular footprints of selective pressure in the neuraminidase gene of currently circulating human influenza subtypes and lineages. Virology 2018, 522, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Prachanronarong, K.L.; Canale, A.S.; Liu, P.; Somasundaran, M.; Hou, S.; Poh, Y.-P.; Han, T.; Zhu, Q.; Renzette, N.; Zeldovich, K.B.; et al. Mutations in Influenza a Virus Neuraminidase and Hemagglutinin Confer Resistance against a Broadly Neutralizing Hemagglutinin Stem Antibody. J. Virol. 2019, 93, e01639-18. [Google Scholar] [CrossRef]
- Lau, S.-Y.; Wang, P.; Mok, B.W.-Y.; Zhang, A.J.; Chu, H.; Lee, A.C.-Y.; Deng, S.; Chen, P.; Chan, K.-H.; Song, W.; et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020, 9, 837–842. [Google Scholar] [CrossRef]
- Ning, T.; Nie, J.; Huang, W.; Li, C.; Li, X.; Liu, Q.; Zhao, H.; Wang, Y. Antigenic drift of influenza A(H7N9) virus hemagglutinin. J. Infect. Dis. 2019, 219, 19–25. [Google Scholar] [CrossRef]
- Yewdell, J.W. Antigenic drift: Understanding COVID-19. Immunity 2021, 54, 2681–2687. [Google Scholar] [CrossRef]
- Carey, L.B. RNA polymerase errors cause splicing defects and can be regulated by differential expression of RNA polymerase subunits. eLife 2015, 4, e09945. [Google Scholar] [CrossRef]
- Fisun, A.Y.; Cherkashin, D.V.; Tyrenko, V.V.; Zhdanov, C.V.; Kozlov, C.V. Role of renin-angiotensin-aldosterone system in the interaction with coronavirus SARS-CoV-2 and in the development of strategies for prevention and treatment of new coronavirus infection (COVID-19). Arterial’naya Gipertenz. Arter. Hypertens. 2020, 26, 248–262. [Google Scholar] [CrossRef]
- Yin, X.; Popa, H.; Stapon, A.; Bouda, E.; Garcia-Diaz, M. Fidelity of Ribonucleotide Incorporation by the SARS-CoV-2 Replication Complex. J. Mol. Biol. 2023, 435, 167973. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.D.; Beck, A.S.; Widen, S.G.; Wood, T.G.; Higgs, S.; Barrett, A.D.T. Structural and nonstructural genes contribute to the genetic diversity of RNA viruses. MBio 2018, 9, e01871-18. [Google Scholar] [CrossRef] [PubMed]
- Cruz-González, A.; Muñoz-Velasco, I.; Cottom-Salas, W.; Becerra, A.; Campillo-Balderas, J.A.; Hernández-Morales, R.; Vázquez-Salazar, A.; Jácome, R.; Lazcano, A. Structural analysis of viral ExoN domains reveals polyphyletic hijacking events. PLoS ONE 2021, 16, e0246981. [Google Scholar] [CrossRef] [PubMed]
- Villa, T.G.; Abril, A.G.; Sánchez, S.; de Miguel, T.; Sánchez-Pérez, A. Animal and human RNA viruses: Genetic variability and ability to overcome vaccines. Arch. Microbiol. 2021, 203, 443–464. [Google Scholar] [CrossRef]
- Mattenberger, F.; Vila-Nistal, M.; Geller, R. Increased RNA virus population diversity improves adaptability. Sci. Rep. 2021, 11, 6824. [Google Scholar] [CrossRef]
- Bouvet, M.; Imbert, I.; Subissi, L.; Gluais, L.; Canard, B.; Decroly, E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA 2012, 109, 9372–9377. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell 2020, 79, 710–727. [Google Scholar] [CrossRef]
- Moeller, N.H.; Shi, K.; Demir, Ö.; Belica, C.; Banerjee, S.; Yin, L.; Durfee, C.; Amaro, R.E.; Aihara, H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. USA 2022, 119, e2106379119. [Google Scholar] [CrossRef]
