AgRP Neuron-Specific Ablation Represses Appetite, Energy Intake, and Somatic Growth in Larval Zebrafish
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation and Maintenance of Transgenic Zebrafish
2.2. Promoter Cloning
2.3. Transcription Factor-Binding Site Prediction
2.4. MTZ Treatment
2.5. Quantitative Reverse Transcription PCR (RT-qPCR)
2.6. Whole Mount In Situ Hybridization (WISH)
2.7. Quantification of Food Intake
2.8. Whole-Mount Oil Red O Staining
2.9. Statistical Analysis
3. Results
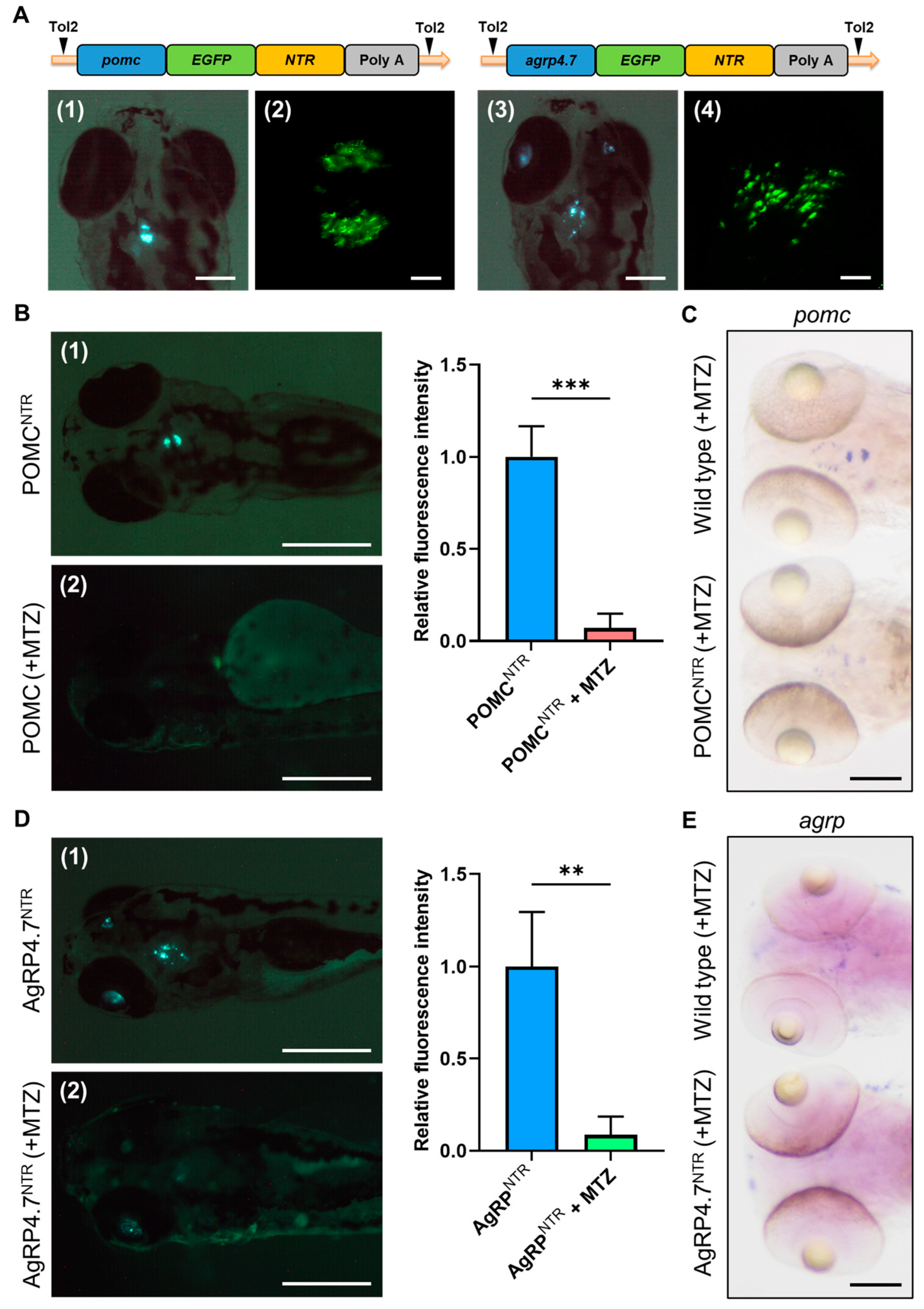
3.1. Generation of Transgenic AgRP4.7NTR Zebrafish Lines
3.2. EGFP-NTR Fusion Protein Causes Target Neural Cell Ablation
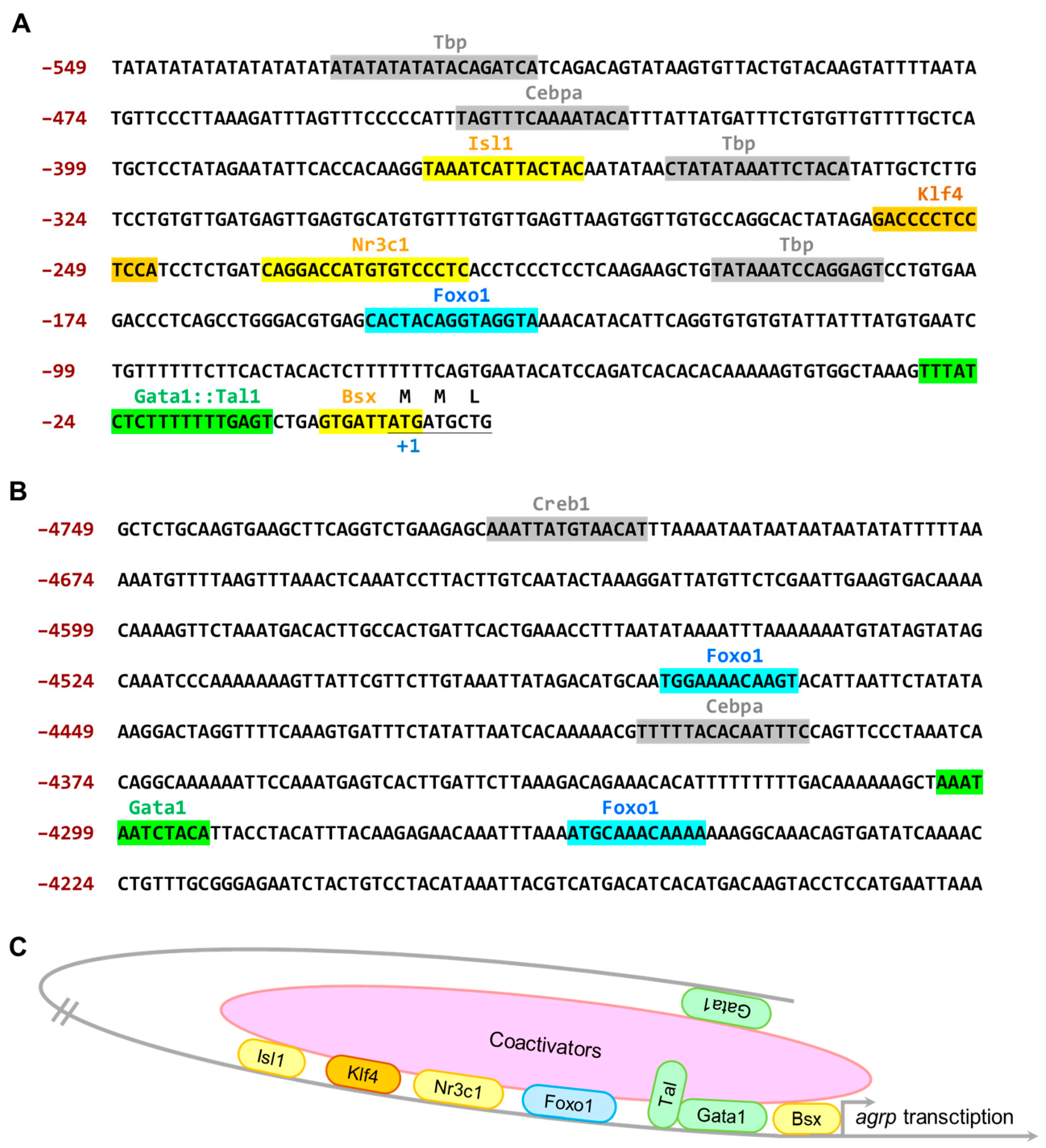
3.3. The 4.7 kb Zebrafish agrp Promoter Has Several Essential Transcription Factor-Binding Sites That Are Exclusively Expressed in the AgRP Neuron
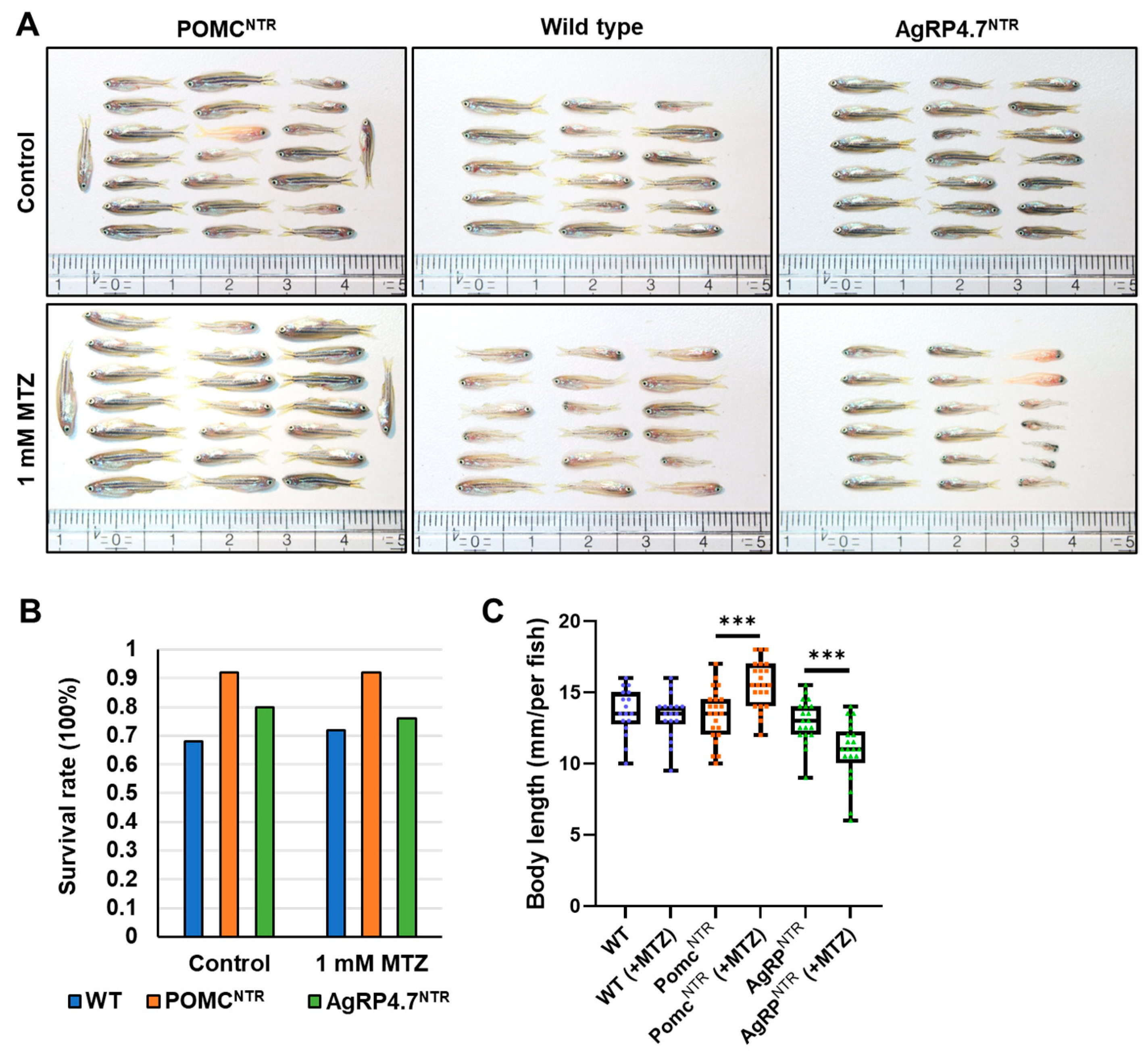
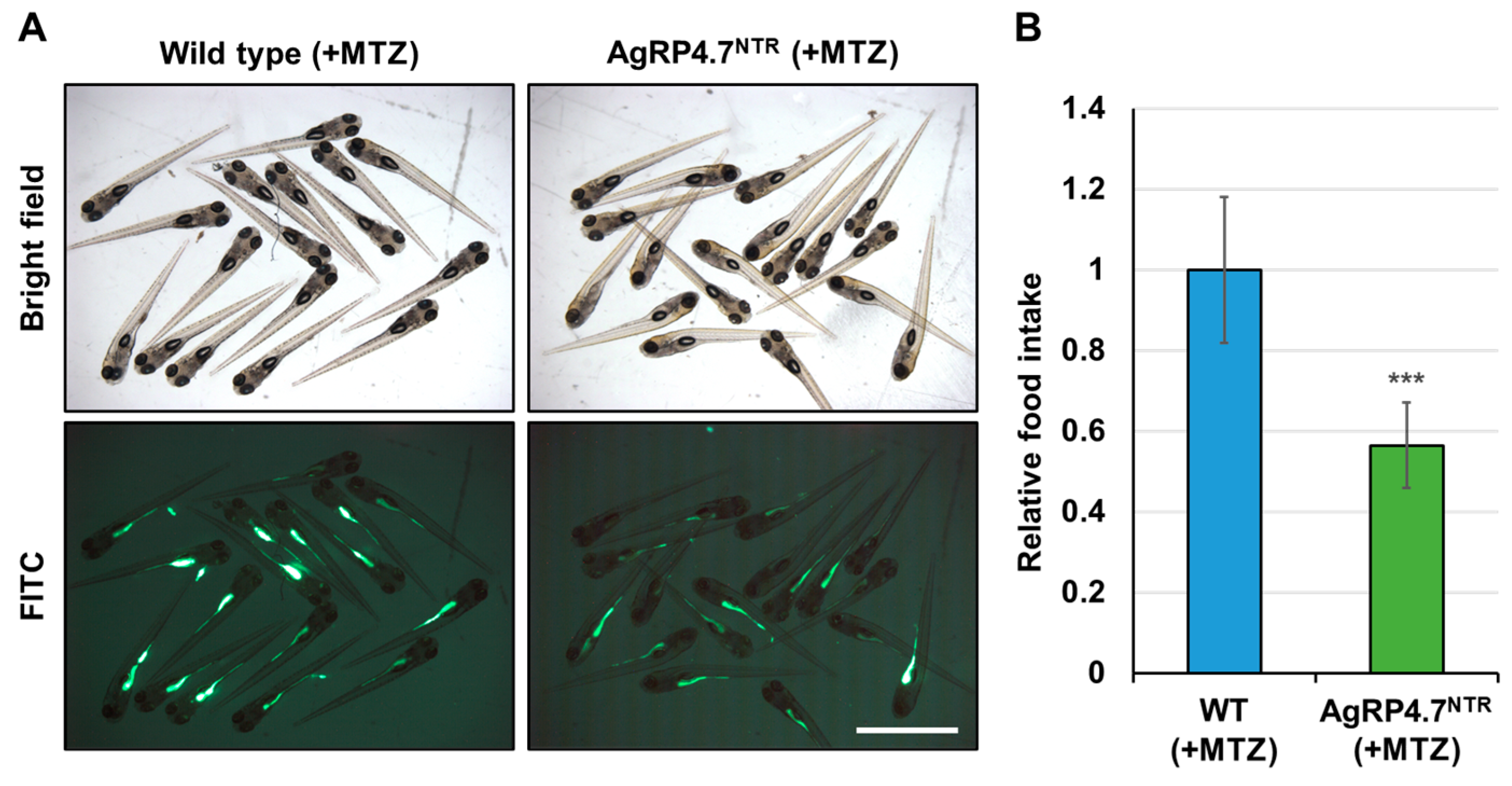
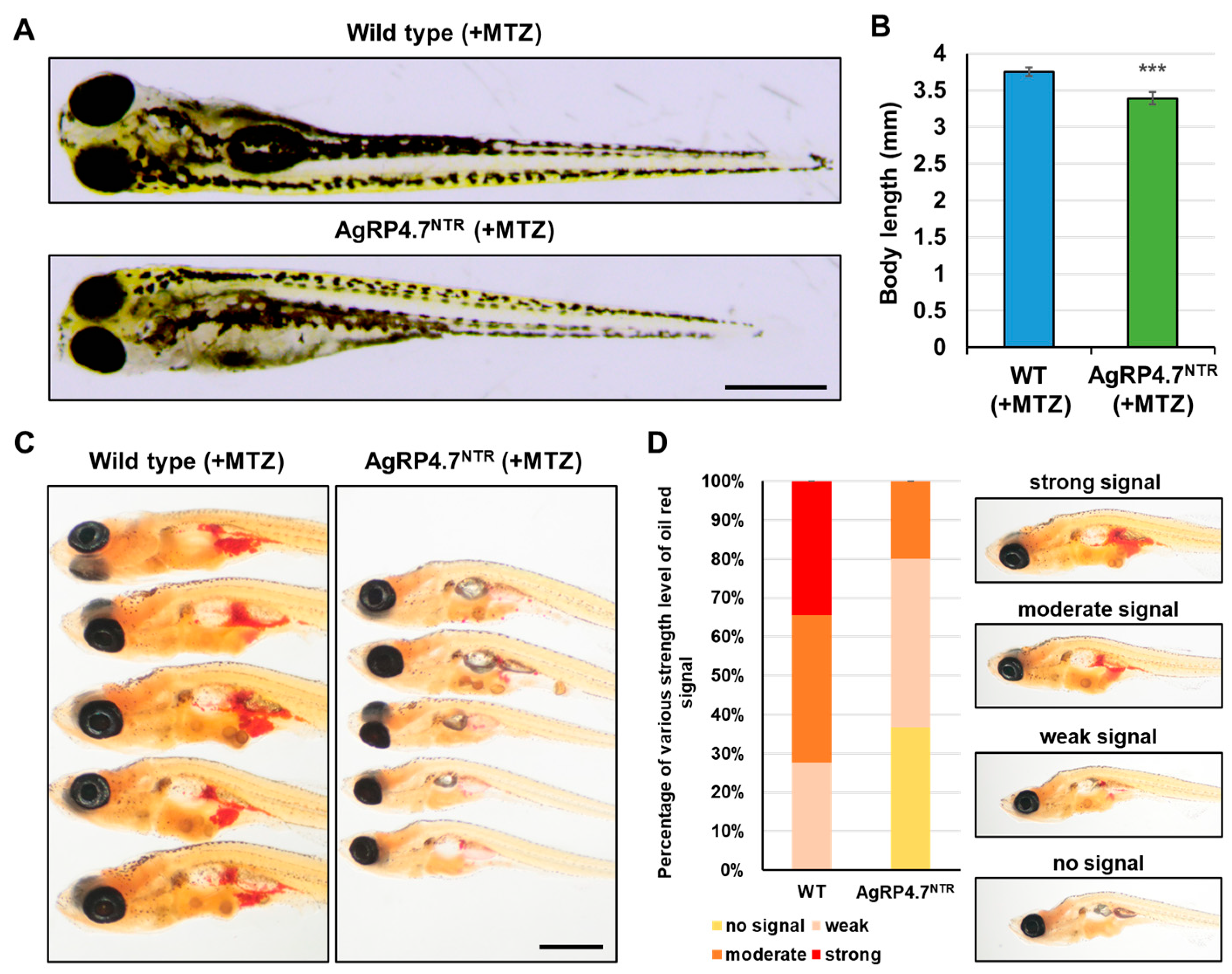
3.4. Targeted Ablation of AgRP Neuron Represses Food Intake and Body Growth in Zebrafish Larvae
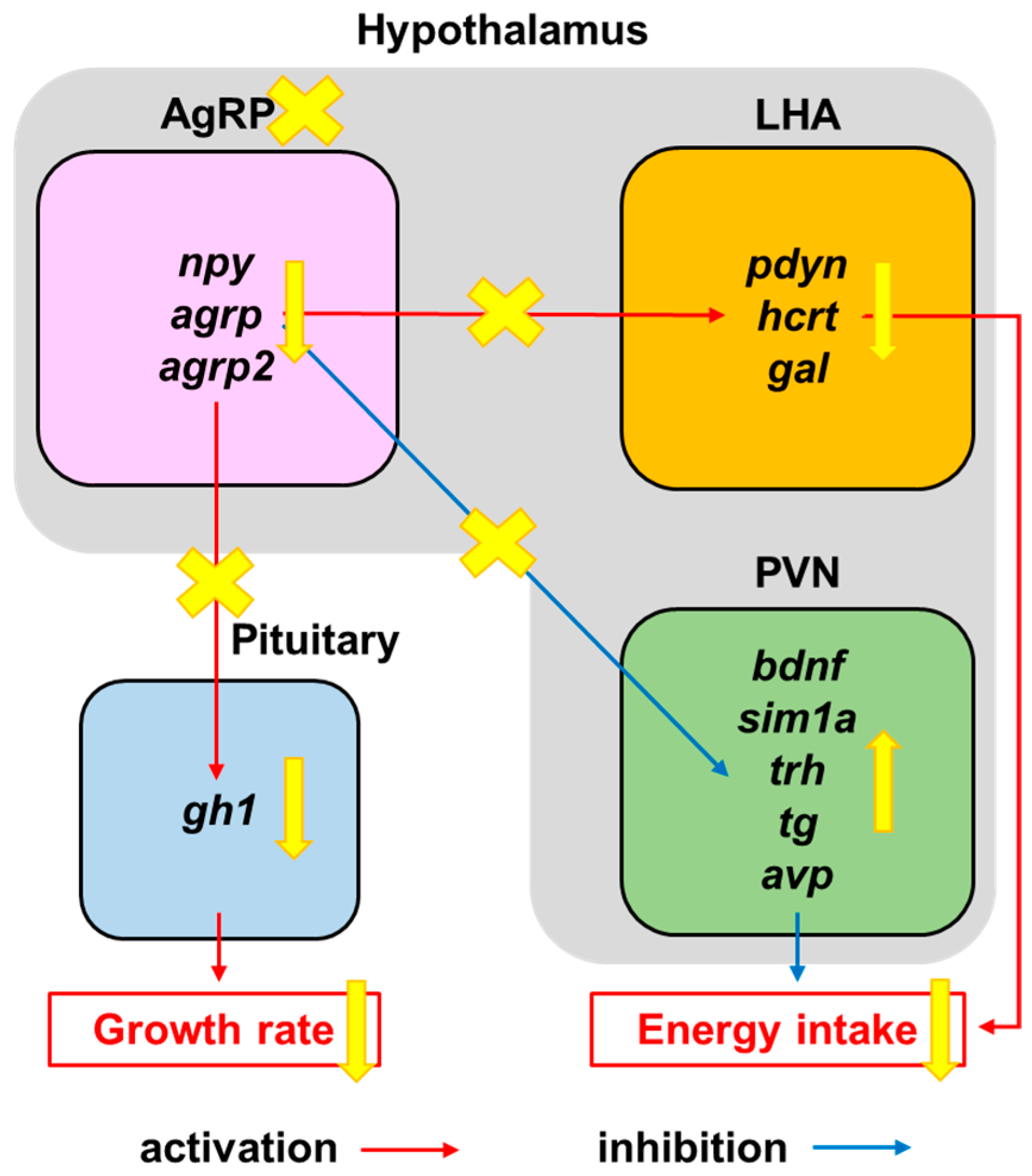
3.5. Defective AgRP Neuron Suppresses the Expression of Orexigenic Genes and Growth Hormones While Increasing the Anorexigenic Gene Expression in Zebrafish Larvae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Funahashi, H.; Hori, T.; Shimoda, Y.; Mizushima, H.; Ryushi, T.; Katoh, S.; Shioda, S. Morphological evidence for neural interactions between leptin and orexin in the hypothalamus. Regul. Pept. 2000, 92, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Wilding, J.P. Neuropeptides and appetite control. Diabet. Med. 2002, 19, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Coll, A.P.; Farooqi, I.S.; O’Rahilly, S. The hormonal control of food intake. Cell 2007, 129, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Palmiter, R.D. GABAergic signaling by AgRP neurons prevents anorexia via a melanocortin-independent mechanism. Eur. J. Pharmacol. 2011, 660, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.M. Leptin and the endocrine control of energy balance. Nat. Metab. 2019, 1, 754–764. [Google Scholar] [CrossRef]
- Cheung, C.C.; Clifton, D.K.; Steiner, R.A. Proopiomelanocortin neurons are direct targets for leptin in the hypothalamus. Endocrinology 1997, 138, 4489–4492. [Google Scholar] [CrossRef]
