Abstract
The visual process begins with the absorption of photons by photopigments of cone and rod photoreceptors in the retina. In this process, the signal is first amplified by a cyclic guanosine monophosphate (cGMP)-based signaling cascade and then converted into an electrical signal by cyclic nucleotide-gated (CNG) channels. CNG channels are purely ligand-gated channels whose activity can be controlled by cGMP, which induces a depolarizing Na+/Ca2+ current upon binding to the channel. Structurally, CNG channels belong to the superfamily of pore-loop cation channels and share structural similarities with hyperpolarization-activated cyclic nucleotide (HCN) and voltage-gated potassium (KCN) channels. Cone and rod photoreceptors express distinct CNG channels encoded by homologous genes. Mutations in the genes encoding the rod CNG channel (CNGA1 and CNGB1) result in retinitis-pigmentosa-type blindness. Mutations in the genes encoding the cone CNG channel (CNGA3 and CNGB3) lead to achromatopsia. Here, we review the molecular properties of CNG channels and describe their physiological and pathophysiological roles in the retina. Moreover, we summarize recent activities in the field of gene therapy aimed at developing the first gene therapies for CNG channelopathies.
1. Introduction
Cyclic nucleotides, such as cAMP and cGMP, are second messengers that regulate important signaling pathways in our body by controlling the activity of several effector proteins, including cyclic nucleotide-binding domain (CNBD)-containing cation channels. Among CNBD-containing ion channels, cyclic nucleotide-gated (CNG) channels are the only strictly ligand-gated channels because their opening requires binding of cAMP or cGMP [1]. In vertebrates, the CNG channel gene family includes six homologous members. CNGA1, CNGA2, and CNGA3 encode subunits that confer key channel properties and have been shown to form functional homotetrameric ion channels in heterologous expression systems [1]. CNGA4, CNGB1, and CNGB3 encode structurally similar subunits that cannot form functional ion channels by themselves, but are important for the correct localization of native channel complexes and confer specific biophysical properties to the channel [1]. Four of the CNG channel genes are linked to inherited retinal disorders (IRD): mutations in CNGA1 and CNGB1 are known to cause retinitis pigmentosa (RP) and mutations in CNGA3 and CNGB3 cause achromatopsia (ACHM).
2. Insights on Structure and Activation of CNG Channels
Each of the six CNG channel genes encode a membrane protein with six α-helical transmembrane segments (S1–S6), a channel core consisting of a reentrant pore (P) loop between S5 and S6, and cytosolic N- and C-termini. S5, S6, and the intervening reentrant pore (P) loop form the actual pore domain [2] (Figure 1). Similar to classical voltage-gated channels, S4 hosts multiple positively charged residues and S1–S4 form a voltage-sensor-like domain (VSLD). However, unlike canonical voltage-gated channels, the VSLD structure is segmented and the positively charged amino acids are not regularly spaced [3]. This hinders appropriate charge movement and helps to explain why the function of the CNG channel does not depend on voltage [3]. The C-terminus harbors the CNBD and is connected to the S6 via the C-linker (Figure 1). Single particle cryo-electron microscopy structures of native human rod and cone CNG channels confirmed the heterotetrameric 3:1 stoichiometry of the native rod [4] and cone [5] channel complex previously postulated on the basis of biophysical and biochemical experiments [6,7,8,9] (Figure 1). The native CNG channels in the outer segments of rod photoreceptors are heterotetramers consisting of three CNGA1 and one CNGB1 subunit. The CNG channel in the outer segments of cone photoreceptors is formed by three CNGA3 and one CNGB3 subunit. Unlike other members of the voltage-gated channel superfamily, but similar to HCN channels, the four subunits of the tetrameric CNG channel complex are arranged in a non-swapped configuration where the VSLD interacts only with the pore domain of the same subunit [3,10].
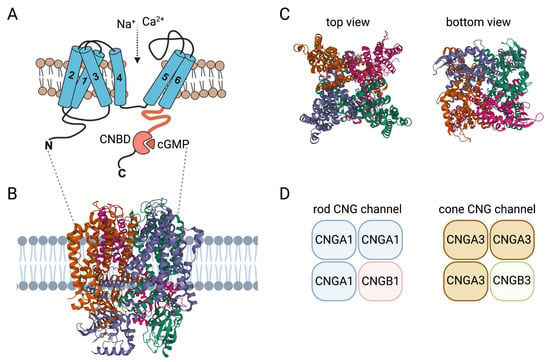
Figure 1.
Structure and activation of CNG channels. (A) Membrane topology of CNG channel subunits: 1–6, transmembrane segment 1–6; C, carboxy-terminus; CNBD, cyclic nucleotide-binding domain; N, amino-terminus. (B) Model of the CNG channel complex embedded in the plasma membrane based on the human rod CNGA1/CNGB1 channel structure (PDB 7RHH). (C) Top and bottom views of the heterotetrameric human rod CNGA1/CNGB1 channel complex. (D) Subunit composition of the CNG channels from rods and cones. Structures in this figure were generated with the RSCB PDB 3D View tool (www.rcsb.org/3d-view/ accessed on 19 Dec 2022) based on PDB 7RHH.
CNG channel structures captured at different activation states in the presence of cGMP and/or pharmacological blockers revealed details about the architecture of the ion-conducting pore and contributed to our understanding of channel function and the effects of pathogenic mutations on channel function [4,5,11,12,13,14]. Based on comparisons of available structures in the open and closed states and previous mutagenesis studies [15,16,17,18], activation of the CNG channel is thought to involve coordinated movements of at least three basic elements, the CNBD, the C-linker with its gating ring, and the channel gate [10,19]. The binding of cyclic nucleotides to the CNBD results in a rotational change of the entire C-terminus relative to the pore. The C-linker, a domain that allosterically couples the binding of cyclic nucleotides to the channel gate via its gating ring, also follows this rotation and moves partially upward. The channel gate, located in the intracellular part of the S6 segment, is constricted and presumably kept in a closed state by the constant forces of the C-linker. After binding of the ligand, the movements described above cause the inhibitory forces of the C-linker to subside and the channel pore is opened, allowing ions to permeate. Despite a sequence identity of only about 35%, the structures of CNGB1 and CNGA1 align well and exhibit similar domain arrangements, which results in a quite symmetrical pore of the closed heterotetrameric CNGA1/CNGB1 channel [4]. In homotetrameric CNG channels, all four A subunits show symmetrical rotational movements that lead to pore opening. However, opening of the heterotetrameric CNGA1/CNGB1 channel is asymmetrical [4]. Only the two CNGA1 subunits left and opposite of the CNGB1 subunit show the movements known from the homomeric channel, whereas CNGB1 and the other CNGA1 subunit barely move, resulting in an asymmetrical open-pore geometry [4]. Structures of the heteromeric cone CNG channel in different activation states are still missing. However, the solved structures revealed an asymmetrical pore architecture already at the closed state with an arginine residue of the CNGB1 S6 projecting directly into and occluding the ion conduction pathway of the pore [5,14]. Mutation of this arginine to a glycine led to a higher single channel conductance [4]. Despite these significant advances in our understanding of the structure and function of CNG channels, there are still gaps in our knowledge, particularly with respect to the gating mechanism of the cone CNG channel or the N- and C-terminal regions, which, for technical reasons, could so far only be modeled for the bovine rod CNG channel [4,5,14]. Further insights are expected from studies aimed at determining the structure of specific pathogenic ACHM- or RP-causing CNG channel mutants with yet unexplained effects on channel structure and function.
3. Role of CNG Channels in Signal Transduction in Photoreceptors
Vertebrates have two types of highly specialized photoreceptors, rods and cones, which have similar but distinct phototransduction signaling cascades and enable the detection of light under different ambient conditions (Figure 2). Rods mediate vision in low light, whereas daylight vision is conferred by cones and only to a lesser extent by rods. The cone visual system also enables color vision because it can discriminate between wavelengths by comparing inputs from two (in most vertebrates) or three (in humans and some nonhuman primates) types of cones, which are equipped with different cone opsin variants with varying spectral sensitivities [20,21]. In both rods and cones, signal transduction follows the same principle and is facilitated by enzymes that control the concentration of cyclic guanosine monophosphate (cGMP). In turn, cGMP controls activation of the CNG channel in the plasma membrane of outer segments (Figure 2). In the dark, the constant activity of transmembrane guanylyl cyclases results in high cGMP concentrations that maintain the CNG channels in an open conformation [22,23]. CNG channels conduct a constant, non-inactivating Na+ and Ca2+ current (“dark current”) that depolarizes the photoreceptor and promotes glutamate release at the photoreceptor synaptic terminals. In response to a light-triggered conformational change, opsins, being G-protein-coupled receptors, release their G protein transducin, which in turn binds to and activates PDE6-type cGMP phosphodiesterases [24]. PDE6 enzymes hydrolyze cGMP, leading to CNG channel closure and photoreceptor hyperpolarization, thus reducing synaptic glutamate release. Ca2+ influx into the outer segments is mediated exclusively by the CNG channels [1,25,26] and balanced by Ca2+ outflow via the Na+/Ca2+, K+ exchangers [25,27,28,29,30]. The closure of CNG channels upon light stimulation, together with the constant activity of Na+/Ca2+, K+ exchangers, leads to a decrease in intracellular Ca2+ concentration. This reduced Ca2+ contributes to recovery from the light response by modulating the activities of PDE6 and guanylyl cyclases [24,31,32,33].
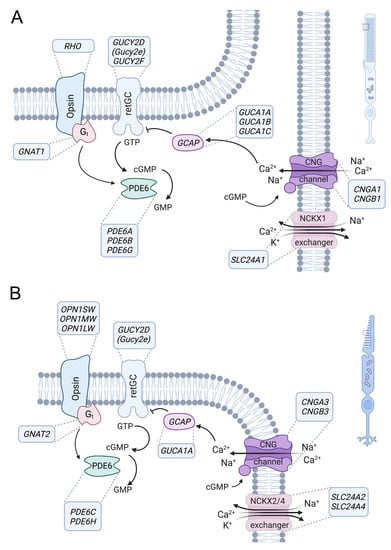
Figure 2.
Role of CNG channels in rod and cone photoreceptor signaling. Phototransduction in outer segments of rod (A) and cone (B) photoreceptors. The principle of the phototransduction is similar in both cell types, but key proteins are encoded by distinct but homologous genes (human gene names are indicated in the boxes next to the proteins). In the dark, the cyclic nucleotide-gated (CNG) channel (CNGA1/B1 in rods and CNGA3/B3 in cones) of the outer membrane is kept open by high concentrations of cyclic guanosine monophosphate (cGMP) produced by retinal guanylyl cyclase (retGC) (note: GUCY2E is a pseudogene in humans, whereas Gucy2e is functional in rodents and the major retGC encoding gene). The resulting influx of Na+ and Ca2+ depolarizes the plasma membrane. Light activates the opsin, which in turn activates transducin (Gt), whose alpha subunit activates a phosphodiesterase (PDE6) that leads to hydrolysis of cGMP. The decrease in the cGMP concentration leads to the closure of the CNG channel, resulting in membrane hyperpolarization. Ca2+ is an important regulator of phototransduction. At high concentrations, Ca2+ binds to guanylyl-cyclase-activating proteins (GCAP), leading to the inhibition of retGC. High Ca2+ concentrations also lead to a slight reduction in the cGMP affinity of the CNG channel through Ca2+/calmodulin-mediated feedback inhibition (not illustrated). Ca2+ is cleared from the outer segment via a Na+-Ca2+-K+-exchanger (NCKX1 in rods, NCKX2/4 in cones). At low Ca2+ levels, Ca2+-free GCAPs can bind and activate retGC to stimulate cGMP production and reopen the CNG channel.