- Amicone, M.; Borges, V.; Alves, M.J.; Isidro, J.; Zé-Zé, L.; Duarte, S.; Vieira, L.; Guiomar, R.; Gomes, J.P.; Gordo, I. Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evol. Med. Public Health 2022, 10, 142–155. [Google Scholar] [CrossRef]
- Koyama, T.; Platt, D.; Parida, L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020, 98, 495–504. [Google Scholar] [CrossRef]
- Yeh, T.Y.; Contreras, G.P. Emerging Viral Mutants in Australia Suggest RNA Recombination Event in the SARS-CoV-2 Genome. Med. J. Aust. 2020, 1. Available online: https://www.mja.com.au/journal/2020/213/1/emerging-viral-mutants-australia-suggest-rna-recombination-event-sars-cov-2 (accessed on 7 February 2023). [CrossRef]
- da Silva Francisco Junior, R.; de Almeida, L.G.P.; Lamarca, A.P.; Cavalcante, L.; Martins, Y.; Gerber, A.L.; de C. Guimarães, A.P.; Salviano, R.B.; dos Santos, F.L.; de Oliveira, T.H.; et al. Emergence of within-Host SARS-CoV-2 Recombinant Genome after Coinfection by Gamma and Delta Variants: A Case Report. Front. Public Health 2022, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Doxey, A.C.; Benjamin, J.M.T.; Mansfield, M.J.; Subudhi, S.; Hirota, J.A.; Miller, M.S.; Andrew, G.M.; Mubareka, S.; Mossman, K. Predicting the recombination potential of severe acute respiratory syndrome coronavirus 2 and Middle East respiratory syndrome coronavirus. J. Gen. Virol. 2021, 101, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Ball, P. Pattern Formation in Nature: Physical Constraints and Self-Organising Characteristics. Archit. Des. 2012, 82, 22–27. [Google Scholar] [CrossRef]
- Ball, P. Pattern of life. Nature 2000. [Google Scholar] [CrossRef]
- Rodriguez, S. Hardy–Weinberg Law. In Brenner’s Encyclopedia of Genetics; Elsevier: Amsterdam, The Netherlands, 2013; pp. 396–398. ISBN 9780080961569. [Google Scholar]
- Bhole, G.; Shukla, A.; Mahesh, T.S. Benford Analysis: A useful paradigm for spectroscopic analysis. Chem. Phys. Lett. 2014, 639, 36–40. [Google Scholar] [CrossRef]
- Berger, A.; Hill, T.P. The mathematics of Benford’s law: A primer. Stat. Methods Appt. 2021, 30, 779–795. [Google Scholar] [CrossRef]
- Das, J.K.; Das, P.; Ray, K.K.; Choudhury, P.P.; Jana, S.S. Mathematical Characterization of Protein Sequences Using Patterns as Chemical Group Combinations of Amino Acids. PLoS ONE 2016, 11, e0167651. [Google Scholar] [CrossRef]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bolton, E.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012, 35, D5–D12. [Google Scholar] [CrossRef]
- O’Toole, Á.; Pybus, O.G.; Abram, M.E.; Kelly, E.J.; Rambaut, A. Pango lineage designation and assignment using SARS-CoV-2 spike gene nucleotide sequences. BMC Genom. 2022, 23, 121. [Google Scholar] [CrossRef]
- Embarak, D.O. Data Analysis and Visualization Using Python; Apress: Berkeley, CA, USA, 2018; ISBN 978-1-4842-4108-0. [Google Scholar]
- Ghosh, S.; Neha, K.; Praveen Kumar, Y. Data Wrangling Using Python. Int. J. Recent Technol. Eng. 2019, 8, 3491–3495. [Google Scholar] [CrossRef]