- Cowley, M.A.; Smart, J.L.; Rubinstein, M.; Cerdan, M.G.; Diano, S.; Horvath, T.L.; Cone, R.D.; Low, M.J. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 2001, 411, 480–484. [Google Scholar] [CrossRef]
- Konner, A.C.; Janoschek, R.; Plum, L.; Jordan, S.D.; Rother, E.; Ma, X.; Xu, C.; Enriori, P.; Hampel, B.; Barsh, G.S.; et al. Insulin action in AgRP-expressing neurons is required for suppression of hepatic glucose production. Cell Metab. 2007, 5, 438–449. [Google Scholar] [CrossRef]
- Broberger, C.; Johansen, J.; Johansson, C.; Schalling, M.; Hokfelt, T. The neuropeptide Y/agouti gene-related protein (AGRP) brain circuitry in normal, anorectic, and monosodium glutamate-treated mice. Proc. Natl. Acad. Sci. USA 1998, 95, 15043–15048. [Google Scholar] [CrossRef]
- Shutter, J.R.; Graham, M.; Kinsey, A.C.; Scully, S.; Luthy, R.; Stark, K.L. Hypothalamic expression of ART, a novel gene related to agouti, is up-regulated in obese and diabetic mutant mice. Genes Dev. 1997, 11, 593–602. [Google Scholar] [CrossRef]
- Ollmann, M.M.; Wilson, B.D.; Yang, Y.K.; Kerns, J.A.; Chen, Y.; Gantz, I.; Barsh, G.S. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science 1997, 278, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Werther, G.A.; Hogg, A.; Oldfield, B.J.; McKinley, M.J.; Figdor, R.; Allen, A.M.; Mendelsohn, F.A. Localization and characterization of insulin receptors in rat brain and pituitary gland using in vitro autoradiography and computerized densitometry. Endocrinology 1987, 121, 1562–1570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Proenca, R.; Maffei, M.; Barone, M.; Leopold, L.; Friedman, J.M. Positional cloning of the mouse obese gene and its human homologue. Nature 1994, 372, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Sternson, S.M.; Atasoy, D. Agouti-related protein neuron circuits that regulate appetite. Neuroendocrinology 2014, 100, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Vong, L.; Ye, C.; Yang, Z.; Choi, B.; Chua, S., Jr.; Lowell, B.B. