Association between SMAD4 Mutations and GATA6 Expression in Paired Pancreatic Ductal Adenocarcinoma Tumor Specimens: Data from Two Independent Molecularly-Characterized Cohorts
Abstract
:1. Introduction
2. Methods
Statistical Analysis
3. Results
3.1. Patient Cohort
3.2. Association between SMAD4 Alteration Status and Basal Subtype
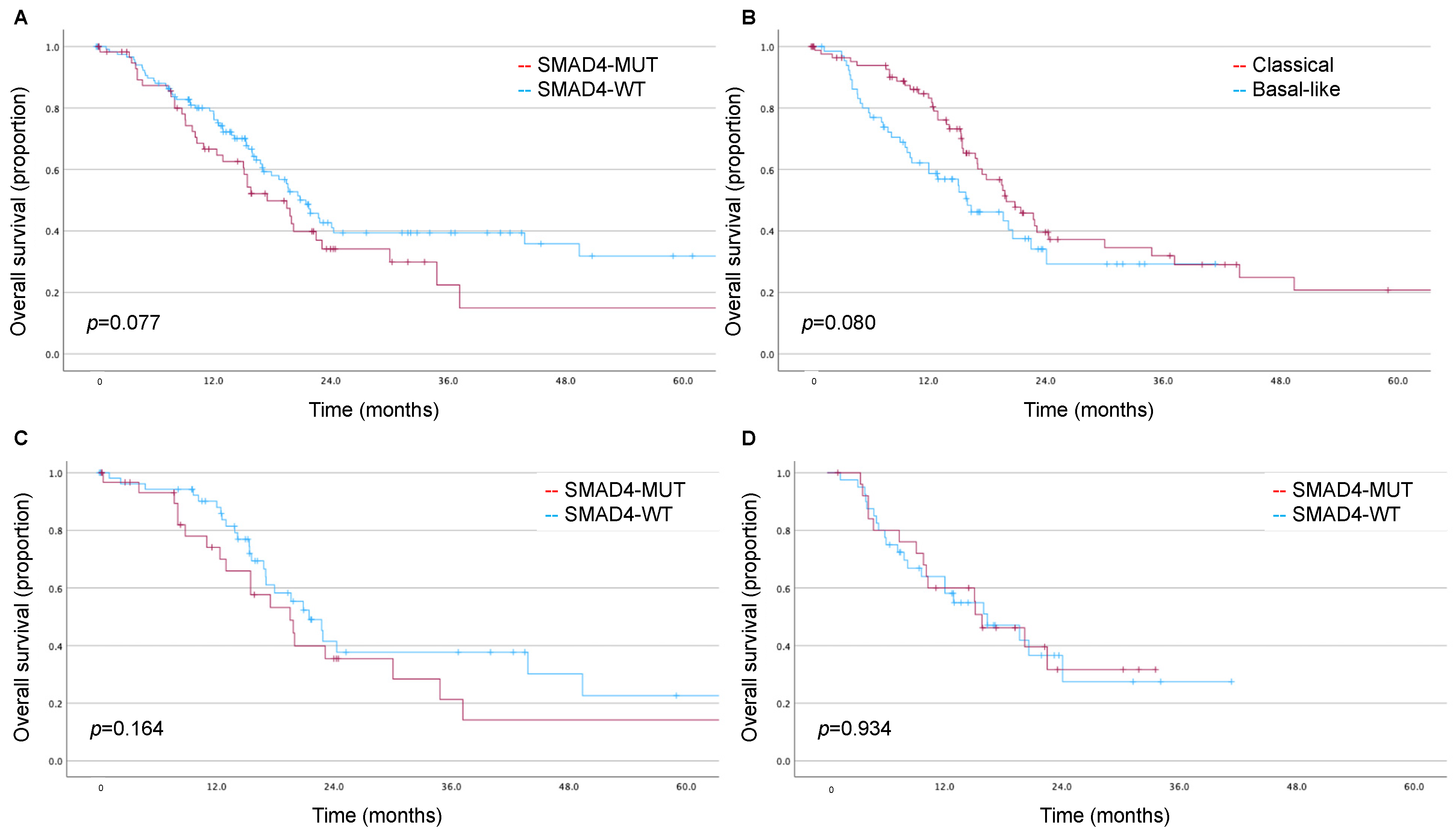
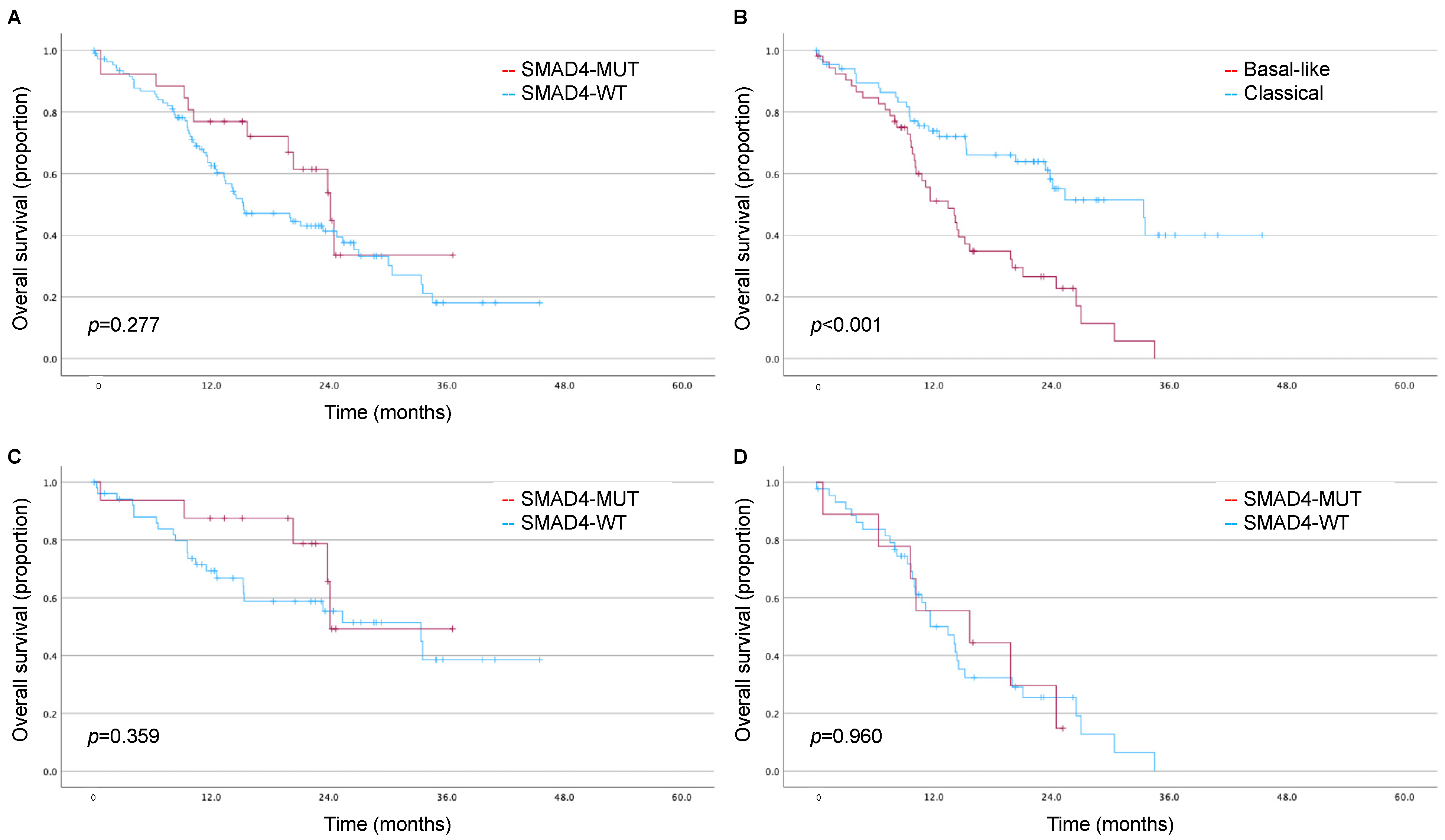
3.3. Impact of SMAD4 and Moffitt Subtype on Long-Term Survival
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Chawla, A.; O’Reilly, E.M. Pancreatic Cancer: A Review. JAMA 2021, 326, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.A.; Duong, M.; Sohal, D.P.S.; Gandhi, N.S.; Beg, M.S.; Wang-Gillam, A.; Wade, J.L., 3rd; Chiorean, E.G.; Guthrie, K.A.; Lowy, A.M.; et al. Surgical Outcome Results from SWOG S1505: A Randomized Clinical Trial of mFOLFIRINOX versus Gemcitabine/Nab-paclitaxel for Perioperative Treatment of Resectable Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2020, 272, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.L.; Chone, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Tempero, M.A.; Pelzer, U.; O’Reilly, E.M.; Winter, J.; Oh, D.Y.; Li, C.P.; Tortora, G.; Chang, H.M.; Lopez, C.D.; Bekaii-Saab, T.; et al. Adjuvant nab-Paclitaxel + Gemcitabine in Resected Pancreatic Ductal Adenocarcinoma: Results From a Randomized, Open-Label, Phase III Trial. J. Clin. Oncol. 2023, 41, 2007–2019. [Google Scholar] [CrossRef]
- Wang, F.; Xia, X.; Yang, C.; Shen, J.; Mai, J.; Kim, H.C.; Kirui, D.; Kang, Y.; Fleming, J.B.; Koay, E.J.; et al. SMAD4 Gene Mutation Renders Pancreatic Cancer Resistance to Radiotherapy through Promotion of Autophagy. Clin. Cancer Res. 2018, 24, 3176–3185. [Google Scholar] [CrossRef]
- Ecker, B.L.; Court, C.M.; Janssen, Q.P.; Tao, A.J.; D’Angelica, M.I.; Drebin, J.A.; Gonen, M.; O’Reilly, E.M.; Jarnagin, W.R.; Wei, A.C.; et al. Alterations in Somatic Driver Genes Are Associated with Response to Neoadjuvant FOLFIRINOX in Patients with Localized Pancreatic Ductal Adenocarcinoma. J. Am. Coll. Surg. 2022, 235, 342–349. [Google Scholar] [CrossRef]
- Ecker, B.L.; Tao, A.J.; Janssen, Q.P.; Walch, H.S.; Court, C.M.; Balachandran, V.P.; Crane, C.H.; D’Angelica, M.I.; Drebin, J.A.; Kingham, T.P.; et al. Genomic Biomarkers Associated with Response to Induction Chemotherapy in Patients with Localized Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2023, 29, 1368–1374. [Google Scholar] [CrossRef]