- Löytynoja, A.; Goldman, N. webPRANK: A phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinform. 2010, 11, 579. [Google Scholar] [CrossRef] [PubMed]
- Holland, L.A.; Kaelin, E.A.; Maqsood, R.; Estifanos, B.; Wu, L.I.; Varsani, A.; Halden, R.U.; Hogue, B.G.; Scotch, M.; Lim, E.S. An 81 base-pair deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (Jan-Mar 2020). medRxiv Prepr. Serv. Health Sci. 2020. [Google Scholar] [CrossRef]
- Yuan, F.; Wang, L.; Fang, Y.; Wang, L. Global SNP analysis of 11,183 SARS-CoV-2 strains reveals high genetic diversity. Transbound. Emerg. Dis. 2021, 68, 3288–3304. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Delerce, J.; Burel, E.; Beye, M.; Fournier, P.-E.; Levasseur, A.; Lagier, J.-C.; Raoult, D. Occurrence of a substitution or deletion of SARS-CoV-2 spike amino acid 677 in various lineages in Marseille, France. Virus Genes 2022, 58, 53–58. [Google Scholar] [CrossRef]
- Kemp, S.A.; Meng, B.; Ferriera, I.A.T.M.; Datir, R.; Harvey, W.T.; Collier, D.A.; Lytras, S.; Papa, G.; Carabelli, A.; Kenyon, J.; et al. Recurrent Emergence and Transmission of a SARS-CoV-2 Spike Deletion H69/V70. SSRN Electron. J. 2021. [Google Scholar] [CrossRef]
- Benedetti, F.; Snyder, G.A.; Giovanetti, M.; Angeletti, S.; Gallo, R.C.; Ciccozzi, M.; Zella, D. Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J. Transl. Med. 2020, 18, 329. [Google Scholar] [CrossRef]
- Loureiro, C.L.; Jaspe, R.C.; D’Angelo, P.; Zambrano, J.L.; Rodriguez, L.; Alarcon, V.; Delgado, M.; Aguilar, M.; Garzaro, D.; Rangel, H.R.; et al. SARS-CoV-2 genetic diversity in Venezuela: Predominance of D614G variants and analysis of one outbreak. PLoS ONE 2021, 16, e0247196. [Google Scholar] [CrossRef]
- Zanchi, F.B.; Mariúba, L.A.; Nascimento, V.; Souza, V.; Corado, A.; Nascimento, F.; Costa, Á.K.; Duarte, D.; Silva, G.; Mejía, M.; et al. Structural analysis of SARS-Cov-2 nonstructural protein 1 polymorphisms found in the Brazilian Amazon. Exp. Biol. Med. 2021, 246, 2332–2337. [Google Scholar] [CrossRef]
- Lesbon, J.C.C.; Poleti, M.D.; de Mattos Oliveira, E.C.; Patané, J.S.L.; Clemente, L.G.; Viala, V.L.; Ribeiro, G.; Giovanetti, M.; de Alcantara, L.C.J.; de Lima, L.P.O.; et al. Nucleocapsid (N) gene mutations of sars-cov-2 can affect real-time rt-pcr diagnostic and impact false-negative results. Viruses 2021, 13, 2474. [Google Scholar] [CrossRef]
- Finkel, Y.; Mizrahi, O.; Nachshon, A.; Weingarten-Gabbay, S.; Morgenstern, D.; Yahalom-Ronen, Y.; Tamir, H.; Achdout, H.; Stein, D.; Israeli, O.; et al. The coding capacity of SARS-CoV-2. Nature 2021, 589, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Firth, A.E. A putative new SARS-CoV protein, 3c, encoded in an ORF overlapping ORF3a. J. Gen. Virol. 2020, 101, 1085–1089. [Google Scholar] [CrossRef] [PubMed]
- Patten, M.L. The Mean and Standard Deviation. In Understanding Research Methods; Routledge: New York, NY, USA, 2017; pp. 137–138. [Google Scholar] [CrossRef]