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 2011, 71, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Enriori, P.J.; Evans, A.E.; Sinnayah, P.; Jobst, E.E.; Tonelli-Lemos, L.; Billes, S.K.; Glavas, M.M.; Grayson, B.E.; Perello, M.; Nillni, E.A.; et al. Diet-induced obesity causes severe but reversible leptin resistance in arcuate melanocortin neurons. Cell Metab. 2007, 5, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Creemers, J.W.; Pritchard, L.E.; Gyte, A.; Le Rouzic, P.; Meulemans, S.; Wardlaw, S.L.; Zhu, X.; Steiner, D.F.; Davies, N.; Armstrong, D.; et al. Agouti-related protein is posttranslationally cleaved by proprotein convertase 1 to generate agouti-related protein (AGRP)83-132: Interaction between AGRP83-132 and melanocortin receptors cannot be influenced by syndecan-3. Endocrinology 2006, 147, 1621–1631. [Google Scholar] [CrossRef]
- Zagmutt, S.; Mera, P.; Soler-Vazquez, M.C.; Herrero, L.; Serra, D. Targeting AgRP neurons to maintain energy balance: Lessons from animal models. Biochem. Pharmacol. 2018, 155, 224–232. [Google Scholar] [CrossRef]
- Ollmann, M.M.; Lamoreux, M.L.; Wilson, B.D.; Barsh, G.S. Interaction of Agouti protein with the melanocortin 1 receptor in vitro and in vivo. Genes Dev. 1998, 12, 316–330. [Google Scholar] [CrossRef]
- Yeo, G.S.H.; Chao, D.H.M.; Siegert, A.M.; Koerperich, Z.M.; Ericson, M.D.; Simonds, S.E.; Larson, C.M.; Luquet, S.; Clarke, I.; Sharma, S.; et al. The melanocortin pathway and energy homeostasis: From discovery to obesity therapy. Mol. Metab. 2021, 48, 101206. [Google Scholar] [CrossRef]
- Yeo, G.S.; Heisler, L.K. Unraveling the brain regulation of appetite: Lessons from genetics. Nat. Neurosci. 2012, 15, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Baldini, G.; Phelan, K.D. The melanocortin pathway and control of appetite-progress and therapeutic implications. J. Endocrinol. 2019, 241, R1–R33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Forlano, P.M.; Cone, R.D. AgRP and POMC neurons are hypophysiotropic and coordinately regulate multiple endocrine axes in a larval teleost. Cell Metab. 2012, 15, 256–264. [Google Scholar] [CrossRef]
- Qian, S.; Chen, H.; Weingarth, D.; Trumbauer, M.E.; Novi, D.E.; Guan, X.; Yu, H.; Shen, Z.; Feng, Y.; Frazier, E.; et al. Neither agouti-related protein nor neuropeptide Y is critically required for the regulation of energy homeostasis in mice. Mol. Cell. Biol. 2002, 22, 5027–5035. [Google Scholar] [CrossRef] [PubMed]