- Martinelli, P.; Carrillo-de Santa Pau, E.; Cox, T.; Sainz, B., Jr.; Dusetti, N.; Greenhalf, W.; Rinaldi, L.; Costello, E.; Ghaneh, P.; Malats, N.; et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 2017, 66, 1665–1676. [Google Scholar] [CrossRef]
- Eyres, M.; Lanfredini, S.; Xu, H.; Burns, A.; Blake, A.; Willenbrock, F.; Goldin, R.; Hughes, D.; Hughes, S.; Thapa, A.; et al. TET2 Drives 5hmc Marking of GATA6 and Epigenetically Defines Pancreatic Ductal Adenocarcinoma Transcriptional Subtypes. Gastroenterology 2021, 161, 653–668.e6. [Google Scholar] [CrossRef]
- Williams, H.L.; Dias Costa, A.; Zhang, J.; Raghavan, S.; Winter, P.S.; Kapner, K.S.; Ginebaugh, S.P.; Väyrynen, S.A.; Väyrynen, J.P.; Yuan, C.; et al. Spatially Resolved Single-Cell Assessment of Pancreatic Cancer Expression Subtypes Reveals Co-expressor Phenotypes and Extensive Intratumoral Heterogeneity. Cancer Res. 2023, 83, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- O’Kane, G.M.; Grunwald, B.T.; Jang, G.H.; Masoomian, M.; Picardo, S.; Grant, R.C.; Denroche, R.E.; Zhang, A.; Wang, Y.; Lam, B.; et al. GATA6 Expression Distinguishes Classical and Basal-like Subtypes in Advanced Pancreatic Cancer. Clin. Cancer Res. 2020, 26, 4901–4910. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Huang, C.; Cui Zhou, D.; Hu, Y.; Lih, T.M.; Savage, S.R.; Krug, K.; Clark, D.J.; Schnaubelt, M.; Chen, L.; et al. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell 2021, 184, 5031–5052 e5026. [Google Scholar] [CrossRef]
- de Santiago, I.; Yau, C.; Heij, L.; Middleton, M.R.; Markowetz, F.; Grabsch, H.I.; Dustin, M.L.; Sivakumar, S. Immunophenotypes of pancreatic ductal adenocarcinoma: Meta-analysis of transcriptional subtypes. Int. J. Cancer 2019, 145, 1125–1137. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef]
- Aung, K.L.; Fischer, S.E.; Denroche, R.E.; Jang, G.-H.; Dodd, A.; Creighton, S.; Southwood, B.; Liang, S.-B.; Chadwick, D.; Zhang, A.; et al. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clin. Cancer Res. 2018, 24, 1344–1354. [Google Scholar] [CrossRef]