The key activities in this phototransduction cascade are the same in rods and cones but are often mediated by functionally equivalent proteins encoded by distinct genes. This is also the case for the rod and cone CNG channels, which have basically the same functional properties and differ only in some specific features. Notable examples are the higher Ca2+ permeability of the cone CNG channel and the stronger Ca2+-dependent inhibition of ligand sensitivity in the rod CNG channel [1,34]. However, these differences cannot fully explain the different sensitivity and kinetics of rods and cones [1,35].
4. Genetics and Biology of the Rod and Cone CNG Channel
4.1. The Rod CNGA1/CNGB1 Channel
Dysfunction of the rod CNG channel causes autosomal recessive retinitis pigmentosa (RP) [36,37]. RP comprises a genetically diverse group of progressive degenerative retinal diseases primarily affecting the photoreceptors of the retina [38]. Common symptoms of RP include night blindness, progressive constriction of the visual field, and abnormal migration and accumulation of pigment in the retina [39]. The disease is characterized by a primary loss of rod function followed by degeneration and loss of rod photoreceptors, which can vary from patient to patient depending on the underlying gene mutation. As rod loss progresses, cone photoreceptor morphology and function become impaired, affecting daylight vision. This leads to a gradual constriction of the visual field under daylight conditions. In advanced stages, RP leads to impaired visual acuity and may lead to blindness in the final stage. More than 70 genes have already been linked to RP, with different forms of inheritance [40] (see also https://web.sph.uth.edu/RetNet/ accessed on 19 December 2022). Many RP genes encode proteins involved in the phototransduction cascade (Figure 2) or proteins required for the maintenance of photoreceptor architecture. CNGA1 (OMIM #123825) and CNGB1 (OMIM #600724) are linked to autosomal recessive RP (arRP). The prevalence of mutations that cause CNGA1-RP varies in different geographic regions ranging from 1–8% of arRP [37,38,41,42,43,44]. Most of the identified CNGA1 mutations cause deletions of key functional domains or result in impaired membrane trafficking [1,2,37,45]. Mutations in CNGB1 account for 1–4% of arRP cases, and again the prevalence is expected to vary in different geographic areas [36,38,40,42,46,47,48,49]. Although the known CNGB1 mutations cause only minor deletions or single amino acid substitutions, the phenotype is comparable to the RP phenotype in CNGA1-RP patients. Functional characterization studies of some CNGB1 mutations have been performed, showing that some mutations affect rod CNG channel stability or transport, whereas others do not affect expression but result in functionally inactive CNG channels [36,48,50,51].
Today, animal models exist for both CNGA1-RP and CNGB1-RP [52,53,54,55,56,57,58] (Table 1). However, most phenotypic data come from Cngb1 animal models, some of which have been available for more than two decades and have been extensively characterized [54,55,56,57,59,60,61,62].
A naturally occurring Cnga1 mutation has been identified in a Shetland sheepdog breed with progressive retinal atrophy [52]. However, detailed information on the retinal phenotype is still missing. Recently, a mouse model with a targeted deletion in exon 2 of Cnga1 was reported [53]. Cnga1 knockout mice carry a 65-bp frame-shift deletion that, although not experimentally verified at the protein level, should lead to a premature stop codon and loss of most of the Cnga1 protein shortly after deletion. Homozygous mice show loss of most photoreceptors at 16 weeks of age [53]. Dark-adapted electroretinogram (ERG) responses to a single flash of 3 cd*s/m2 were greatly reduced in these mice after 3 weeks, which further decreased after 10 weeks [53]. More recently, an N-ethyl-N-nitrosourea (ENU)-induced Cnga1 mutant mouse model was generated and characterized [63]. The mutant mice carry a c.1526 A > G mutation in Cnga1 that leads to a Y509C exchange in the CNBD of the Cnga1 protein. Y509 corresponds to Y513 in the human CNGA1 protein and participates in the formation of the b3 strand of the CNBD [1]. The Y509C mutation appears to impair the stability of the rod CNG channel complex, resulting in a complete loss of Cnga1 and Cngb1 proteins despite largely unchanged mRNA levels. As a result, rod-driven ERG responses were diminished by 3 weeks of age. Non-functional rods degenerated over time, and from the sixth month of life, secondary progressive degeneration of cones was observed, which was completed by 1 year of age, a time point at which ERG responses were no longer measurable [63].
Nearly two decades ago, a first Cngb1 knockout mouse model was described [54]. This mouse model carries a deletion of exon 26 of the Cngb1 gene which encodes S6. The deletion also leads to a reading frame shift that generates a stop codon at the first triplet of exon 27, thus terminating translation [54]. This leads to a loss of Cngb1 protein expression, but also to degradation of the Cnga1 protein, which appears to require Cngb1 for proper expression. These Cngb1-X26 knockout mice completely lack rod CNG channel function which manifests in diminished responses of rods to light and reduced scotopic ERG responses. The dysfunction is paralleled by the progressive degeneration of the rods and the secondary degeneration of the primarily unaffected cones. Degeneration of the cone photoreceptors begins at 6 months of age, when approximately 50% of the rods have been lost. At about 1 year of age, only 10–20% of the photoreceptors are left in the retina [54]. Overall, the phenotypes of Cngb1- and Cnga1-deficient mice are very similar in terms of disease manifestation and progression. This is most likely because the loss of either CNG channel subunit leads to the secondary degradation and loss of the remaining CNG channel subunit protein. In both cases, this results in a complete loss of the rod CNG channel complex and its functions, which explains the phenotypic similarities observed in the corresponding mouse models.
A spontaneous mutation in CNGB1 was also found in a Papillon dog breed with markedly reduced or absent rod function and slowly progressive retinal degeneration [55]. Interestingly, this c.2387delA;2389_2390insAGCTAC mutation found in this naturally occurring dog model leads to premature termination of the Cngb1 protein at almost the same position as in the engineered Cngb1-X26 mice [54,55,58]. Comparative analyses revealed that CNGB1-RP patients and mouse and dog models with Cngb1 deficiency have a similar phenotype characterized by early loss of rod function and slow degeneration of rod photoreceptors along with a secondary decrease in cone function [59]. The existence of this canine model for CNGB1-RP is of great importance for evaluating the translational potential of future gene therapies. This is due to the structural similarities of canine and human eyes in terms of size and, to some extent, in terms of the spatial distribution of photoreceptor subtypes (e.g., the canine eye has a cone-rich visual stripe that partially mimics the human macula). Moreover, the fact that CNGB1 mutant dogs and Cngb1-X26 mice have genetic alterations that result in the termination of the transcript at an almost identical position increases confidence in extrapolating results from the two models to the situation in humans.
The CNGB1 locus encodes multiple transcripts with distinct expression patterns. The longest transcript uses all 33 exons and produces the CNGB1 (also termed CNGB1a) subunit of the rod CNG channel [64]. The first 11 or 16 exons (including a unique alternative exon) [64] give rise to separate cytosolic proteins corresponding to portions of the glutamic-acid-rich protein (GARP) in the N terminus of CNGB1a. Another mouse model with a genetic modification in exon 1 of Cngb1 (Cngb1-X1 mice) was generated to study the effect of GARP deletion [56]. In principle, Cngb1-X1 mice exhibited similar functional defects as Cngb1-X26 mice, but they showed more dramatically impaired outer rod segment morphology, suggesting that soluble and channel-bound GARP proteins are essential for rod disc morphogenesis and outer segment integrity. Additional studies have shown that channel-attached and soluble GARPs are inherently unfolded [65] and play distinct roles in shaping the morphology of the outer rod segment, transport, and function of the CNG rod channel [50,61,64,66,67,68,69]. These findings are relevant to CNGB1-RP patients who harbor mutations in parts of the CNGB1 locus that give rise to GARP proteins.
4.2. The Cone CNGA3/CNGB3 Channel
Dysfunction of the cone CNG channel causes achromatopsia (ACHM), a rare retinal disease that is inherited in an autosomal recessive manner and affects approximately one in 30,000 individuals [70]. Unlike color blindness, in which mutations in genes encoding the various cone photopigments affect only spectral sensitivity [71], ACHM has severe consequences for all aspects of daylight vision. Symptoms include poor visual acuity, photophobia, nystagmus, and lack of color discrimination [72]. The symptoms reflect a functional defect of the cone photoreceptors that occurs in early infancy and is characterized by a lack of light-adapted ERG but preserved scotopic ERG signal [73,74]. In addition to the functional deficits, structural changes can be observed in the cone-rich central portion of the retina, ranging from loss of cone outer segments to profound atrophy of the retina [72]. Up to 90% of ACHM cases are due to mutations in CNGA3 (OMIM #216900) and CNGB3 (OMIM #262300) [75,76,77]. The remaining cases are due to mutations in ATF6 (OMIM #616517), GNAT2 (OMIM #613856), PDE6C (OMIM #613093), PDE6H (OMIM #610024), or yet unknown genes [78,79].
To date, more than 250 mutations in CNGA3 [80,81,82,83,84,85,86,87,88] and more than 160 mutations in CNGB3 [76,77,80,83,89,90,91,92,93,94] were found to cause ACHM in humans. Mutations in CNGB3 are more common in Europe and the United States and account for 50–60% of ACHM cases [76,89], and in the Netherlands even close to 90% [83]. Most CNGB3 mutations are nonsense, frameshift, or splice mutations [77,80]. A missense mutation in the CNGB3 gene (S435F) was identified in colorblind individuals originating from the Pingelap atoll of Micronesia [90]. In this small island ACHM is very frequent and affects nearly 10% of the native population [90,91,95,96,97]. An estimated 28–36% of patients in the Western population carry mutations in CNGA3 (ACHM2) [81,84]. In the Middle East and Chinese populations, mutations in CNGA3 account for approximately 80% of ACHM cases [42,80,98,99]. Interestingly, a digenic and triallelic inheritance pattern with mutations in both CNGA3 and CNGB3 was also found in a subset of ACHM patients [100]. The majority of CNGA3 mutations are missense mutations affecting only single amino acid residues of the protein [81,82,83,84,85,86,88]. Folding, intracellular processing, and transport are thought to be impaired [101]. While some insights have been gained, the precise mechanisms linking specific amino acid substitutions to the ACHM phenotype are still poorly understood. The effects of individual amino acid substitutions on CNGA3 protein function have been studied largely in vitro [85,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116].
Several genetically modified and naturally occurring animal models of ACHM exist (Table 1) that have helped to elucidate disease mechanisms and serve as disease models for the preclinical development of emerging gene therapies [58,70]. The first animal model of ACHM described was the Cnga3 knockout mouse, which carries a homozygous deletion of exon 7, leading to the deletion of all channel domains downstream of the third transmembrane segment (S3), including the pore and the CNBD [117]. This results in a complete loss of cone CNG channel function. As a consequence, Cnga3 knockout mice show a selective deficiency of cone-mediated light responses from birth [117], followed by the progressive degeneration and cell death of cones [117,118]. Cone degeneration affects M- and S-cones differentially and cell death proceeds significantly faster in ventral and nasal (S-cone-rich) than in dorsal and temporal (M-cone-rich) parts of the retina. Ventral cones are almost completely missing after the third postnatal month, whereas residual dorsal cones are present even in aged knockout mice [118]. In addition, a naturally occurring mouse model of ACHM has been described. The mouse line designated cpfl5 (cone photoreceptor function loss 5) carries a point mutation in Cnga3 leading to a p.T203A substitution in the cytoplasmic loop between S2 and S3. To date, no missense mutation affecting this phylogenetically conserved threonine, corresponding to p.T224 in human CNGA3 protein, has been reported. Although the exact mechanism remains unclear, this mutation results in the loss of Cnga3 protein expression and a phenotype similar to that observed in the Cnga3 knockout mouse [119].