- Wortley, K.E.; Anderson, K.D.; Yasenchak, J.; Murphy, A.; Valenzuela, D.; Diano, S.; Yancopoulos, G.D.; Wiegand, S.J.; Sleeman, M.W. Agouti-related protein-deficient mice display an age-related lean phenotype. Cell Metab. 2005, 2, 421–427. [Google Scholar] [CrossRef]
- Wu, Q.; Howell, M.P.; Palmiter, R.D. Ablation of neurons expressing agouti-related protein activates fos and gliosis in postsynaptic target regions. J. Neurosci. 2008, 28, 9218–9226. [Google Scholar] [CrossRef]
- Wu, Q.; Whiddon, B.B.; Palmiter, R.D. Ablation of neurons expressing agouti-related protein, but not melanin concentrating hormone, in leptin-deficient mice restores metabolic functions and fertility. Proc. Natl. Acad. Sci. USA 2012, 109, 3155–3160. [Google Scholar] [CrossRef]
- Shainer, I.; Michel, M.; Marquart, G.D.; Bhandiwad, A.A.; Zmora, N.; Ben-Moshe Livne, Z.; Zohar, Y.; Hazak, A.; Mazon, Y.; Forster, D.; et al. Agouti-Related Protein 2 Is a New Player in the Teleost Stress Response System. Curr. Biol. 2019, 29, 2009–2019.e7. [Google Scholar] [CrossRef]
- Lohr, H.; Hess, S.; Pereira, M.M.A.; Reinoss, P.; Leibold, S.; Schenkel, C.; Wunderlich, C.M.; Kloppenburg, P.; Bruning, J.C.; Hammerschmidt, M. Diet-Induced Growth Is Regulated via Acquired Leptin Resistance and Engages a Pomc-Somatostatin-Growth Hormone Circuit. Cell Rep. 2018, 23, 1728–1741. [Google Scholar] [CrossRef]
- Song, Y.; Cone, R.D. Creation of a genetic model of obesity in a teleost. FASEB J. 2007, 21, 2042–2049. [Google Scholar] [CrossRef]
- Hsieh, Y.W.; Tsai, Y.W.; Lai, H.H.; Lai, C.Y.; Lin, C.Y.; Her, G.M. Depletion of Alpha-Melanocyte-Stimulating Hormone Induces Insatiable Appetite and Gains in Energy Reserves and Body Weight in Zebrafish. Biomedicines 2021, 9, 941. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, C.; Uchida, A.; Chuang, J.C.; Walker, A.; Liu, T.; Osborne-Lawrence, S.; Mason, B.L.; Mosher, C.; Berglund, E.D.; et al. Arcuate AgRP neurons mediate orexigenic and glucoregulatory actions of ghrelin. Mol. Metab. 2014, 3, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Luquet, S.; Perez, F.A.; Hnasko, T.S.; Palmiter, R.D. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science 2005, 310, 683–685. [Google Scholar] [CrossRef] [PubMed]