- Zhou, X.; An, J.; Kurilov, R.; Brors, B.; Hu, K.; Peccerella, T.; Roessler, S.; Pfütze, K.; Schulz, A.; Wolf, S.; et al. Persister cell phenotypes contribute to poor patient outcomes after neoadjuvant chemotherapy in PDAC. Nat. Cancer 2023, 4, 1362–1381. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardière, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Aguirre, A.J.; Nowak, J.A.; Camarda, N.D.; Moffitt, R.A.; Ghazani, A.A.; Hazar-Rethinam, M.; Raghavan, S.; Kim, J.; Brais, L.K.; Ragon, D.; et al. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Discov. 2018, 8, 1096–1111. [Google Scholar] [CrossRef]
- David, C.J.; Huang, Y.H.; Chen, M.; Su, J.; Zou, Y.; Bardeesy, N.; Iacobuzio-Donahue, C.A.; Massagué, J. TGF-β Tumor Suppression through a Lethal EMT. Cell 2016, 164, 1015–1030. [Google Scholar] [CrossRef]
- Grossberg, A.J.; Chu, L.C.; Deig, C.R.; Fishman, E.K.; Hwang, W.L.; Maitra, A.; Marks, D.L.; Mehta, A.; Nabavizadeh, N.; Simeone, D.M. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J. Clin. 2020, 70, 375–403. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.M.; Tang, L.H.; Klimstra, D.S.; Liu, W.; Linkov, I.; Brennan, M.F.; D’Angelica, M.I.; DeMatteo, R.P.; Fong, Y.; Jarnagin, W.R.; et al. Failure patterns in resected pancreas adenocarcinoma: Lack of predicted benefit to SMAD4 expression. Ann. Surg. 2013, 258, 331–335. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, C.A.; Lawrence, S.A.; Richards, A.L.; Chou, J.F.; Wong, W.; Capanu, M.; Berger, M.F.; Donoghue, M.T.A.; Yu, K.H.; Varghese, A.M.; et al. Alterations in driver genes are predictive of survival in patients with resected pancreatic ductal adenocarcinoma. Cancer 2020, 126, 3939–3949. [Google Scholar] [CrossRef] [PubMed]
- Iacobuzio-Donahue, C.A.; Fu, B.; Yachida, S.; Luo, M.; Abe, H.; Henderson, C.M.; Vilardell, F.; Wang, Z.; Keller, J.W.; Banerjee, P.; et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J. Clin. Oncol. 2009, 27, 1806–1813. [Google Scholar] [CrossRef] [PubMed]
- Yokose, T.; Kitago, M.; Matsuda, S.; Sasaki, Y.; Masugi, Y.; Nakamura, Y.; Shinoda, M.; Yagi, H.; Abe, Y.; Oshima, G.; et al. Combination of KRAS and SMAD4 mutations in formalin-fixed paraffin-embedded tissues as a biomarker for pancreatic cancer. Cancer Sci. 2020, 111, 2174–2182. [Google Scholar] [CrossRef]