Moreover, a naturally occurring sheep model of ACHM was reported with congenital visual impairment characterized by diminished cone, but normal rod function [120,121]. Affected lambs were found to be homozygous for a nonsense mutation in Cnga3 (p.R236X) [121]. Subsequently, a Cnga3 missense mutation (p.G540S) was identified in another breed, causing a similar phenotype of day blindness [120]. In addition, two spontaneous canine models of ACHM have been described [122]. One is a Labrador retriever with a p.V644del mutation that removes a conserved valine within a C-terminal domain shown to be important for heterotetrameric channel assembly and stability [6]. The second model is a German shepherd, carrying a p.R424W mutation. This amino acid is found in the gating ring within the C-linker that connects transmembrane domain S6 with the CNBD and is conserved in eukaryotes. Importantly, a mutation that leads to a R-to-W substitution of the corresponding human CNGA3 sequence (p.R410W) was also found in achromatopsia patients [82]. Recent cryo-electron microscopy (cryo-EM) studies with the C. elegans channel tax-4 version (CNGA3/p.R421W) have shed light on the potential pathogenic mechanism of this missense mutation [11]. Careful analysis of cryo-EM data in conjunction with electrophysiological and biochemical data led to the conclusion that this R-to-W substitution in the gating ring destabilizes the closed state of the channel and favors spontaneous channel opening in the absence of the ligand [11]. Thus, if the channel is expressed in the cone outer segments of CNGA3/p.R421W patients, its excessive activity could induce (e.g., Ca2+-mediated) cell death and cone degeneration.
Various animal models also exist for CNGB3-linked ACHM (Table 1). A Cngb3 knockout mouse with a genetic deletion that causes a frame shift and removes part of S1 and all other channel domains has been described [123]. These knockout mice lack Cngb3 protein expression and show strongly reduced Cnga3 protein levels. The lack of the cone CNG channel severely impairs cone function and leads to progressive cone degeneration reminiscent of the Cnga3 knockout mouse phenotype [123,124]. Residual cone function is observed in this model, most likely conferred by irregular homomeric CNGA3 channels. In addition to the Cngb3 knockout mouse, a naturally occurring mouse model designated cpfl10 has been described [125]. The mice carry a c.692G>A point mutation leading to p.R231H [125]. Basic characterization of the mouse line revealed a loss of cone-driven ERG responses and slow progressive degeneration of the cones [125].
Two naturally occurring canine cone degeneration models with mutations in Cngb3 have been identified in Alaskan malamute and German shorthaired pointer breeds [126]. Genetic analysis has shown that in the Alaskan malamute, the complete gene is deleted, while in the German shorthaired pointers the disease is caused by a missense mutation c.784G > A;p.D262N affecting a conserved aspartate residue in S2 [126]. Affected Alaskan malamute pups develop day blindness and photophobia resembling the clinical phenotype of human ACHM patients. Symptoms are present only in bright light, while vision in dim light is normal. The cone ERG signals begin to diminish a few weeks after birth and are extinguished in older affected dogs [127]. Recently, a viral vector-delivered CRISPR-Cas9 strategy was used to generate an in situ knockout model of CNGB3-ACHM in cynomolgus monkeys [128]. This acute model can provide valuable information about the pathobiology of CNGB3 deficiency in a non-human primate retina with a foveo-macular structure similar to that in the human eye.
The existence of the numerous animal models of CNGA3- and CNGB3-ACHM not only contributes to a better understanding of the biology of the cone CNG channel, but also greatly facilitates the development and testing of potential treatments. As mentioned for the rod CNG channel, the availability of the large animal models with morphological similarities to the human eye (in terms of size and cell distribution) is of paramount importance for translational studies. Other than for rod CNG channel models, Cnga3- and Cngb3-deficient mouse models exhibit some phenotypic differences, possibly due to cone-specific morphologic features that, in the absence of Cngb3, still allow a more efficient transport of homotetrameric Cnga3 channels to the outer segment, thereby supporting residual CNG channel function. However, in the absence of Cnga3, Cngb3 cannot support channel function on its own.

Table 1.
Overview of retinal CNG genes, associated human diseases, animal models, and preclinical studies. NCT ID, www.clinicaltrials.gov identifier (accessed on 19 December 2022). OMIM, Online Mendelian Inheritance in Man. POC, prove of concept.
Table 1.
Overview of retinal CNG genes, associated human diseases, animal models, and preclinical studies. NCT ID, www.clinicaltrials.gov identifier (accessed on 19 December 2022). OMIM, Online Mendelian Inheritance in Man. POC, prove of concept.
| Gene | Chromosomal Location | Phenotype, OMIM | Animal Models | POC Studies | Preclinical Safety Studies | Clinical Trials (NCT ID) |
|---|---|---|---|---|---|---|
| CNGA1 | 4q12 | RP49, 613756 | knockout mouse [53] mutant mouse [63] canine model [52] | - | - | - |
| CNGB1 | 16q21 | RP45, 613767 | knockout mouse [54,56] canine model [55,57,59] | Refs. [59,129,130] | - | - |
| CNGA3 | 2q11.2 | ACHM2, 600053 | knockout mouse [117] mutant mouse [131] canine model [122] ovine model [121] | Refs. [119,132,133,134,135,136,137,138] | Refs. [137,138,139,140,141,142] | 02610582 02935517 03758404 03278873 |
| CNGB3 | 8q21.3 | ACHM3, 605080 | knockout mouse [123] mutant mouse [125] canine model [126] in situ NHP model [128] | Refs. [143,144,145] | Refs. [146,147,148] | 02599922 03001310 03278873 |
5. Gene Therapy for the Treatment of CNG Channelopathies
To date, there is no curative treatment for any CNG channelopathy, and clinical treatment is currently limited to specialized genetic counseling, use of visual aids, and tinted contact lenses or glasses to reduce symptoms of photophobia. Our improved understanding of CNG channel biology, the availability of suitable animal models, and the emergence of efficient and safe adeno-associated virus (AAV) vectors led to the initiation of several gene therapy programs for potential treatment of CNG-channel-related retinopathies (Table 1).
ACHM and RP caused by mutations in CNG channel genes are inherited in an autosomal recessive manner. Unlike most autosomal inherited retinopathies, it is not necessary to remove a (dominant) pathogenic variant, and the addition of a functional gene copy would be sufficient for therapeutic benefit. Therefore, the gene therapy approaches aim at adding a healthy copy of the disease-causing gene into the affected cells (in this case, the cone or rod photoreceptors). Some of these so-called gene supplementation (or augmentation) approaches have already reached the clinical phase of development. The following sections provide an introduction into the AAV vector technology and summarize the key findings of preclinical studies and publicly available data from clinical trials.
5.1. The AAV Vector Technology
AAVs are small (diameter of 25 nm), non-enveloped, non-pathogenic DNA viruses that can only replicate in the presence of adeno, papilloma, or herpes viruses. Multiple naturally occurring AAV serotypes exist and have been explored for their use as vectors for gene transfer [149,150]. The AAV vector platform has already been clinically validated, and five gene therapy products have been approved for ophthalmic, CNS, and other indications in recent years (www.ema.europa.eu and www.fda.gov accessed on 19 December 2022). More than three decades ago, AAVs were vectorized by the replacement of the viral genes rep and cap with a gene expression cassette of choice [151]. The AAV rep and cap genes are provided in trans during recombinant AAV production process in form of helper plasmids [152]. AAVs consist of a 60-mer capsid of structural viral proteins (VP1, VP2, and VP3) assembled in a 5:5:50 ratio and an approximately 4.7 kb long single-stranded DNA genome containing the desired gene expression cassette for the gene of interest, flanked by two inverted terminal repeats (ITR) [153]. Such AAV-derived viral vectors can deliver their genome into the target cell nucleus where it remains episomal and transcribes the gene of interest without integrating into the host genome. In recent years, AAV vectors have evolved as the gold standard gene delivery vector for retinal photoreceptors and retinal pigment epithelial cells with proven tropism and efficacy. Their success is based on the fact that they are easy to produce at large scale and show a generally good safety profile with limited immunogenicity and dose-dependent toxicity [154,155].
5.2. Gene Therapy for CNG-Channel-Linked RP
To date, there are no gene therapy approaches for CNGA1-linked RP. For CNGB1-linked RP, successful proof-of-concept studies for AAV-based gene supplementation have been reported in both the Cngb1 knockout mouse model [129] and the Cngb1 mutant dog model [59]. To enable the efficient packaging and rod-specific expression of the relatively large full-length Cngb1 cDNA (~4 kb), the two studies used an AAV expression cassette with a short rod- [129] or photoreceptor-specific [59] promoter to drive expression of a species-matched Cngb1 cDNA (e.g., mouse or canine). In both species, subretinal injection of therapeutic AAV gene supplementation vectors (serotype 5 or 8) led to efficient expression of the Cngb1 protein and the restoration of CNG channel expression and localization. This resulted in the improvement of rod-mediated retinal function, preservation of retinal structure, and delay of secondary cone degeneration. Finally, treated Cngb1 knockout mice as well as CNGB1 mutant dogs performed significantly better than untreated controls in rod-dependent vision-guided behavior tests [59,129]. These promising results facilitated the initiation of translational studies with a humanized vector version (AAV5-RHO-CNGB1) in which a short human rhodopsin promoter drives expression of the full-length human CNGB1 [130]. When administered via single subretinal injection in 4-week-old Cngb1 knockout mice, AAV5-RHO-CNGB1 led to efficient expression of the human CNGB1 protein in mouse rods and restored the expression of the endogenous mouse Cngb1 protein [130]. The treatment resulted in a dose-dependent recovery of rod-driven ERG responses and the preservation of retinal structure [130]. Studies in large animal models are currently underway to support the implementation of this gene therapy approach for the future treatment of CNGB1-RP patients.
5.3. Gene Therapy for CNG-Channel-Linked ACHM
5.3.1. ACHM Gene Therapy: Preclinical Proof-of-Concept Studies
An AAV5 vector expressing human CNGB3 cDNA under control of one of three different truncated versions of the human M/L opsin promoter was evaluated in a gene augmentation approach in dogs affected by achromatopsia due to mutations in CNGB3 [143]. The dogs were injected unilaterally into the subretinal space at 3 to 81 weeks of age. Improvement in cone function was observed as early as 4 weeks after treatment under photopic conditions using ERG and behavior and persisted for at least 14 months. The best treatment results were achieved in 3-week-old-animals, whereas treatment was minimally effective in dogs 1 year of age and older [143]. The exact reasons for the age-dependence of treatment are not known, but could be related to morphological changes observed in later stages of the disease. Accordingly, efficacy in this dog model was improved in older dogs when the AAV gene augmentation was combined with the administration of ciliary neurotrophic factor (CNTF), which is known to cause a temporal deconstruction of photoreceptor outer segments [144].