- Krick, T.; Verstraete, N.; Alonso, L.G.; Shub, D.A.; Ferreiro, D.U.; Shub, M.; Sánchez, I.E. Amino Acid Metabolism Conflicts with Protein Diversity. Mol. Biol. Evol. 2014, 31, 2905–2912. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Mohanta, Y.K.; Avula, S.K.; Nongbet, A.; Al-Harrasi, A. Virtual 2D map of cyanobacterial proteomes. PLoS ONE 2022, 17, e0275148. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Khan, A.; Hashem, A.; Abd_Allah, E.F.; Al-Harrasi, A. The molecular mass and isoelectric point of plant proteomes. BMC Genom. 2019, 20, 631. [Google Scholar] [CrossRef]
- Miseta, A.; Csutora, P. Relationship Between the Occurrence of Cysteine in Proteins and the Complexity of Organisms. Mol. Biol. Evol. 2000, 17, 1232–1239. [Google Scholar] [CrossRef]
- Haque, S.M.; Ashwaq, O.; Sarief, A.; Azad John Mohamed, A.K. A comprehensive review about SARS-CoV-2. Future Virol. 2020, 15, 625–648. [Google Scholar] [CrossRef]
- Masters, P.S. The Molecular Biology of Coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [CrossRef]
- San Martín, C. Structure and Assembly of Complex Viruses. Subcell. Biochem. 2013, 68, 329–360. [Google Scholar] [CrossRef]
- Serwin, K.; Ossowski, A.; Szargut, M.; Cytacka, S.; Urbańska, A.; Majchrzak, A.; Niedźwiedź, A.; Czerska, E.; Pawińska-Matecka, A.; Gołąb, J.; et al. Molecular Evolution and Epidemiological Characteristics of SARS COV-2 in (Northwestern) Poland. Viruses 2021, 13, 1295. [Google Scholar] [CrossRef]
- Hodcroft, E.B.; Zuber, M.; Nadeau, S.; Vaughan, T.G.; Crawford, K.H.D.; Althaus, C.L.; Reichmuth, M.L.; Bowen, J.E.; Walls, A.C.; Corti, D.; et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 2021, 595, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Rafael Ciges-Tomas, J.; Franco, M.L.; Vilar, M. Identification of a guanine-specific pocket in the protein N of SARS-CoV-2. Commun. Biol. 2022, 5, 711. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Wang, S.; Zhang, J.; Zhang, K.; Zhang, F.; Wang, H.; Xie, S.; Hu, W.; Gu, L. Structure-Based Primer Design Minimizes the Risk of PCR Failure Caused by SARS-CoV-2 Mutations. Front. Cell. Infect. Microbiol. 2021, 11, 741147. [Google Scholar] [CrossRef] [PubMed]
- Santiveri, C.M.; Jiménez, M.A. Tryptophan residues: Scarce in proteins but strong stabilizers of β-hairpin peptides. Biopolymers 2010, 94, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Bielecki, M.; Wójtowicz, H.; Olczak, T. Differential roles of tryptophan residues in conformational stability of Porphyromonas gingivalis HmuY hemophore. BMC Biochem. 2014, 15, 2. [Google Scholar] [CrossRef]
- Bansal, K.; Kumar, S. Mutational cascade of SARS-CoV-2 leading to evolution and emergence of omicron variant. Virus Res. 2022, 315, 198765. [Google Scholar] [CrossRef]
- Rajpal, V.R.; Sharma, S.; Kumar, A.; Chand, S.; Joshi, L.; Chandra, A.; Babbar, S.; Goel, S.; Raina, S.N.; Shiran, B. “Is Omicron mild”? Testing this narrative with the mutational landscape of its three lineages and response to existing vaccines and therapeutic antibodies. J. Med. Virol. 2022, 94, 3521–3539. [Google Scholar] [CrossRef]
- Barton, M.I.; MacGowan, S.A.; Kutuzov, M.A.; Dushek, O.; Barton, G.J.; van der Merwe, P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. eLife 2021, 10, e70658. [Google Scholar] [CrossRef]