- Tascilar, M.; Skinner, H.G.; Rosty, C.; Sohn, T.; Wilentz, R.E.; Offerhaus, G.J.; Adsay, V.; Abrams, R.A.; Cameron, J.L.; Kern, S.E.; et al. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2001, 7, 4115–4121. [Google Scholar]
- Biankin, A.V.; Morey, A.L.; Lee, C.S.; Kench, J.G.; Biankin, S.A.; Hook, H.C.; Head, D.R.; Hugh, T.B.; Sutherland, R.L.; Henshall, S.M. DPC4/Smad4 expression and outcome in pancreatic ductal adenocarcinoma. J. Clin. Oncol. 2002, 20, 4531–4542. [Google Scholar] [CrossRef]
- Oshima, M.; Okano, K.; Muraki, S.; Haba, R.; Maeba, T.; Suzuki, Y.; Yachida, S. Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer. Ann. Surg. 2013, 258, 336–346. [Google Scholar] [CrossRef]
- Gu, Y.; Ji, Y.; Jiang, H.; Qiu, G. Clinical Effect of Driver Mutations of KRAS, CDKN2A/P16, TP53, and SMAD4 in Pancreatic Cancer: A Meta-Analysis. Genet. Test. Mol. Biomark. 2020, 24, 777–788. [Google Scholar] [CrossRef]
- Crane, C.H.; Varadhachary, G.R.; Yordy, J.S.; Staerkel, G.A.; Javle, M.M.; Safran, H.; Haque, W.; Hobbs, B.D.; Krishnan, S.; Fleming, J.B.; et al. Phase II trial of cetuximab, gemcitabine, and oxaliplatin followed by chemoradiation with cetuximab for locally advanced (T4) pancreatic adenocarcinoma: Correlation of Smad4(Dpc4) immunostaining with pattern of disease progression. J. Clin. Oncol. 2011, 29, 3037–3043. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.H.; Kim, H.J.; Hwang, D.W.; Lee, J.H.; Song, K.B.; Jun, E.; Shim, I.K.; Hong, S.M.; Kim, H.J.; Park, K.M.; et al. The DPC4/SMAD4 genetic status determines recurrence patterns and treatment outcomes in resected pancreatic ductal adenocarcinoma: A prospective cohort study. Oncotarget 2017, 8, 17945–17959. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.R.; Rubinson, D.A.; Nowak, J.A.; Morales-Oyarvide, V.; Dunne, R.F.; Kozak, M.M.; Welch, M.W.; Brais, L.K.; Da Silva, A.; Li, T.; et al. Association of Alterations in Main Driver Genes with Outcomes of Patients with Resected Pancreatic Ductal Adenocarcinoma. JAMA Oncol. 2018, 4, e173420. [Google Scholar] [CrossRef] [PubMed]
- Maeda, M.; Ohashi, K.; Ohashi-Kobayashi, A. Further extension of mammalian GATA-6. Dev. Growth Differ. 2005, 47, 591–600. [Google Scholar] [CrossRef]
- Martinelli, P.; Cañamero, M.; del Pozo, N.; Madriles, F.; Zapata, A.; Real, F.X. Gata6 is required for complete acinar differentiation and maintenance of the exocrine pancreas in adult mice. Gut 2013, 62, 1481–1488. [Google Scholar] [CrossRef]