A positive proof-of-concept for gene supplementation therapy with an AAV5 vector driving expression of mouse Cnga3 cDNA under control of a short mouse S opsin promoter was also achieved in the Cnga3 knockout mouse model of achromatopsia [132]. Two-week-old mice treated with subretinal injection showed cone-driven ERG responses, normalization of cGMP levels and expression of cone CNG channel complexes and opsins, and delay of cone cell death. In addition, ganglion cells from treated but not untreated Cnga3 knockout mice showed cone-driven light-evoked spiking activity, suggesting that signals generated in the outer retina are transmitted to the brain. Finally, it was demonstrated that the newly acquired sensory information was translated into cone-mediated, vision-guided behavior [132]. The therapeutic effect was stable for at least 12 months and was also seen with an AAV8 serotype vector or with treatment at 3 months of age [133]. Similar effects were obtained in different Cnga3 mouse models after subretinal administration of an AAV5 vector and the human M/L opsin promoter [119] as well as intravitreal delivery of engineered AAV8 (Y447, 733F) [134] or AAV2.GL [135] vectors. In line with the dog studies, an AAV8 vector driving expression of the human CNGB3 cDNA under control of a short human ARR3 promoter was shown to efficiently rescue cone-driven ERG responses and visual acuity in the Cngb3 knockout mouse model of achromatopsia [145]. Successful AAV-based gene supplementation therapy has also been described in the Awassi sheep model of CNGA3 achromatopsia [136]. Significant long-term improvement in cone function was demonstrated for at least 6 years after a single dose of an AAV vector expressing human CNGA3 [137,138].
These promising preclinical studies led to the initiation of a total of five independent gene therapy programs for CNGA3- and CNGB3-linked achromatopsia. Safety studies in sheep and non-human primates revealed some inflammation after subretinal CNGA3 gene delivery, but overall showed an acceptable safety profile for at least two different translatable CNGA3 gene therapy products [138,139,140,156]. Safety data have not yet been published for the third CNGA3 gene therapy product. For one of the two CNGB3 programs, safety data obtained in mice, dogs, and cynomolgus monkeys were published showing acceptable safety with vector- and dose-dependent inflammation and toxicity [146,147,148].
5.3.2. ACHM Gene Therapy: Clinical Studies
All of the aforementioned translational programs for CNGA3- and CNGB3-linked achromatopsia have already reached the clinical phase [70] (Table 1). The German academic research consortium RD-CURE initiated the first clinical trial which evaluated the effect of three different doses (1 × 1010, 5 × 1010, and 1 × 1011 total vector genomes per eye) of AAV8.CNGA3 administered via subretinal injection into one eye. The study enrolled nine patients in three dose groups. Despite the highly invasive delivery procedure, which involved vitrectomy and the detachment of the foveo-macular retina, the treatment was well tolerated and resulted in dose-independent mild and transient procedure- or drug-related adverse events [157,158] and transient subclinical induction of inflammatory markers [140,142]. Treatment led to improvement in secondary end points related to cone function, including improvement in visual acuity and contrast sensitivity from baseline in all treated patients [158], which showed a tendency to be dose-dependent and persisted until at least 3 years after treatment [157]. A phase IIb clinical trial targeting treatment of the second eye of the first patients and treatment of children aged 6 to 12 years is currently ongoing (Table 1).
Four other programs are currently in phase I/II of clinical trials, two on CNGA3-ACHM and an additional two on CNGB3-ACHM (Table 1). Preliminary safety and efficacy data from industry-sponsored clinical trials testing the safety and efficacy of the gene therapy products AGTC-401 and AGTC-402 in CNGB3- and CNGA3-related achromatopsia, respectively, were presented at the annual meeting of the American Society for Vision Research and Ophthalmology (ARVO). Both gene therapy products used a modified AAV2 capsid with three surface-exposed Y to F mutations (Y275F, Y447F, and Y733F), and designated AAV2tYF and a 1.7 kb human M/L opsin promoter driving expression of either CNGA3 or CNGB3. The AGTC-401 study enrolled 21 adult and 10 pediatric CNGB3 achromatopsia patients in six dose groups (1.2 × 1011 vector genomes (vg)/mL to 3.2 × 1012 vg/mL). The AGTC-402 trial included 16 adult and 8 pediatric CNGA3 achromatopsia patients distributed into five dose groups (4 × 1010 vg/mL to 3.2 × 1012 vg/mL). In both studies, dose-limiting toxicity was noted at the highest dose (3.2 × 1012 vg/mL) in children, which included uveitis and posterior segment changes. The highest dose was better tolerated in adult patients. Gene therapy improved photosensitivity in some CNGB3-ACHM patients but less in CNGA3-ACHM patients. Long-term follow-up studies were initiated for both programs (Table 1), but recently, the company announced that the CNGA3-ACHM program will not be developed further. The other industry-sponsored CNGA3 and CNGB3 gene therapy programs have released only limited safety data at clinicaltrials.gov (https://clinicaltrials.gov/ct2/show/results/NCT03758404, accessed on 19 December 2022), but communicated plans to initiate late-stage clinical studies.
6. Conclusions and Outlook
The four retinal CNG channelopathies are severe inherited retinopathies leading to either achromatopsia or a retinitis pigmentosa phenotype. Although they are rare disorders, the estimated total number of affected patients is a quarter of a million [42]. Several well-characterized small and large animal models have contributed to a better understanding of the underlying pathomechanisms and are important for preclinical testing of novel therapies based on AAV vectors. Gene therapy programs targeting CNGA3- or CNGB3-linked ACHM are already in early clinical development but need to demonstrate clinical proof-of-concept in pivotal clinical trials before marketing approval can be granted. Additional programs for CNG-linked RP are expected to follow in the near future to address the remaining gaps in the treatment of CNGA1- and CNGB1-linked forms of RP.
Author Contributions
Conceptualization, M.B., M.J.G., S.M.; investigation, M.B., M.J.G., S.M.; methodology, S.M. software, S.M.; validation, M.B., M.J.G., S.M., S.G.P.; visualization, S.M.; writing—original draft, S.M., M.J.G.; writing—review and editing, M.B., M.J.G., S.M., S.G.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
Figures were created with BioRender (https://app.biorender.com/ accessed on 19 December 2022).
Conflicts of Interest
S.M. received speaker’s honoraria of Novartis and is co-founder of the gene therapy company ViGeneron GmbH. M.B. is co-founder of the gene therapy company ViGeneron GmbH. M.J.G. received speaker’s honoraria from Novartis and Bayer. S.G.P. received speaker’s honoraria from Allergan, Alcon, BVI, B&L, Bayer, Novartis, Örtli, Roche, and Zeiss and is on the advisory board of Allergan, Alcon, Bayer, B&L, Roche, and Zeiss.
References
- Kaupp, U.B.; Seifert, R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 2002, 82, 769–824. [Google Scholar] [CrossRef] [PubMed]
- Biel, M.; Michalakis, S. Cyclic nucleotide-gated channels. Handb. Exp. Pharmacol. 2009, 191, 111–136. [Google Scholar] [CrossRef]
- Li, M.; Zhou, X.; Wang, S.; Michailidis, I.; Gong, Y.; Su, D.; Li, H.; Li, X.; Yang, J. Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature 2017, 542, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Han, Y.; Zeng, W.; Jiang, Y. Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel. Neuron 2022, 110, 86–95.e85. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Hu, Z.; Li, H.; Yang, J. Structure of the human cone photoreceptor cyclic nucleotide-gated channel. Nat. Struct Mol. Biol. 2022, 29, 40–46. [Google Scholar] [CrossRef]
- Zhong, H.; Molday, L.L.; Molday, R.S.; Yau, K.W. The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature 2002, 420, 193–198. [Google Scholar] [CrossRef]
- Zheng, J.; Trudeau, M.C.; Zagotta, W.N. Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron 2002, 36, 891–896. [Google Scholar] [CrossRef]
- Weitz, D.; Ficek, N.; Kremmer, E.; Bauer, P.J.; Kaupp, U.B. Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron 2002, 36, 881–889. [Google Scholar] [CrossRef]
- Shuart, N.G.; Haitin, Y.; Camp, S.S.; Black, K.D.; Zagotta, W.N. Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels. Nat. Commun. 2011, 2, 457. [Google Scholar] [CrossRef]
- James, Z.M.; Zagotta, W.N. Structural insights into the mechanisms of CNBD channel function. J. Gen. Physiol. 2018, 150, 225–244. [Google Scholar] [CrossRef]
- Zheng, X.; Li, H.; Hu, Z.; Su, D.; Yang, J. Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel. Commun. Biol. 2022, 5, 190. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Han, Y.; Zeng, W.; Wang, Y.; Jiang, Y. Structural mechanisms of gating and selectivity of human rod CNGA1 channel. Neuron 2021, 109, 1302–1313.e1304. [Google Scholar] [CrossRef] [PubMed]
- Barret, D.C.A.; Schertler, G.F.X.; Kaupp, U.B.; Marino, J. Structural basis of the partially open central gate in the human CNGA1/CNGB1 channel explained by additional density for calmodulin in cryo-EM map. J. Struct. Biol. 2022, 214, 107828. [Google Scholar] [CrossRef] [PubMed]