- Suster, M.L.; Abe, G.; Schouw, A.; Kawakami, K. Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 2011, 6, 1998–2021. [Google Scholar] [CrossRef] [PubMed]
- Suster, M.L.; Sumiyama, K.; Kawakami, K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genom. 2009, 10, 477. [Google Scholar] [CrossRef]
- Shainer, I.; Buchshtab, A.; Hawkins, T.A.; Wilson, S.W.; Cone, R.D.; Gothilf, Y. Novel hypophysiotropic AgRP2 neurons and pineal cells revealed by BAC transgenesis in zebrafish. Sci. Rep. 2017, 7, 44777. [Google Scholar] [CrossRef]
- Hagerman, G.F.; Noel, N.C.; Cao, S.Y.; DuVal, M.G.; Oel, A.P.; Allison, W.T. Rapid Recovery of Visual Function Associated with Blue Cone Ablation in Zebrafish. PLoS ONE 2016, 11, e0166932. [Google Scholar] [CrossRef]
- Stoddard, M.; Huang, C.; Enyedi, B.; Niethammer, P. Live imaging of leukocyte recruitment in a zebrafish model of chemical liver injury. Sci. Rep. 2019, 9, 28. [Google Scholar] [CrossRef]
- Liu, N.A.; Huang, H.; Yang, Z.; Herzog, W.; Hammerschmidt, M.; Lin, S.; Melmed, S. Pituitary corticotroph ontogeny and regulation in transgenic zebrafish. Mol. Endocrinol. 2003, 17, 959–966. [Google Scholar] [CrossRef]
- Jowett, T. Double in situ hybridization techniques in zebrafish. Methods 2001, 23, 345–358. [Google Scholar] [CrossRef]
- Shimada, Y.; Hirano, M.; Nishimura, Y.; Tanaka, T. A high-throughput fluorescence-based assay system for appetite-regulating gene and drug screening. PLoS ONE 2012, 7, e52549. [Google Scholar] [CrossRef] [PubMed]
- Suster, M.L.; Kikuta, H.; Urasaki, A.; Asakawa, K.; Kawakami, K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol. Biol. 2009, 561, 41–63. [Google Scholar] [CrossRef] [PubMed]
- Curado, S.; Stainier, D.Y.; Anderson, R.M. Nitroreductase-mediated cell/tissue ablation in zebrafish: A spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc. 2008, 3, 948–954. [Google Scholar] [CrossRef]
- Song, Y.; Golling, G.; Thacker, T.L.; Cone, R.D. Agouti-related protein (AGRP) is conserved and regulated by metabolic state in the zebrafish, Danio rerio. Endocrine 2003, 22, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, S.; Lee, S.K.; Lee, J.W. The LIM-homeobox transcription factor Isl1 plays crucial roles in the development of multiple arcuate nucleus neurons. Development 2016, 143, 3763–3773. [Google Scholar] [CrossRef]