- Liu, H.; Wei, P.; Zhang, Q.; Chen, Z.; Aviszus, K.; Downing, W.; Peterson, S.; Reynoso, L.; Downey, G.P.; Frankel, S.K.; et al. 501Y.V2 and 501Y.V3 variants of SARS-CoV-2 lose binding to bamlanivimab in vitro. MAbs 2021, 13, 1919285. [Google Scholar] [CrossRef]
- Teng, S.; Sobitan, A.; Rhoades, R.; Liu, D.; Tang, Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief. Bioinform. 2021, 22, 1239–1253. [Google Scholar] [CrossRef]
- Arora, P.; Pöhlmann, S.; Hoffmann, M. Mutation D614G increases SARS-CoV-2 transmission. Signal Transduct. Target. Ther. 2021, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Aleem, A.; Akbar Samad, A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 and Novel Therapeutics against Coronavirus (COVID-19); StatPearls: Treasure Island, FL, USA, 2021. [Google Scholar]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Wolter, N.; Jassat, W.; Walaza, S.; Welch, R.; Moultrie, H.; Groome, M.J.; Amoako, D.G.; Everatt, J.; Bhiman, J.N.; Scheepers, C.; et al. Clinical severity of SARS-CoV-2 Omicron BA.4 and BA.5 lineages compared to BA.1 and Delta in South Africa. Nat. Commun. 2022, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D.G.; et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022, 28, 1785–1790. [Google Scholar] [CrossRef]
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Du, Q.; Wei, D.; Chou, K.-C. Correlations of amino acids in proteins. Peptides 2003, 24, 1863–1869. [Google Scholar] [CrossRef]
- Rehman, S.; Mahmood, T.; Aziz, E.; Batool, R. Identification of novel mutations in SARS-COV-2 isolates from Turkey. Arch. Virol. 2020, 165, 2937–2944. [Google Scholar] [CrossRef]
- Nasser, H.; Shimizu, R.; Ito, J.; Matsuno, K.; Nao, N.; Sawa, H.; Kishimoto, M.; Tanaka, S.; Tsuda, M.; Wang, L.; et al. Monitoring fusion kinetics of viral and target cell membranes in living cells using a SARS-CoV-2 spike-protein-mediated membrane fusion assay. STAR Protoc. 2022, 3, 101773. [Google Scholar] [CrossRef]
- Gong, Y.-N.; Tsao, K.-C.; Hsiao, M.-J.; Huang, C.-G.; Huang, P.-N.; Huang, P.-W.; Lee, K.-M.; Liu, Y.-C.; Yang, S.-L.; Kuo, R.-L.; et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microbes Infect. 2020, 9, 1457–1466. [Google Scholar] [CrossRef]
- Su, Y.C.F.; Anderson, D.E.; Young, B.E.; Linster, M.; Zhu, F.; Jayakumar, J.; Zhuang, Y.; Kalimuddin, S.; Low, J.G.H.; Tan, C.W.; et al. Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. MBio 2020, 11, 4. [Google Scholar] [CrossRef]
- Young, B.E.; Fong, S.-W.; Chan, Y.-H.; Mak, T.-M.; Ang, L.W.; Anderson, D.E.; Lee, C.Y.-P.; Amrun, S.N.; Lee, B.; Goh, Y.S.; et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet 2020, 396, 603–611. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, W. Fast-spreading SARS-CoV-2 variants: Challenges to and new design strategies of COVID-19 vaccines. Signal Transduct. Target. Ther. 2021, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Zinzula, L. Lost in deletion: The enigmatic ORF8 protein of SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021, 538, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.-M.; Tsai, J.-L.; Hung, J.-N.; Chen, I.-H.; Chen, S.-T.; Tsai, M.-H. The ORF8 Protein of SARS-CoV-2 Modulates the Spike Protein and Its Implications in Viral Transmission. Front. Microbiol. 2022, 13, 883597. [Google Scholar] [CrossRef] [PubMed]