- Barret, D.C.A.; Schertler, G.F.X.; Benjamin Kaupp, U.; Marino, J. The structure of the native CNGA1/CNGB1 CNG channel from bovine retinal rods. Nat. Struct. Mol. Biol. 2022, 29, 32–39. [Google Scholar] [CrossRef]
- Zhou, L.; Siegelbaum, S.A. Gating of HCN Channels by cyclic nucleotides: Residue contacts that underlie ligand binding, selectivity, and efficacy. Structure 2007, 15, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Zagotta, W.N.; Olivier, N.B.; Black, K.D.; Young, E.C.; Olson, R.; Gouaux, E. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 2003, 425, 200–205. [Google Scholar] [CrossRef]
- Taraska, J.W.; Zagotta, W.N. Cyclic nucleotide-regulated ion channels: Spotlight on symmetry. Structure 2007, 15, 1023–1024. [Google Scholar] [CrossRef]
- Flynn, G.E.; Black, K.D.; Islas, L.D.; Sankaran, B.; Zagotta, W.N. Structure and rearrangements in the carboxy-terminal region of SpIH channels. Structure 2007, 15, 671–682. [Google Scholar] [CrossRef]
- Napolitano, L.M.R.; Torre, V.; Marchesi, A. CNG channel structure, function, and gating: A tale of conformational flexibility. Pflugers Arch. 2021, 473, 1423–1435. [Google Scholar] [CrossRef]
- Ahnelt, P.K.; Kolb, H. The mammalian photoreceptor mosaic-adaptive design. Prog. Retinal Eye Res. 2000, 19, 711–777. [Google Scholar] [CrossRef]
- Yokoyama, S. Molecular evolution of vertebrate visual pigments. Prog. Retinal Eye Res. 2000, 19, 385–419. [Google Scholar] [CrossRef]
- Pugh, E.N., Jr.; Duda, T.; Sitaramayya, A.; Sharma, R.K. Photoreceptor guanylate cyclases: A review. Biosci Rep. 1997, 17, 429–473. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.B.; Robinson, S.W.; Xiong, W.H.; Yau, K.W.; Birch, D.G.; Garbers, D.L. Disruption of a retinal guanylyl cyclase gene leads to cone-specific dystrophy and paradoxical rod behavior. Journal Neurosci. 1999, 19, 5889–5897. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Getter, T.; Salom, D.; Wu, D.; Quetschlich, D.; Chorev, D.S.; Palczewski, K.; Robinson, C.V. Capturing a rhodopsin receptor signalling cascade across a native membrane. Nature 2022, 604, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, A.L.; McNaughton, P.A.; Nunn, B.J. The ionic selectivity and calcium dependence of the light-sensitive pathway in toad rods. J. Physiol. 1985, 358, 447–468. [Google Scholar] [CrossRef]
- Yau, K.W.; Nakatani, K. Cation selectivity of light-sensitive conductance in retinal rods. Nature 1984, 309, 352–354. [Google Scholar] [CrossRef]
- Cervetto, L.; Lagnado, L.; Perry, R.J.; Robinson, D.W.; McNaughton, P.A. Extrusion of calcium from rod outer segments is driven by both sodium and potassium gradients. Nature 1989, 337, 740–743. [Google Scholar] [CrossRef]
- Schnetkamp, P.P. The SLC24 Na+/Ca2+-K+ exchanger family: Vision and beyond. Pflugers Arch. 2004, 447, 683–688. [Google Scholar] [CrossRef]
- Vinberg, F.; Wang, T.; De Maria, A.; Zhao, H.; Bassnett, S.; Chen, J.; Kefalov, V.J. The Na(+)/Ca(2+), K(+) exchanger NCKX4 is required for efficient cone-mediated vision. eLife 2017, 6, e24550. [Google Scholar] [CrossRef]
- Yau, K.W.; Nakatani, K. Light-induced reduction of cytoplasmic free calcium in retinal rod outer segment. Nature 1985, 313, 579–582. [Google Scholar] [CrossRef]
- Irwin, M.J.; Gupta, R.; Gao, X.Z.; Cahill, K.B.; Chu, F.; Cote, R.H. The molecular architecture of photoreceptor phosphodiesterase 6 (PDE6) with activated G protein elucidates the mechanism of visual excitation. J. Biological Chem. 2019, 294, 19486–19497. [Google Scholar] [CrossRef]
- Gao, Y.; Eskici, G.; Ramachandran, S.; Poitevin, F.; Seven, A.B.; Panova, O.; Skiniotis, G.; Cerione, R.A. Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6. Mol. Cell 2020, 80, 237–245.e234. [Google Scholar] [CrossRef] [PubMed]
- Ames, J.B. Structural Insights into Retinal Guanylate Cyclase Activator Proteins (GCAPs). Int. J. Mol. Sci. 2021, 22, 8731. [Google Scholar] [CrossRef]
- Ohyama, T.; Hackos, D.H.; Frings, S.; Hagen, V.; Kaupp, U.B.; Korenbrot, J.I. Fraction of the dark current carried by Ca(2+) through cGMP-gated ion channels of intact rod and cone photoreceptors. J. Gen. Physiol. 2000, 116, 735–754. [Google Scholar] [CrossRef] [PubMed]
- Korenbrot, J.I. Speed, sensitivity, and stability of the light response in rod and cone photoreceptors: Facts and models. Prog. Retin. Eye Res. 2012, 31, 442–466. [Google Scholar] [CrossRef] [PubMed]
- Bareil, C.; Hamel, C.P.; Delague, V.; Arnaud, B.; Demaille, J.; Claustres, M. Segregation of a mutation in CNGB1 encoding the beta-subunit of the rod cGMP-gated channel in a family with autosomal recessive retinitis pigmentosa. Hum. Genet. 2001, 108, 328–334. [Google Scholar] [CrossRef]
- Dryja, T.P.; Finn, J.T.; Peng, Y.W.; McGee, T.L.; Berson, E.L.; Yau, K.W. Mutations in the gene encoding the alpha subunit of the rod cGMP-gated channel in autosomal recessive retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 1995, 92, 10177–10181. [Google Scholar] [CrossRef]
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- Kalloniatis, M.; Fletcher, E.L. Retinitis pigmentosa: Understanding the clinical presentation, mechanisms and treatment options. Clin. Exp. Optom. 2004, 87, 65–80. [Google Scholar] [CrossRef]
- Xu, Y.; Guan, L.; Shen, T.; Zhang, J.; Xiao, X.; Jiang, H.; Li, S.; Yang, J.; Jia, X.; Yin, Y.; et al. Mutations of 60 known causative genes in 157 families with retinitis pigmentosa based on exome sequencing. Hum. Genet. 2014, 133, 1255–1271. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, K.; Sheng, X.; Li, Y.; Gao, X.; Zhang, X.; Kang, X.; Pan, X.; Liu, Y.; Jiang, C.; et al. Targeted sequencing of 179 genes associated with hereditary retinal dystrophies and 10 candidate genes identifies novel and known mutations in patients with various retinal diseases. Investig. Ophthalmol. Vis. Sci. 2013, 54, 2186–2197. [Google Scholar] [CrossRef]
- Hanany, M.; Rivolta, C.; Sharon, D. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc. Natl. Acad. Sci. USA 2020, 117, 2710–2716. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, S.; Akahori, M.; Sergeev, Y.; Yoshitake, K.; Ikeo, K.; Furuno, M.; Hayashi, T.; Kondo, M.; Ueno, S.; Tsunoda, K.; et al. Whole exome analysis identifies frequent CNGA1 mutations in Japanese population with autosomal recessive retinitis pigmentosa. PloS ONE 2014, 9, e108721. [Google Scholar] [CrossRef]
- Paloma, E.; Martinez-Mir, A.; Garcia-Sandoval, B.; Ayuso, C.; Vilageliu, L.; Gonzalez-Duarte, R.; Balcells, S. Novel homozygous mutation in the alpha subunit of the rod cGMP gated channel (CNGA1) in two Spanish sibs affected with autosomal recessive retinitis pigmentosa. J. Med. Genet. 2002, 39, E66. [Google Scholar] [CrossRef] [PubMed]
- Mallouk, N.; Ildefonse, M.; Pages, F.; Ragno, M.; Bennett, N. Basis for intracellular retention of a human mutant of the retinal rod channel alpha subunit. J. Membr. Biol. 2002, 185, 129–136. [Google Scholar] [CrossRef]
- Ge, Z.; Bowles, K.; Goetz, K.; Scholl, H.P.; Wang, F.; Wang, X.; Xu, S.; Wang, K.; Wang, H.; Chen, R. NGS-based Molecular diagnosis of 105 eyeGENE((R)) probands with Retinitis Pigmentosa. Sci. Rep. 2015, 5, 18287. [Google Scholar] [CrossRef]
- Nassisi, M.; Smirnov, V.M.; Solis Hernandez, C.; Mohand-Said, S.; Condroyer, C.; Antonio, A.; Kuhlewein, L.; Kempf, M.; Kohl, S.; Wissinger, B.; et al. CNGB1-related rod-cone dystrophy: A mutation review and update. Hum. Mutat 2021, 42, 641–666. [Google Scholar] [CrossRef]
- Kondo, H.; Qin, M.; Mizota, A.; Kondo, M.; Hayashi, H.; Hayashi, K.; Oshima, K.; Tahira, T.; Hayashi, K. A homozygosity-based search for mutations in patients with autosomal recessive retinitis pigmentosa, using microsatellite markers. Investig. Ophthalmol. Vis. Sci. 2004, 45, 4433–4439. [Google Scholar] [CrossRef]
- Simpson, D.A.; Clark, G.R.; Alexander, S.; Silvestri, G.; Willoughby, C.E. Molecular diagnosis for heterogeneous genetic diseases with targeted high-throughput DNA sequencing applied to retinitis pigmentosa. J. Med. Genet. 2011, 48, 145–151. [Google Scholar] [CrossRef]
- Michalakis, S.; Zong, X.; Becirovic, E.; Hammelmann, V.; Wein, T.; Wanner, K.T.; Biel, M. The glutamic acid-rich protein is a gating inhibitor of cyclic nucleotide-gated channels. J. Neurosci. 2011, 31, 133–141. [Google Scholar] [CrossRef]
- Becirovic, E.; Nakova, K.; Hammelmann, V.; Hennel, R.; Biel, M.; Michalakis, S. The retinitis pigmentosa mutation c.3444+1G>A in CNGB1 results in skipping of exon 32. PloS ONE 2010, 5, e8969. [Google Scholar] [CrossRef] [PubMed]
- Wiik, A.C.; Ropstad, E.O.; Ekesten, B.; Karlstam, L.; Wade, C.M.; Lingaas, F. Progressive retinal atrophy in Shetland sheepdog is associated with a mutation in the CNGA1 gene. Anim Genet. 2015, 46, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Xiao, Y.; Li, X.; Ruan, S.; Luo, X.; Wan, X.; Wang, F.; Sun, X. Retinal degeneration in mice lacking the cyclic nucleotide-gated channel subunit CNGA1. FASEB J. 2021, 35, e21859. [Google Scholar] [CrossRef] [PubMed]
- Hüttl, S.; Michalakis, S.; Seeliger, M.; Luo, D.-G.; Acar, N.; Geiger, H.; Hudl, K.; Mader, R.; Haverkamp, S.; Moser, M.; et al. Impaired Channel Targeting and Retinal Degeneration in Mice Lacking the Cyclic Nucleotide-Gated Channel Subunit CNGB1. J. Neurosci. 2005, 25, 130–138. [Google Scholar] [CrossRef]
- Winkler, P.A.; Ekenstedt, K.J.; Occelli, L.M.; Frattaroli, A.V.; Bartoe, J.T.; Venta, P.J.; Petersen-Jones, S.M. A large animal model for CNGB1 autosomal recessive retinitis pigmentosa. PloS ONE 2013, 8, e72229. [Google Scholar] [CrossRef]