- Lee, B.; Kim, S.G.; Kim, J.; Choi, K.Y.; Lee, S.; Lee, S.K.; Lee, J.W. Brain-specific homeobox factor as a target selector for glucocorticoid receptor in energy balance. Mol. Cell. Biol. 2013, 33, 2650–2658. [Google Scholar] [CrossRef]
- Schredelseker, T.; Veit, F.; Dorsky, R.I.; Driever, W. Bsx Is Essential for Differentiation of Multiple Neuromodulatory Cell Populations in the Secondary Prosencephalon. Front. Neurosci. 2020, 14, 525. [Google Scholar] [CrossRef]
- Deng, W.; Lee, J.; Wang, H.; Miller, J.; Reik, A.; Gregory, P.D.; Dean, A.; Blobel, G.A. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 2012, 149, 1233–1244. [Google Scholar] [CrossRef]
- Kitamura, T.; Feng, Y.; Kitamura, Y.I.; Chua, S.C., Jr.; Xu, A.W.; Barsh, G.S.; Rossetti, L.; Accili, D. Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat. Med. 2006, 12, 534–540. [Google Scholar] [CrossRef]
- Imbernon, M.; Sanchez-Rebordelo, E.; Gallego, R.; Gandara, M.; Lear, P.; Lopez, M.; Dieguez, C.; Nogueiras, R. Hypothalamic KLF4 mediates leptin’s effects on food intake via AgRP. Mol. Metab. 2014, 3, 441–451. [Google Scholar] [CrossRef]
- Nakajima, K.; Cui, Z.; Li, C.; Meister, J.; Cui, Y.; Fu, O.; Smith, A.S.; Jain, S.; Lowell, B.B.; Krashes, M.J.; et al. Gs-coupled GPCR signalling in AgRP neurons triggers sustained increase in food intake. Nat. Commun. 2016, 7, 10268. [Google Scholar] [CrossRef] [PubMed]
- Ilnytska, O.; Stutz, A.M.; Park-York, M.; York, D.A.; Ribnicky, D.M.; Zuberi, A.; Cefalu, W.T.; Argyropoulos, G. Molecular mechanisms for activation of the agouti-related protein and stimulation of appetite. Diabetes 2011, 60, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Nasif, S.; de Souza, F.S.; Gonzalez, L.E.; Yamashita, M.; Orquera, D.P.; Low, M.J.; Rubinstein, M. Islet 1 specifies the identity of hypothalamic melanocortin neurons and is critical for normal food intake and adiposity in adulthood. Proc. Natl. Acad. Sci. USA 2015, 112, E1861–E1870. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, E.; Azuma, M.; Nishiguchi, R.; Kang, K.S.; Kamijo, M.; Uchiyama, M.; Matsuda, K. Neuropeptide Y stimulates food intake in the Zebrafish, Danio rerio. J. Neuroendocrinol. 2012, 24, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Mercer, R.E.; Chee, M.J.; Colmers, W.F. The role of NPY in hypothalamic mediated food intake. Front. Neuroendocrinol. 2011, 32, 398–415. [Google Scholar] [CrossRef] [PubMed]