- Erster, O.; Mendelson, E.; Kabat, A.; Levy, V.; Mannasse, B.; Assraf, H.; Azar, R.; Ali, Y.; Bucris, E.; Bar-Ilan, D.; et al. Specific Detection of SARS-CoV-2 Variants B.1.1.7 (Alpha) and B.1.617.2 (Delta) Using a One-Step Quantitative PCR Assay. Microbiol. Spectr. 2022, 10, 2. [Google Scholar] [CrossRef]
- Azad, G.K.; Khan, P.K. Variations in Orf3a protein of SARS-CoV-2 alter its structure and function. Biochem. Biophys. Rep. 2021, 26, 100933. [Google Scholar] [CrossRef]
- Hassan, S.S.; Attrish, D.; Ghosh, S.; Choudhury, P.P.; Roy, B. Pathogenic perspective of missense mutations of ORF3a protein of SARS-CoV-2. Virus Res. 2021, 300, 198441. [Google Scholar] [CrossRef]
- Sengupta, A.; Hassan, S.S.; Choudhury, P.P. Clade GR and clade GH isolates of SARS-CoV-2 in Asia show highest amount of SNPs. Infect. Genet. Evol. 2021, 89, 104724. [Google Scholar] [CrossRef]
- Abbas, Q.; Kusakin, A.; Sharrouf, K.; Jyakhwo, S.; Komissarov, A.S. Follow-up investigation and detailed mutational characterization of the SARS-CoV-2 Omicron variant lineages (BA.1, BA.2, BA.3 and BA.1.1). bioRxiv 2022. [Google Scholar] [CrossRef]
- Zeng, H.-L.; Dichio, V.; Rodríguez Horta, E.; Thorell, K.; Aurell, E. Global analysis of more than 50,000 SARS-CoV-2 genomes reveals epistasis between eight viral genes. Proc. Natl. Acad. Sci. USA 2020, 117, 31519–31526. [Google Scholar] [CrossRef]
- Velazquez-Salinas, L.; Zarate, S.; Eberl, S.; Gladue, D.P.; Novella, I.; Borca, M.V. Positive Selection of ORF1ab, ORF3a, and ORF8 Genes Drives the Early Evolutionary Trends of SARS-CoV-2 during the 2020 COVID-19 Pandemic. Front. Microbiol. 2020, 11, 550674. [Google Scholar] [CrossRef] [PubMed]

| Nextstrain Clade | Pango Lineage | WHO Label | Number of Files | Remarks | |
|---|---|---|---|---|---|
| Before Cleaning | After Cleaning | ||||
| 20I (Alpha, V1) | B.1.1.7 | Alpha | 830 | 706 | - |
| 20H (Beta, V2) | B.1.351 | Beta | 16 | 4 | - |
| 20J (Gamma, V3) | P.1 | Gamma | 79 | 73 | - |
| 21A (Delta) | B.1.617.2 | Delta | 100 | 43 | - |
| 21B (Kappa) | B.1.617.1 | Kappa | 1 | - | Part of B.1.617.2 |
| 21C (Epsilon) | B.1.427, B.1.429 | Epsilon | 0 | - | - |
| 21D (Eta) | B.1.525 | Eta | 13 | 13 | - |
| 21F (Iota) | B.1.526 | Iota | 3085 | 773 | - |
| 21G (Lambda) | C.37 | Lambda | 19 | 8 | - |
| 21H (Mu) | B.1.621 | Mu | 0 | - | - |
| 21K (Omicron) | BA.1 | Omicron | 26 | 13 | - |
| 21L (Omicron) | BA.2 | Omicron | 23 | - | Part of BA.1 |
| 22A (Omicron) | BA.4 | Omicron | 3 | 2 | - |
| 22B (Omicron) | BA.5 | Omicron | 23 | 0 | - |
| 22C (Omicron) | BA.2.12.1 | Omicron | 0 | - | - |
| 22D (Omicron) | BA.2.75 | Omicron | 0 | - | - |
| 20E (EU1) | B.1.177 | 1 | 1 | - | |
| 20B/S:732A | B.1.1.519 | 0 | - | - | |
| 20A/S:126A | B.1.620 | 0 | - | - | |
| 20A.EU2 | B.1.160 | 1 | 1 | - | |
| 20A/S:439K | B.1.258 | 0 | - | - | |
| 20A/S:98F | B.1.221 | 0 | - | - | |
| 20C/S:80Y | B.1.367 | 0 | - | - | |
| 20B/S:626S | B.1.1.277 | 0 | - | - | |
| 20B/S:1122L | B.1.1.302 | 0 | - | - | |