- Zhang, Y.; Molday, L.L.; Molday, R.S.; Sarfare, S.S.; Woodruff, M.L.; Fain, G.L.; Kraft, T.W.; Pittler, S.J. Knockout of GARPs and the beta-subunit of the rod cGMP-gated channel disrupts disk morphogenesis and rod outer segment structural integrity. J. Cell Sci. 2009, 122, 1192–1200. [Google Scholar] [CrossRef]
- Ahonen, S.J.; Arumilli, M.; Lohi, H. A CNGB1 frameshift mutation in Papillon and Phalene dogs with progressive retinal atrophy. PloS ONE 2013, 8, e72122. [Google Scholar] [CrossRef]
- Winkler, P.A.; Occelli, L.M.; Petersen-Jones, S.M. Large Animal Models of Inherited Retinal Degenerations: A Review. Cells 2020, 9, 882. [Google Scholar] [CrossRef]
- Petersen-Jones, S.M.; Occelli, L.M.; Winkler, P.A.; Lee, W.; Sparrow, J.R.; Tsukikawa, M.; Boye, S.L.; Chiodo, V.; Capasso, J.E.; Becirovic, E.; et al. Patients and animal models of CNGbeta1-deficient retinitis pigmentosa support gene augmentation approach. J. Clin. Investig. 2018, 128, 190–206. [Google Scholar] [CrossRef]
- Blank, T.; Goldmann, T.; Koch, M.; Amann, L.; Schon, C.; Bonin, M.; Pang, S.; Prinz, M.; Burnet, M.; Wagner, J.E.; et al. Early Microglia Activation Precedes Photoreceptor Degeneration in a Mouse Model of CNGB1-Linked Retinitis Pigmentosa. Front. Immunol. 2017, 8, 1930. [Google Scholar] [CrossRef]
- DeRamus, M.L.; Stacks, D.A.; Zhang, Y.; Huisingh, C.E.; McGwin, G.; Pittler, S.J. GARP2 accelerates retinal degeneration in rod cGMP-gated cation channel beta-subunit knockout mice. Sci. Rep. 2017, 7, 42545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rubin, G.R.; Fineberg, N.; Huisingh, C.; McGwin, G.; Pittler, S.J.; Kraft, T.W. Age-related changes in Cngb1-X1 knockout mice: Prolonged cone survival. Doc. Ophthalmol 2012, 124, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Kandaswamy, S.; Zobel, L.; John, B.; Santhiya, S.T.; Bogedein, J.; Przemeck, G.K.H.; Gailus-Durner, V.; Fuchs, H.; Biel, M.; de Angelis, M.H.; et al. Mutations within the cGMP-binding domain of CNGA1 causing autosomal recessive retinitis pigmentosa in human and animal model. Cell Death Discov. 2022, 8, 387. [Google Scholar] [CrossRef]
- Ardell, M.D.; Bedsole, D.L.; Schoborg, R.V.; Pittler, S.J. Genomic organization of the human rod photoreceptor cGMP-gated cation channel beta-subunit gene. Gene 2000, 245, 311–318. [Google Scholar] [CrossRef]
- Batra-Safferling, R.; Abarca-Heidemann, K.; Korschen, H.G.; Tziatzios, C.; Stoldt, M.; Budyak, I.; Willbold, D.; Schwalbe, H.; Klein-Seetharaman, J.; Kaupp, U.B. Glutamic acid-rich proteins of rod photoreceptors are natively unfolded. J. Biological Chem. 2006, 281, 1449–1460. [Google Scholar] [CrossRef]
- Ritter, L.M.; Khattree, N.; Tam, B.; Moritz, O.L.; Schmitz, F.; Goldberg, A.F. In situ visualization of protein interactions in sensory neurons: Glutamic acid-rich proteins (GARPs) play differential roles for photoreceptor outer segment scaffolding. J. Neurosci. 2011, 31, 11231–11243. [Google Scholar] [CrossRef]
- Chakraborty, D.; Conley, S.M.; DeRamus, M.L.; Pittler, S.J.; Naash, M.I. Varying the GARP2-to-RDS Ratio Leads to Defects in Rim Formation and Rod and Cone Function. Investig. Ophthalmology Visual Sci. 2015, 56, 8187–8198. [Google Scholar] [CrossRef] [PubMed]
- Pearring, J.N.; Martinez-Marquez, J.; Willer, J.R.; Lieu, E.C.; Salinas, R.Y.; Arshavsky, V.Y. The GARP Domain of the Rod CNG Channel’s beta1-Subunit Contains Distinct Sites for Outer Segment Targeting and Connecting to the Photoreceptor Disk Rim. J. Neurosci. 2021, 41, 3094–3104. [Google Scholar] [CrossRef]
- Ba-Abbad, R.; Holder, G.E.; Robson, A.G.; Neveu, M.M.; Waseem, N.; Arno, G.; Webster, A.R. Isolated rod dysfunction associated with a novel genotype of CNGB1. Am. J. Ophthalmol Case Rep. 2019, 14, 83–86. [Google Scholar] [CrossRef]
- Michalakis, S.; Gerhardt, M.; Rudolph, G.; Priglinger, S.; Priglinger, C. Achromatopsia: Genetics and Gene Therapy. Mol. Diagn Ther 2022, 26, 51–59. [Google Scholar] [CrossRef]
- Neitz, J.; Neitz, M. The genetics of normal and defective color vision. Vision Res. 2011, 51, 633–651. [Google Scholar] [CrossRef]
- Hirji, N.; Aboshiha, J.; Georgiou, M.; Bainbridge, J.; Michaelides, M. Achromatopsia: Clinical features, molecular genetics, animal models and therapeutic options. Ophthalmic Genet. 2018, 39, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Brunetti-Pierri, R.; Karali, M.; Melillo, P.; Di Iorio, V.; De Benedictis, A.; Iaccarino, G.; Testa, F.; Banfi, S.; Simonelli, F. Clinical and Molecular Characterization of Achromatopsia Patients: A Longitudinal Study. Int J. Mol. Sci 2021, 22, 1681. [Google Scholar] [CrossRef]
- Andreasson, S.; Tornqvist, K. Electroretinograms in patients with achromatopsia. Acta Ophthalmol. 1991, 69, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Felden, J.; Baumann, B.; Ali, M.; Audo, I.; Ayuso, C.; Bocquet, B.; Casteels, I.; Garcia-Sandoval, B.; Jacobson, S.G.; Jurklies, B.; et al. Mutation spectrum and clinical investigation of achromatopsia patients with mutations in the GNAT2 gene. Hum. Mutat 2019, 40, 1145–1155. [Google Scholar] [CrossRef]
- Kohl, S.; Varsanyi, B.; Antunes, G.A.; Baumann, B.; Hoyng, C.B.; Jagle, H.; Rosenberg, T.; Kellner, U.; Lorenz, B.; Salati, R.; et al. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur J. Hum. Genet. 2005, 13, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.K.; Van Cauwenbergh, C.; Rother, C.; Baumann, B.; Reuter, P.; De Baere, E.; Wissinger, B.; Kohl, S.; Group, A.S. CNGB3 mutation spectrum including copy number variations in 552 achromatopsia patients. Hum. Mutat 2017, 38, 1579–1591. [Google Scholar] [CrossRef]
- Georgiou, M.; Fujinami, K.; Michaelides, M. Inherited retinal diseases: Therapeutics, clinical trials and end points-A review. Clin. Exp. Ophthalmol. 2021, 49, 270–288. [Google Scholar] [CrossRef]
- Weisschuh, N.; Obermaier, C.D.; Battke, F.; Bernd, A.; Kuehlewein, L.; Nasser, F.; Zobor, D.; Zrenner, E.; Weber, E.; Wissinger, B.; et al. Genetic architecture of inherited retinal degeneration in Germany: A large cohort study from a single diagnostic center over a 9-year period. Hum. Mutat 2020, 41, 1514–1527. [Google Scholar] [CrossRef]
- Sun, W.; Li, S.; Xiao, X.; Wang, P.; Zhang, Q. Genotypes and phenotypes of genes associated with achromatopsia: A reference for clinical genetic testing. Molecular vision 2020, 26, 588–602. [Google Scholar]
- Solaki, M.; Baumann, B.; Reuter, P.; Andreasson, S.; Audo, I.; Ayuso, C.; Balousha, G.; Benedicenti, F.; Birch, D.; Bitoun, P.; et al. Comprehensive variant spectrum of the CNGA3 gene in patients affected by achromatopsia. Hum. Mutat 2022, 43, 832–858. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.; Marx, T.; Giddings, I.; Jagle, H.; Jacobson, S.G.; Apfelstedt-Sylla, E.; Zrenner, E.; Sharpe, L.T.; Wissinger, B. Total colourblindness is caused by mutations in the gene encoding the alpha-subunit of the cone photoreceptor cGMP-gated cation channel. Nature genetics 1998, 19, 257–259. [Google Scholar] [CrossRef] [PubMed]
- Thiadens, A.A.; Slingerland, N.W.; Roosing, S.; van Schooneveld, M.J.; van Lith-Verhoeven, J.J.; van Moll-Ramirez, N.; van den Born, L.I.; Hoyng, C.B.; Cremers, F.P.; Klaver, C.C. Genetic etiology and clinical consequences of complete and incomplete achromatopsia. Ophthalmology 2009, 116, 1984–1989.e1981. [Google Scholar] [CrossRef] [PubMed]
- Wissinger, B.; Gamer, D.; Jagle, H.; Giorda, R.; Marx, T.; Mayer, S.; Tippmann, S.; Broghammer, M.; Jurklies, B.; Rosenberg, T.; et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am. J. Hum. Genet. 2001, 69, 722–737. [Google Scholar] [CrossRef]
- Tränkner, D.; Jagle, H.; Kohl, S.; Apfelstedt-Sylla, E.; Sharpe, L.T.; Kaupp, U.B.; Zrenner, E.; Seifert, R.; Wissinger, B. Molecular basis of an inherited form of incomplete achromatopsia. J. Neurosci. 2004, 24, 138–147. [Google Scholar] [CrossRef]
- Nishiguchi, K.M.; Sandberg, M.A.; Gorji, N.; Berson, E.L.; Dryja, T.P. Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum. Mutat 2005, 25, 248–258. [Google Scholar] [CrossRef]
- Varsanyi, B.; Wissinger, B.; Kohl, S.; Koeppen, K.; Farkas, A. Clinical and genetic features of Hungarian achromatopsia patients. Molecular vision 2005, 11, 996–1001. [Google Scholar]
- Goto-Omoto, S.; Hayashi, T.; Gekka, T.; Kubo, A.; Takeuchi, T.; Kitahara, K. Compound heterozygous CNGA3 mutations (R436W, L633P) in a Japanese patient with congenital achromatopsia. Vis. Neurosci 2006, 23, 395–402. [Google Scholar] [CrossRef]
- Wiszniewski, W.; Lewis, R.A.; Lupski, J.R. Achromatopsia: The CNGB3 p.T383fsX mutation results from a founder effect and is responsible for the visual phenotype in the original report of uniparental disomy 14. Hum. Genet. 2007, 121, 433–439. [Google Scholar] [CrossRef]
- Sundin, O.H.; Yang, J.M.; Li, Y.; Zhu, D.; Hurd, J.N.; Mitchell, T.N.; Silva, E.D.; Maumenee, I.H. Genetic basis of total colourblindness among the Pingelapese islanders. Nat. Genet. 2000, 25, 289–293. [Google Scholar] [CrossRef]
- Kohl, S.; Baumann, B.; Broghammer, M.; Jagle, H.; Sieving, P.; Kellner, U.; Spegal, R.; Anastasi, M.; Zrenner, E.; Sharpe, L.T.; et al. Mutations in the CNGB3 gene encoding the beta-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum. Mol. Genet. 2000, 9, 2107–2116. [Google Scholar] [CrossRef] [PubMed]
- Michaelides, M.; Aligianis, I.A.; Ainsworth, J.R.; Good, P.; Mollon, J.D.; Maher, E.R.; Moore, A.T.; Hunt, D.M. Progressive cone dystrophy associated with mutation in CNGB3. Investig. Ophthalmol. Vis. Sci. 2004, 45, 1975–1982. [Google Scholar]