- Garfield, A.S.; Shah, B.P.; Burgess, C.R.; Li, M.M.; Li, C.; Steger, J.S.; Madara, J.C.; Campbell, J.N.; Kroeger, D.; Scammell, T.E.; et al. Dynamic GABAergic afferent modulation of AgRP neurons. Nat. Neurosci. 2016, 19, 1628–1635. [Google Scholar] [CrossRef]
- Krashes, M.J.; Shah, B.P.; Madara, J.C.; Olson, D.P.; Strochlic, D.E.; Garfield, A.S.; Vong, L.; Pei, H.; Watabe-Uchida, M.; Uchida, N.; et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 2014, 507, 238–242. [Google Scholar] [CrossRef]
- Merkle, F.T.; Maroof, A.; Wataya, T.; Sasai, Y.; Studer, L.; Eggan, K.; Schier, A.F. Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Development 2015, 142, 633–643. [Google Scholar] [CrossRef]
- Qualls-Creekmore, E.; Yu, S.; Francois, M.; Hoang, J.; Huesing, C.; Bruce-Keller, A.; Burk, D.; Berthoud, H.R.; Morrison, C.D.; Munzberg, H. Galanin-Expressing GABA Neurons in the Lateral Hypothalamus Modulate Food Reward and Noncompulsive Locomotion. J. Neurosci. 2017, 37, 6053–6065. [Google Scholar] [CrossRef]
- Benarroch, E.E. Brain-derived neurotrophic factor: Regulation, effects, and potential clinical relevance. Neurology 2015, 84, 1693–1704. [Google Scholar] [CrossRef]
- Marosi, K.; Mattson, M.P. BDNF mediates adaptive brain and body responses to energetic challenges. Trends Endocrinol. Metab. 2014, 25, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Xi, D.; Roizen, J.; Lai, M.; Gandhi, N.; Kublaoui, B. Paraventricular nucleus Sim1 neuron ablation mediated obesity is resistant to high fat diet. PLoS ONE 2013, 8, e81087. [Google Scholar] [CrossRef] [PubMed]
- Mullur, R.; Liu, Y.Y.; Brent, G.A. Thyroid hormone regulation of metabolism. Physiol. Rev. 2014, 94, 355–382. [Google Scholar] [CrossRef]
- Pei, H.; Sutton, A.K.; Burnett, K.H.; Fuller, P.M.; Olson, D.P. AVP neurons in the paraventricular nucleus of the hypothalamus regulate feeding. Mol. Metab. 2014, 3, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Madara, J.C.; Wu, C.; Andermann, M.L.; Lowell, B.B. Neural basis for regulation of vasopressin secretion by anticipated disturbances in osmolality. eLife 2021, 10, e66609. [Google Scholar] [CrossRef]