| Residues | SARS-CoV-2 Variants | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B.1.1.7 | B.1.351 | P.1 | B.1.617.2 | B.1.525 | B.1.526 | C.37 | BA.1 | BA.4 | B.1.177 | B.1.160 | |
| M | 311.508 | 312 | 311.973 | 312.512 | 312 | 310.979 | 312 | 311.923 | 312 | 312 | 309 |
| E | 679.016 | 680 | 678.123 | 677.721 | 680 | 680.049 | 681 | 680.385 | 680 | 681 | 680 |
| S | 950.745 | 952.75 | 951.849 | 952.535 | 950.615 | 952.779 | 956.75 | 950.846 | 949 | 955 | 955 |
| L | 1363.72 | 1365 | 1366.75 | 1366.58 | 1365.69 | 1364.88 | 1363.12 | 1363.12 | 1362 | 1370 | 1365 |
| V | 1148.64 | 1153 | 1150.74 | 1147.14 | 1152.62 | 1152.57 | 1154.62 | 1152.77 | 1152 | 1154 | 1151 |
| P | 553.214 | 556 | 555.438 | 549.953 | 557 | 556.211 | 555 | 552.923 | 551.5 | 557 | 557 |
| G | 838.057 | 840 | 837.781 | 839.465 | 840 | 839.507 | 833 | 836.615 | 835 | 840 | 841 |
| F | 707.078 | 707 | 708.301 | 703.884 | 707 | 707.248 | 703 | 710.385 | 709.5 | 709 | 710 |
| N | 763.834 | 767.5 | 764.932 | 765.395 | 765.154 | 765.133 | 767.5 | 763 | 763 | 765 | 766 |
| K | 836.659 | 834 | 837.63 | 836.535 | 838.846 | 838.477 | 838.75 | 841.615 | 842 | 836 | 838 |
| T | 1060.65 | 1059 | 1061.75 | 1056.91 | 1061 | 1058.85 | 1055 | 1056.46 | 1052 | 1064 | 1064 |
| H | 263.683 | 265 | 263.192 | 262.047 | 264 | 265.706 | 264 | 265.846 | 267 | 267 | 265 |
| Q | 514.037 | 515 | 517.534 | 514.14 | 514 | 513.877 | 517 | 513.538 | 513 | 511 | 515 |
| R | 479.565 | 483 | 480.959 | 480.837 | 482 | 481.034 | 480 | 482.923 | 483 | 481 | 481 |
| D | 718.683 | 718.5 | 720.493 | 716.093 | 720 | 719.699 | 720 | 719.769 | 719.5 | 722 | 722 |
| A | 964.416 | 965.75 | 966.329 | 965.698 | 964.923 | 965.577 | 967.375 | 965.462 | 967 | 963 | 965 |
| C | 432.669 | 434 | 433.904 | 432.047 | 434 | 433.875 | 435 | 434.154 | 435 | 434 | 434 |
| Y | 640.779 | 644 | 645.589 | 642.93 | 643 | 642.849 | 642 | 644.538 | 645.5 | 642 | 643 |
| I | 727.377 | 731.75 | 728.986 | 727.116 | 729.154 | 732.816 | 733.875 | 734 | 737 | 729 | 731 |
| W | 156.469 | 156.75 | 156.959 | 156.721 | 157 | 156.951 | 157 | 157 | 157 | 157 | 157 |
| Residues | Average Amino Acid Composition (AAC) of SARS-CoV-2 Variants | Avg AAC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B.1.1.7 | B.1.351 | P.1 | B.1.617.2 | B.1.525 | B.1.526 | C.37 | BA.1 | BA.4 | B.1.177 | B.1.160 | ||
| M | 2.208 | 2.207 | 2.206 | 2.215 | 2.207 | 2.199 | 2.207 | 2.206 | 2.208 | 2.205 | 2.184 | 2.205 |
| E | 4.812 | 4.810 | 4.796 | 4.804 | 4.810 | 4.810 | 4.817 | 4.813 | 4.812 | 4.813 | 4.806 | 4.809 |
| S | 6.738 | 6.738 | 6.732 | 6.753 | 6.724 | 6.739 | 6.768 | 6.726 | 6.715 | 6.750 | 6.750 | 6.739 |
| L | 9.664 | 9.653 | 9.666 | 9.688 | 9.660 | 9.653 | 9.643 | 9.647 | 9.638 | 9.683 | 9.647 | 9.658 |
| V | 8.140 | 8.154 | 8.139 | 8.132 | 8.153 | 8.152 | 8.168 | 8.154 | 8.152 | 8.156 | 8.135 | 8.149 |
| P | 3.921 | 3.932 | 3.928 | 3.899 | 3.940 | 3.934 | 3.926 | 3.911 | 3.902 | 3.937 | 3.937 | 3.924 |
| G | 5.939 | 5.941 | 5.925 | 5.951 | 5.941 | 5.938 | 5.893 | 5.918 | 5.909 | 5.937 | 5.944 | 5.930 |
| F | 5.011 | 5 | 5.009 | 4.990 | 5.001 | 5.002 | 4.973 | 5.025 | 5.021 | 5.011 | 5.018 | 5.005 |