- Okada, A.; Ueyama, H.; Toyoda, F.; Oda, S.; Ding, W.G.; Tanabe, S.; Yamade, S.; Matsuura, H.; Ohkubo, I.; Kani, K. Functional role of hCngb3 in regulation of human cone cng channel: Effect of rod monochromacy-associated mutations in hCNGB3 on channel function. Investig. Ophthalmol. Vis. Sci. 2004, 45, 2324–2332. [Google Scholar] [CrossRef]
- Rojas, C.V.; Maria, L.S.; Santos, J.L.; Cortes, F.; Alliende, M.A. A frameshift insertion in the cone cyclic nucleotide gated cation channel causes complete achromatopsia in a consanguineous family from a rural isolate. Eur J. Hum. Genet. 2002, 10, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Brody, J.A.; Hussels, I.; Brink, E.; Torres, J. Hereditary blindness among Pingelapese people of Eastern Caroline Islands. Lancet 1970, 1, 1253–1257. [Google Scholar] [CrossRef]
- Sacks, O.W. The Island of the Colorblind, 1st ed.; A.A. Knopf: New York, NY, USA, 1997; p. 298. [Google Scholar]
- Winick, J.D.; Blundell, M.L.; Galke, B.L.; Salam, A.A.; Leal, S.M.; Karayiorgou, M. Homozygosity mapping of the Achromatopsia locus in the Pingelapese. Am. J. Human Gene. 1999, 64, 1679–1685. [Google Scholar] [CrossRef]
- Zelinger, L.; Cideciyan, A.V.; Kohl, S.; Schwartz, S.B.; Rosenmann, A.; Eli, D.; Sumaroka, A.; Roman, A.J.; Luo, X.; Brown, C.; et al. Genetics and Disease Expression in the CNGA3 Form of Achromatopsia: Steps on the Path to Gene Therapy. Ophthalmology 2015, 122, 997–1007. [Google Scholar] [CrossRef]
- Liang, X.; Dong, F.; Li, H.; Li, H.; Yang, L.; Sui, R. Novel CNGA3 mutations in Chinese patients with achromatopsia. Br. J. Ophthalmol. 2015, 99, 571–576. [Google Scholar] [CrossRef]
- Burkard, M.; Kohl, S.; Kratzig, T.; Tanimoto, N.; Brennenstuhl, C.; Bausch, A.E.; Junger, K.; Reuter, P.; Sothilingam, V.; Beck, S.C.; et al. Accessory heterozygous mutations in cone photoreceptor CNGA3 exacerbate CNG channel-associated retinopathy. J. Clinical Investig. 2018, 128, 5663–5675. [Google Scholar] [CrossRef]
- Täger, J.; Wissinger, B.; Kohl, S.; Reuter, P. Identification of Chemical and Pharmacological Chaperones for Correction of Trafficking-Deficient Mutant Cyclic Nucleotide-Gated A3 Channels. Mol. Pharmacol 2021, 99, 460–468. [Google Scholar] [CrossRef]
- Brennenstuhl, C.; Tanimoto, N.; Burkard, M.; Wagner, R.; Bolz, S.; Trifunovic, D.; Kabagema-Bilan, C.; Paquet-Durand, F.; Beck, S.C.; Huber, G.; et al. Targeted ablation of the Pde6h gene in mice reveals cross-species differences in cone and rod phototransduction protein isoform inventory. J. Biol. Chem. 2015, 290, 10242–10255. [Google Scholar] [CrossRef] [PubMed]
- Buena-Atienza, E.; Ruther, K.; Baumann, B.; Bergholz, R.; Birch, D.; De Baere, E.; Dollfus, H.; Greally, M.T.; Gustavsson, P.; Hamel, C.P.; et al. De novo intrachromosomal gene conversion from OPN1MW to OPN1LW in the male germline results in Blue Cone Monochromacy. Sci. Rep. 2016, 6, 28253. [Google Scholar] [CrossRef] [PubMed]
- Dai, G.; Varnum, M.D. CNGA3 achromatopsia-associated mutation potentiates the phosphoinositide sensitivity of cone photoreceptor CNG channels by altering intersubunit interactions. Am. J. Physiol. Cell Physiol. 2013, 305, C147–159. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Duricka, D.L.; Brown, R.L.; Varnum, M.D. Defective trafficking of cone photoreceptor CNG channels induces the unfolded protein response and ER-stress-associated cell death. Biochem J. 2012, 441, 685–696. [Google Scholar] [CrossRef]
- Koeppen, K.; Reuter, P.; Kohl, S.; Baumann, B.; Ladewig, T.; Wissinger, B. Functional analysis of human CNGA3 mutations associated with colour blindness suggests impaired surface expression of channel mutants A3(R427C) and A3(R563C). Eur. J. Neurosci. 2008, 27, 2391–2401. [Google Scholar] [CrossRef]
- Koeppen, K.; Reuter, P.; Ladewig, T.; Kohl, S.; Baumann, B.; Jacobson, S.G.; Plomp, A.S.; Hamel, C.P.; Janecke, A.R.; Wissinger, B. Dissecting the pathogenic mechanisms of mutations in the pore region of the human cone photoreceptor cyclic nucleotide-gated channel. Hum. Mutat. 2010, 31, 830–839. [Google Scholar] [CrossRef]
- Kuniyoshi, K.; Muraki-Oda, S.; Ueyama, H.; Toyoda, F.; Sakuramoto, H.; Ogita, H.; Irifune, M.; Yamamoto, S.; Nakao, A.; Tsunoda, K.; et al. Novel mutations in the gene for alpha-subunit of retinal cone cyclic nucleotide-gated channels in a Japanese patient with congenital achromatopsia. Jpn J. Ophthalmol. 2016, 60, 187–197. [Google Scholar] [CrossRef]
- Liu, C.; Varnum, M.D. Functional consequences of progressive cone dystrophy-associated mutations in the human cone photoreceptor cyclic nucleotide-gated channel CNGA3 subunit. Am. J. Physiol. Cell Physiol. 2005, 289, C187–C198. [Google Scholar] [CrossRef]
- Matveev, A.V.; Fitzgerald, J.B.; Xu, J.; Malykhina, A.P.; Rodgers, K.K.; Ding, X.Q. The disease-causing mutations in the carboxyl terminus of the cone cyclic nucleotide-gated channel CNGA3 subunit alter the local secondary structure and interfere with the channel active conformational change. Biochemistry 2010, 49, 1628–1639. [Google Scholar] [CrossRef]
- Meighan, P.C.; Peng, C.; Varnum, M.D. Inherited macular degeneration-associated mutations in CNGB3 increase the ligand sensitivity and spontaneous open probability of cone cyclic nucleotide-gated channels. Front. Physiol. 2015, 6, 177. [Google Scholar] [CrossRef]
- Muraki-Oda, S.; Toyoda, F.; Okada, A.; Tanabe, S.; Yamade, S.; Ueyama, H.; Matsuura, H.; Ohji, M. Functional analysis of rod monochromacy-associated missense mutations in the CNGA3 subunit of the cone photoreceptor cGMP-gated channel. Biochem. Biophys. Res. Commun. 2007, 362, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.A.; Bartoli, K.M.; Fandino, R.A.; Ngatchou, A.N.; Woch, G.; Carey, J.; Tanaka, J.C. Transmembrane S1 mutations in CNGA3 from achromatopsia 2 patients cause loss of function and impaired cellular trafficking of the cone CNG channel. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2282–2290. [Google Scholar] [CrossRef] [PubMed]
- Reuter, P.; Koeppen, K.; Ladewig, T.; Kohl, S.; Baumann, B.; Wissinger, B.; Achromatopsia Clinical Study, G. Mutations in CNGA3 impair trafficking or function of cone cyclic nucleotide-gated channels, resulting in achromatopsia. Hum. Mutat. 2008, 29, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, R.S.; Reuter, P.; Sisk, R.A.; Kausar, T.; Shahzad, M.; Maqsood, M.I.; Yousif, A.; Ali, M.; Riazuddin, S.; Wissinger, B.; et al. Homozygous missense variant in the human CNGA3 channel causes cone-rod dystrophy. Eur J. Hum. Genet. 2015, 23, 473–480. [Google Scholar] [CrossRef]
- Täger, J.; Kohl, S.; Birch, D.G.; Wheaton, D.K.H.; Wissinger, B.; Reuter, P. An early nonsense mutation facilitates the expression of a short isoform of CNGA3 by alternative translation initiation. Exp. Eye Res. 2018, 171, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Biel, M.; Seeliger, M.; Pfeifer, A.; Kohler, K.; Gerstner, A.; Ludwig, A.; Jaissle, G.; Fauser, S.; Zrenner, E.; Hofmann, F. Selective loss of cone function in mice lacking the cyclic nucleotide-gated channel CNG3. Proc. Natl. Acad. Sci. USA 1999, 96, 7553–7557. [Google Scholar] [CrossRef]
- Michalakis, S.; Geiger, H.; Haverkamp, S.; Hofmann, F.; Gerstner, A.; Biel, M. Impaired opsin targeting and cone photoreceptor migration in the retina of mice lacking the cyclic nucleotide-gated channel CNGA3. Investig. Ophthalmol. Vis. Sci. 2005, 46, 1516–1524. [Google Scholar] [CrossRef]
- Pang, J.J.; Deng, W.T.; Dai, X.; Lei, B.; Everhart, D.; Umino, Y.; Li, J.; Zhang, K.; Mao, S.; Boye, S.L.; et al. AAV-mediated cone rescue in a naturally occurring mouse model of CNGA3-achromatopsia. PLoS ONE 2012, 7, e35250. [Google Scholar] [CrossRef]
- Reicher, S.; Seroussi, E.; Gootwine, E. A mutation in gene CNGA3 is associated with day blindness in sheep. Genomics 2010, 95, 101–104. [Google Scholar] [CrossRef]
- Shamir, M.H.; Ofri, R.; Bor, A.; Brenner, O.; Reicher, S.; Obolensky, A.; Averbukh, E.; Banin, E.; Gootwine, E. A novel day blindness in sheep: Epidemiological, behavioural, electrophysiological and histopathological studies. Vet. J. 2010, 185, 130–137. [Google Scholar] [CrossRef]
- Tanaka, N.; Dutrow, E.V.; Miyadera, K.; Delemotte, L.; MacDermaid, C.M.; Reinstein, S.L.; Crumley, W.R.; Dixon, C.J.; Casal, M.L.; Klein, M.L.; et al. Canine CNGA3 Gene Mutations Provide Novel Insights into Human Achromatopsia-Associated Channelopathies and Treatment. PloS ONE 2015, 10, e0138943. [Google Scholar] [CrossRef]
- Ding, X.Q.; Harry, C.S.; Umino, Y.; Matveev, A.V.; Fliesler, S.J.; Barlow, R.B. Impaired cone function and cone degeneration resulting from CNGB3 deficiency: Down-regulation of CNGA3 biosynthesis as a potential mechanism. Hum. Mol. Genet. 2009, 18, 4770–4780. [Google Scholar] [CrossRef]
- Xu, J.; Morris, L.; Fliesler, S.J.; Sherry, D.M.; Ding, X.Q. Early-onset, slow progression of cone photoreceptor dysfunction and degeneration in CNG channel subunit CNGB3 deficiency. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3557–3566. [Google Scholar] [CrossRef][Green Version]
- Hassall, M.M.; Barnard, A.R.; Orlans, H.O.; McClements, M.E.; Charbel Issa, P.; Aslam, S.A.; MacLaren, R.E. A Novel Achromatopsia Mouse Model Resulting From a Naturally Occurring Missense Change in Cngb3. Investig. Ophthalmol. Vis. Sci. 2018, 59, 6102–6110. [Google Scholar] [CrossRef]