- Harrington, W.W.; Britt, C.S.; Wilson, J.G.; Milliken, N.O.; Binz, J.G.; Lobe, D.C.; Oliver, W.R.; Lewis, M.C.; Ignar, D.M. The Effect of PPARalpha, PPARdelta, PPARgamma, and PPARpan Agonists on Body Weight, Body Mass, and Serum Lipid Profiles in Diet-Induced Obese AKR/J Mice. PPAR Res. 2007, 2007, 97125. [Google Scholar] [CrossRef]
- Chen, H.C.; Farese, R.V., Jr. Inhibition of triglyceride synthesis as a treatment strategy for obesity: Lessons from DGAT1-deficient mice. Arterioscler Thromb. Vasc. Biol. 2005, 25, 482–486. [Google Scholar] [CrossRef]
- Ruan, H.; Pownall, H.J.; Lodish, H.F. Troglitazone antagonizes tumor necrosis factor-alpha-induced reprogramming of adipocyte gene expression by inhibiting the transcriptional regulatory functions of NF-kappaB. J. Biol. Chem. 2003, 278, 28181–28192. [Google Scholar] [CrossRef]
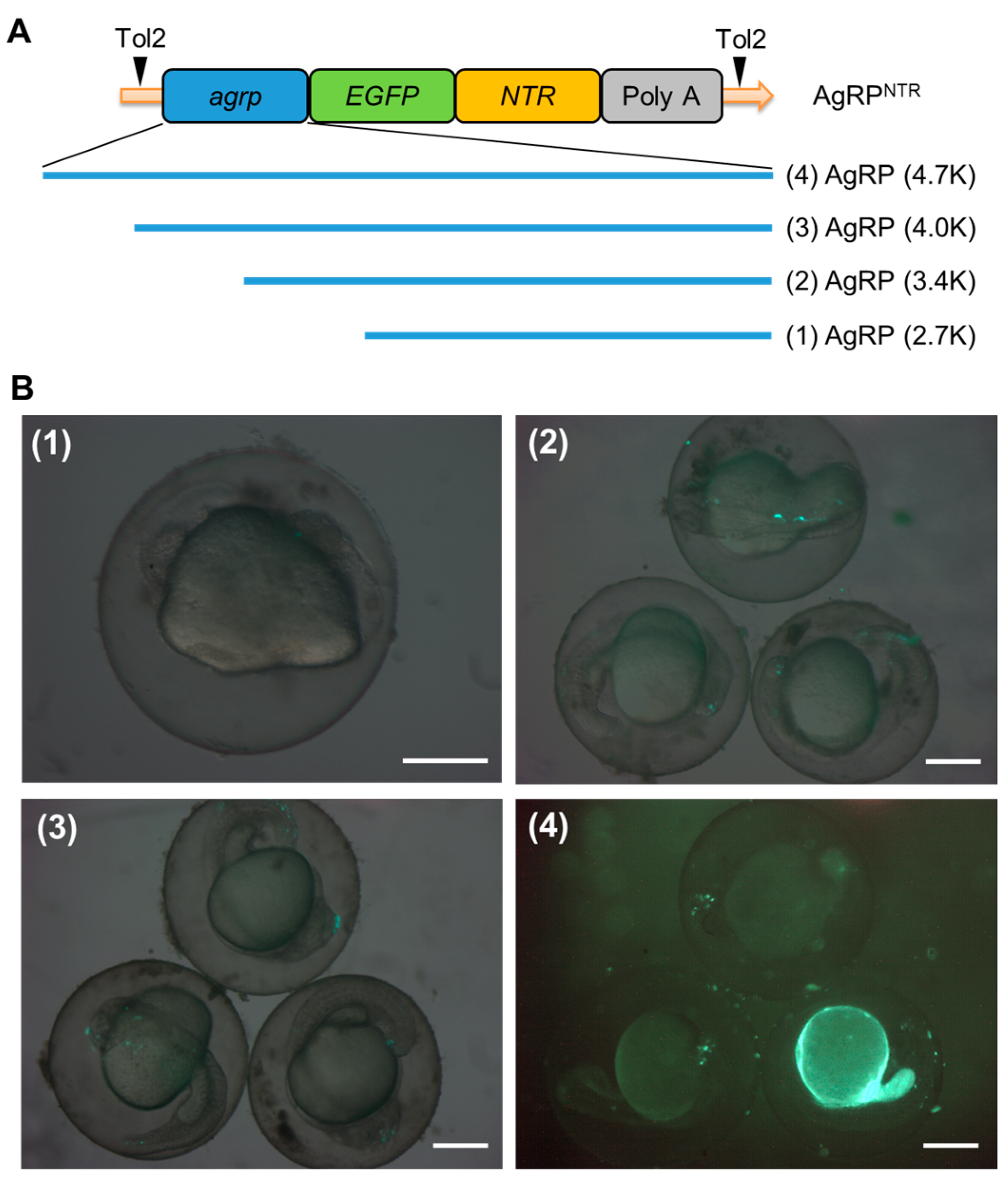

| Construct | No. of Embryo Observed | No. of Embryo with EGFP Expression | Fluorescence Ratio (%) | ||
|---|---|---|---|---|---|
| Head | Trunk | Total | |||
| AgRP (2.7K) | 205 | 0 | 0 | 0 | 0.00 |
| AgRP (3.4K) | 215 | 1 | 4 | 5 | 2.33 |
| AgRP (4.0K) | 221 | 5 | 6 | 11 | 4.98 |
| AgRP (4.7K) | 232 | 32 | 11 | 43 | 18.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-Y.; Yeh, K.-Y.; Lai, H.-H.; Her, G.M. AgRP Neuron-Specific Ablation Represses Appetite, Energy Intake, and Somatic Growth in Larval Zebrafish. Biomedicines 2023, 11, 499. https://doi.org/10.3390/biomedicines11020499
Lin C-Y, Yeh K-Y, Lai H-H, Her GM. AgRP Neuron-Specific Ablation Represses Appetite, Energy Intake, and Somatic Growth in Larval Zebrafish. Biomedicines. 2023; 11(2):499. https://doi.org/10.3390/biomedicines11020499
Chicago/Turabian StyleLin, Chiu-Ya, Kun-Yun Yeh, Hsin-Hung Lai, and Guor Mour Her. 2023. "AgRP Neuron-Specific Ablation Represses Appetite, Energy Intake, and Somatic Growth in Larval Zebrafish" Biomedicines 11, no. 2: 499. https://doi.org/10.3390/biomedicines11020499
APA StyleLin, C.-Y., Yeh, K.-Y., Lai, H.-H., & Her, G. M. (2023). AgRP Neuron-Specific Ablation Represses Appetite, Energy Intake, and Somatic Growth in Larval Zebrafish. Biomedicines, 11(2), 499. https://doi.org/10.3390/biomedicines11020499