| N | 5.413 | 5.428 | 5.410 | 5.426 | 5.412 | 5.411 | 5.429 | 5.397 | 5.310 | 5.407 | 5.414 | 5.413 |
| K | 5.929 | 5.898 | 5.924 | 5.930 | 5.933 | 5.930 | 5.933 | 5.953 | 5.958 | 5.909 | 5.923 | 5.929 |
| T | 7.517 | 7.489 | 7.509 | 7.492 | 7.505 | 7.489 | 7.463 | 7.473 | 7.444 | 7.520 | 7.520 | 7.493 |
| H | 1.869 | 1.874 | 1.861 | 1.858 | 1.867 | 1.879 | 1.868 | 1.880 | 1.889 | 1.887 | 1.873 | 1.873 |
| Q | 3.643 | 3.642 | 3.660 | 3.645 | 3.636 | 3.634 | 3.657 | 3.632 | 3.630 | 3.612 | 3.640 | 3.639 |
| R | 3.399 | 3.416 | 3.402 | 3.409 | 3.409 | 3.402 | 3.396 | 3.416 | 3.418 | 3.400 | 3.400 | 3.406 |
| D | 5.093 | 5.081 | 5.096 | 5.076 | 5.093 | 5.090 | 5.093 | 5.091 | 5.091 | 5.103 | 5.103 | 5.092 |
| A | 6.834 | 6.830 | 6.834 | 6.846 | 6.825 | 6.829 | 6.843 | 6.829 | 6.843 | 6.806 | 6.820 | 6.831 |
| C | 3.066 | 3.069 | 3.069 | 3.063 | 3.070 | 3.069 | 3.077 | 3.071 | 3.078 | 3.067 | 3.067 | 3.070 |
| Y | 4.541 | 4.554 | 4.566 | 4.558 | 4.548 | 4.547 | 4.542 | 4.559 | 4.568 | 4.537 | 4.544 | 4.551 |
| I | 5.155 | 5.175 | 5.156 | 5.155 | 5.157 | 5.183 | 5.192 | 5.192 | 5.215 | 5.152 | 5.166 | 5.173 |
| W | 1.109 | 1.109 | 1.110 | 1.111 | 1.110 | 1.110 | 1.111 | 1.110 | 1.111 | 1.110 | 1.110 | 1.110 |
| Residues | Skewness | Kurtosis | Shapiro Wilk Test | ||
|---|---|---|---|---|---|
| Statistic | p-Value | Test Statistic (W) | p-Value | ||
| M | 2.860 | 0.004 | 2.968 | 0.728 | 0.001 |
| E | −1.834 | 0.067 | 0.978 | 0.906 | 0.217 |
| S | 0.495 | 0.621 | −0.510 | 0.973 | 0.922 |
| L | 1.153 | 0.249 | −0.661 | 0.931 | 0.425 |
| V | 0.040 | 0.968 | −0.772 | 0.922 | 0.336 |
| P | −1.301 | 0.193 | −0.896 | 0.884 | 0.115 |
| G | −1.747 | 0.081 | −0.095 | 0.879 | 0.102 |
| F | −1.463 | 0.143 | 0.167 | 0.936 | 0.473 |
| N | 0.251 | 0.802 | −0.936 | 0.923 | 0.348 |
| K | 0.050 | 0.960 | −0.203 | 0.938 | 0.496 |
| T | −1.128 | 0.259 | −0.643 | 0.923 | 0.341 |
| H | 0.278 | 0.781 | −0.961 | 0.968 | 0.865 |
| Q | −0.562 | 0.574 | 0.210 | 0.947 | 0.610 |
| R | 0.631 | 0.528 | −1.401 | 0.892 | 0.147 |
| D | −0.877 | 0.380 | −0.116 | 0.911 | 0.250 |
| A | −1.226 | 0.220 | 0.102 | 0.938 | 0.493 |
| C | 1.338 | 0.181 | −0.087 | 0.891 | 0.143 |
| Y | 0.568 | 0.570 | −1.220 | 0.937 | 0.481 |
| I | 1.383 | 0.167 | −0.520 | 0.873 | 0.085 |
| W | −1.054 | 0.292 | −0.759 | 0.927 | 0.384 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Broni, E.; Miller, W.A., III. Computational Analysis Predicts Correlations among Amino Acids in SARS-CoV-2 Proteomes. Biomedicines 2023, 11, 512. https://doi.org/10.3390/biomedicines11020512
Broni E, Miller WA III. Computational Analysis Predicts Correlations among Amino Acids in SARS-CoV-2 Proteomes. Biomedicines. 2023; 11(2):512. https://doi.org/10.3390/biomedicines11020512
Chicago/Turabian StyleBroni, Emmanuel, and Whelton A. Miller, III. 2023. "Computational Analysis Predicts Correlations among Amino Acids in SARS-CoV-2 Proteomes" Biomedicines 11, no. 2: 512. https://doi.org/10.3390/biomedicines11020512
APA StyleBroni, E., & Miller, W. A., III. (2023). Computational Analysis Predicts Correlations among Amino Acids in SARS-CoV-2 Proteomes. Biomedicines, 11(2), 512. https://doi.org/10.3390/biomedicines11020512