- Sidjanin, D.J.; Lowe, J.K.; McElwee, J.L.; Milne, B.S.; Phippen, T.M.; Sargan, D.R.; Aguirre, G.D.; Acland, G.M.; Ostrander, E.A. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum. Mol. Genet. 2002, 11, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, G.D.; Rubin, L.F. Pathology of hemeralopia in the Alaskan malamute dog. Investig. Ophthalmol. 1974, 13, 231–235. [Google Scholar] [PubMed]
- Lin, Q.; Lv, J.N.; Wu, K.C.; Zhang, C.J.; Liu, Q.; Jin, Z.B. Generation of Nonhuman Primate Model of Cone Dysfunction through In Situ AAV-Mediated CNGB3 Ablation. Mol. Ther. Methods Clin. Dev. 2020, 18, 869–879. [Google Scholar] [CrossRef]
- Koch, S.; Sothilingam, V.; Garcia Garrido, M.; Tanimoto, N.; Becirovic, E.; Koch, F.; Seide, C.; Beck, S.C.; Seeliger, M.W.; Biel, M.; et al. Gene therapy restores vision and delays degeneration in the CNGB1(-/-) mouse model of retinitis pigmentosa. Hum. Mol. Genet. 2012, 21, 4486–4496. [Google Scholar] [CrossRef]
- Wagner, J.E.; Zobel, L.; Gerhardt, M.J.; O’Riordan, C.R.; Frederick, A.; Petersen-Jones, S.M.; Biel, M.; Michalakis, S. In Vivo Potency Testing of Subretinal rAAV5.hCNGB1 Gene Therapy in the Cngb1 Knockout Mouse Model of Retinitis Pigmentosa. Hum. Gene Ther. 2021, 32, 1158–1170. [Google Scholar] [CrossRef]
- Hawes, N.; Wang, X.; Hurd, R.; Wang, J.; Davisson, M.; Nusinowitz, S.; Heckenlively, J.; Chang, B. A point mutation in the Cnga3 gene causes cone photoreceptor function loss (cpfl5) in mice. Investig. Ophthalmol. Vis. Sci. 2006, 47, 4579. [Google Scholar]
- Michalakis, S.; Mühlfriedel, R.; Tanimoto, N.; Krishnamoorthy, V.; Koch, S.; Fischer, M.D.; Becirovic, E.; Bai, L.; Huber, G.; Beck, S.C.; et al. Restoration of cone vision in the CNGA3-/- mouse model of congenital complete lack of cone photoreceptor function. Mol. Ther. 2010, 18, 2057–2063. [Google Scholar] [CrossRef] [PubMed]
- Mühlfriedel, R.; Tanimoto, N.; Schön, C.; Sothilingam, V.; Garcia Garrido, M.; Beck, S.C.; Huber, G.; Biel, M.; Seeliger, M.W.; Michalakis, S. AAV-Mediated Gene Supplementation Therapy in Achromatopsia Type 2: Preclinical Data on Therapeutic Time Window and Long-Term Effects. Front. Neurosci. 2017, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Tao, Y.; Deng, W.T.; Zhu, P.; Li, J.; Dai, X.; Zhang, Y.; Shi, W.; Liu, X.; Chiodo, V.A.; et al. Vitreal delivery of AAV vectored Cnga3 restores cone function in CNGA3-/-/Nrl-/- mice, an all-cone model of CNGA3 achromatopsia. Hum. Mol. Genet. 2015, 24, 3699–3707. [Google Scholar] [CrossRef] [PubMed]
- Pavlou, M.; Schön, C.; Occelli, L.M.; Rossi, A.; Meumann, N.; Boyd, R.F.; Bartoe, J.T.; Siedlecki, J.; Gerhardt, M.J.; Babutzka, S.; et al. Novel AAV capsids for intravitreal gene therapy of photoreceptor disorders. EMBO Mol. Med. 2021, 13, e13392. [Google Scholar] [CrossRef]
- Banin, E.; Gootwine, E.; Obolensky, A.; Ezra-Elia, R.; Ejzenberg, A.; Zelinger, L.; Honig, H.; Rosov, A.; Yamin, E.; Sharon, D.; et al. Gene Augmentation Therapy Restores Retinal Function and Visual Behavior in a Sheep Model of CNGA3 Achromatopsia. Mol. Ther. 2015, 23, 1423–1433. [Google Scholar] [CrossRef]
- Gootwine, E.; Ofri, R.; Banin, E.; Obolensky, A.; Averbukh, E.; Ezra-Elia, R.; Ross, M.; Honig, H.; Rosov, A.; Yamin, E.; et al. Safety and Efficacy Evaluation of rAAV2tYF-PR1.7-hCNGA3 Vector Delivered by Subretinal Injection in CNGA3 Mutant Achromatopsia Sheep. Hum. Gene Ther. Clin. Dev. 2017, 28, 96–107. [Google Scholar] [CrossRef]
- Ofri, R.; Averbukh, E.; Ezra-Elia, R.; Ross, M.; Honig, H.; Obolensky, A.; Rosov, A.; Hauswirth, W.W.; Gootwine, E.; Banin, E. Six Years and Counting: Restoration of Photopic Retinal Function and Visual Behavior Following Gene Augmentation Therapy in a Sheep Model of CNGA3 Achromatopsia. Human. Gene Ther. 2018, 29, 1376–1386. [Google Scholar] [CrossRef]
- Reichel, F.F.; Peters, T.; Wilhelm, B.; Biel, M.; Ueffing, M.; Wissinger, B.; Bartz-Schmidt, K.U.; Klein, R.; Michalakis, S.; Fischer, M.D.; et al. Humoral Immune Response After Intravitreal But Not After Subretinal AAV8 in Primates and Patients. Investig. Ophthalmol. Vis. Sci. 2018, 59, 1910–1915. [Google Scholar] [CrossRef]
- Seitz, I.P.; Michalakis, S.; Wilhelm, B.; Reichel, F.F.; Ochakovski, G.A.; Zrenner, E.; Ueffing, M.; Biel, M.; Wissinger, B.; Bartz-Schmidt, K.U.; et al. Superior Retinal Gene Transfer and Biodistribution Profile of Subretinal Versus Intravitreal Delivery of AAV8 in Nonhuman Primates. Investig. Ophthalmol. Vis. Sci. 2017, 58, 5792–5801. [Google Scholar] [CrossRef]
- Tobias, P.; Philipp, S.I.; Stylianos, M.; Martin, B.; Barbara, W.; Felix, R.; Alexander, O.G.; Eberhart, Z.; Marius, U.; Birgit, K.; et al. Safety and Toxicology of Ocular Gene Therapy with Recombinant AAV Vector rAAV.hCNGA3 in Nonhuman Primates. Hum. Gene Ther. Clin. Dev. 2019, 30, 50–56. [Google Scholar] [CrossRef]
- Reichel, F.F.; Dauletbekov, D.L.; Klein, R.; Peters, T.; Ochakovski, G.A.; Seitz, I.P.; Wilhelm, B.; Ueffing, M.; Biel, M.; Wissinger, B.; et al. AAV8 Can Induce Innate and Adaptive Immune Response in the Primate Eye. Mol. Therapy. 2017, 25, 2648–2660. [Google Scholar] [CrossRef]
- Komaromy, A.M.; Alexander, J.J.; Rowlan, J.S.; Garcia, M.M.; Chiodo, V.A.; Kaya, A.; Tanaka, J.C.; Acland, G.M.; Hauswirth, W.W.; Aguirre, G.D. Gene therapy rescues cone function in congenital achromatopsia. Hum. Mol. Genet. 2010, 19, 2581–2593. [Google Scholar] [CrossRef] [PubMed]
- Komaromy, A.M.; Rowlan, J.S.; Corr, A.T.; Reinstein, S.L.; Boye, S.L.; Cooper, A.E.; Gonzalez, A.; Levy, B.; Wen, R.; Hauswirth, W.W.; et al. Transient photoreceptor deconstruction by CNTF enhances rAAV-mediated cone functional rescue in late stage CNGB3-achromatopsia. Mol. Therapy 2013, 21, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, L.S.; Xu, J.; Pearson, R.A.; Smith, A.J.; Bainbridge, J.W.; Morris, L.M.; Fliesler, S.J.; Ding, X.Q.; Ali, R.R. Long-term and age-dependent restoration of visual function in a mouse model of CNGB3-associated achromatopsia following gene therapy. Hum. Mol. Genet. 2011, 20, 3161–3175. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.J.; Budzynski, E.; Sonnentag, P.; Nork, T.M.; Miller, P.E.; Sharma, A.K.; Ver Hoeve, J.N.; Smith, L.M.; Arndt, T.; Calcedo, R.; et al. Safety and Biodistribution Evaluation in Cynomolgus Macaques of rAAV2tYF-PR1.7-hCNGB3, a Recombinant AAV Vector for Treatment of Achromatopsia. Hum. Gene Ther Clin. Dev. 2016, 27, 37–48. [Google Scholar] [CrossRef]
- Ye, G.J.; Budzynski, E.; Sonnentag, P.; Nork, T.M.; Miller, P.E.; McPherson, L.; Ver Hoeve, J.N.; Smith, L.M.; Arndt, T.; Mandapati, S.; et al. Safety and Biodistribution Evaluation in CNGB3-Deficient Mice of rAAV2tYF-PR1.7-hCNGB3, a Recombinant AAV Vector for Treatment of Achromatopsia. Hum. Gene Ther. Clin. Dev. 2016, 27, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.J.; Komaromy, A.M.; Zeiss, C.; Calcedo, R.; Harman, C.D.; Koehl, K.L.; Stewart, G.A.; Iwabe, S.; Chiodo, V.A.; Hauswirth, W.W.; et al. Safety and Efficacy of AAV5 Vectors Expressing Human or Canine CNGB3 in CNGB3-Mutant Dogs. Hum. Gene Ther. Clin. Dev. 2017, 28, 197–207. [Google Scholar] [CrossRef]
- Trapani, I.; Puppo, A.; Auricchio, A. Vector platforms for gene therapy of inherited retinopathies. Prog. Retinal Eye Res. 2014, 43, 108–128. [Google Scholar] [CrossRef]
- Mendell, J.R.; Al-Zaidy, S.A.; Rodino-Klapac, L.R.; Goodspeed, K.; Gray, S.J.; Kay, C.N.; Boye, S.L.; Boye, S.E.; George, L.A.; Salabarria, S.; et al. Current Clinical Applications of In Vivo Gene Therapy with AAVs. Mol. Ther. 2021, 29, 464–488. [Google Scholar] [CrossRef]
- Samulski, R.J.; Berns, K.I.; Tan, M.; Muzyczka, N. Cloning of adeno-associated virus into pBR322: Rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. USA 1982, 79, 2077–2081. [Google Scholar] [CrossRef]
- Penaud-Budloo, M.; Francois, A.; Clement, N.; Ayuso, E. Pharmacology of Recombinant Adeno-associated Virus Production. Mol. Ther Methods Clin. Dev. 2018, 8, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, J.; Chan, Y.K.; Samulski, R.J. Adeno-associated Virus (AAV) versus Immune Response. Viruses 2019, 11, 102. [Google Scholar] [CrossRef]
- Verdera, H.C.; Kuranda, K.; Mingozzi, F. AAV Vector Immunogenicity in Humans: A Long Journey to Successful Gene Transfer. Mol. Therapy. 2020, 28, 723–746. [Google Scholar] [CrossRef] [PubMed]
- Rapti, K.; Grimm, D. Adeno-Associated Viruses (AAV) and Host Immunity—A Race Between the Hare and the Hedgehog. Front. Immunol. 2021, 12, 753467. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.; Seitz, I.P.; Michalakis, S.; Biel, M.; Wilhelm, B.; Reichel, F.; Ochakovski, G.A.; Zrenner, E.; Ueffing, M.; Korbmacher, B.; et al. Safety and Toxicology of Ocular Gene Therapy with Recombinant AAV Vector rAAV.hCNGA3 in Nonhuman Primates. Hum. Gene Ther. Clin. Dev. 2019, 30, 50–56. [Google Scholar] [CrossRef]
- Reichel, F.F.; Michalakis, S.; Wilhelm, B.; Zobor, D.; Muehlfriedel, R.; Kohl, S.; Weisschuh, N.; Sothilingam, V.; Kuehlewein, L.; Kahle, N.; et al. Three-year results of phase I retinal gene therapy trial for CNGA3-mutated achromatopsia: Results of a non randomised controlled trial. Bri. J. Ophthalmol. 2021, 106, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.D.; Michalakis, S.; Wilhelm, B.; Zobor, D.; Muehlfriedel, R.; Kohl, S.; Weisschuh, N.; Ochakovski, G.A.; Klein, R.; Schoen, C.; et al. Safety and Vision Outcomes of Subretinal Gene Therapy Targeting Cone Photoreceptors in Achromatopsia: A Nonrandomized Controlled Trial. JAMA Ophthalmol. 2020, 138, 643–651. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).