- Martinelli, P.; Madriles, F.; Cañamero, M.; Pau, E.C.; Pozo, N.D.; Guerra, C.; Real, F.X. The acinar regulator Gata6 suppresses KrasG12V-driven pancreatic tumorigenesis in mice. Gut 2016, 65, 476–486. [Google Scholar] [CrossRef]
- Hermann, P.C.; Sancho, P.; Cañamero, M.; Martinelli, P.; Madriles, F.; Michl, P.; Gress, T.; de Pascual, R.; Gandia, L.; Guerra, C.; et al. Nicotine promotes initiation and progression of KRAS-induced pancreatic cancer via Gata6-dependent dedifferentiation of acinar cells in mice. Gastroenterology 2014, 147, 1119–1133.e4. [Google Scholar] [CrossRef]
- Ocaña, O.H.; Córcoles, R.; Fabra, A.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 2012, 22, 709–724. [Google Scholar] [CrossRef]
- Tsai, J.H.; Donaher, J.L.; Murphy, D.A.; Chau, S.; Yang, J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 2012, 22, 725–736. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Z.; Fu, B.; Pan, F.; Yachida, S.; Dhara, M.; Albesiano, E.; Li, L.; Naito, Y.; Vilardell, F.; et al. GATA6 activates Wnt signaling in pancreatic cancer by negatively regulating the Wnt antagonist Dickkopf-1. PLoS ONE 2011, 6, e22129. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Stocken, D.D.; Bassi, C.; Ghaneh, P.; Cunningham, D.; Goldstein, D.; Padbury, R.; Moore, M.J.; Gallinger, S.; Mariette, C.; et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: A randomized controlled trial. JAMA 2010, 304, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
| SMAD4-WT | SMAD4-MUT | p | |||
|---|---|---|---|---|---|
| (n = 120) | (n = 60) | ||||
| # | % | # | % | ||
| Age | 0.874 | ||||
| Median (IQR) | 65 (54–74) | 65 (58–73) | |||
| Sex | 0.751 | ||||
| Female | 55 | 45.8% | 26 | 43.3% | |
| Male | 65 | 54.2% | 34 | 56.7% | |
| Race a | 0.041 | ||||
| White | 106 | 90.6% | 52 | 88.1% | |
| Black | 6 | 5.1% | 0 | 0.0% | |
| Asian | 5 | 4.3% | 7 | 11.9% | |
| Ethnicity b | 0.588 | ||||
| Hispanic | 3 | 2.5% | 2 | 3.3% | |
| AJCC Stage c | 0.412 | ||||
| I | 16 | 13.6% | 6 | 10.0% | |
| II | 95 | 80.5% | 53 | 88.3% | |
| III | 4 | 3.4% | 0 | 0.0% | |
| IV | 3 | 2.5% | 1 | 1.7% | |
| T Stage | 0.596 | ||||
| T1 | 5 | 4.2% | 3 | 5.0% | |
| T2 | 16 | 13.3% | 7 | 11.7% | |
| T3 | 94 | 78.3% | 50 | 83.3% | |
| T4 | 3 | 2.5% | 0 | 0.0% | |
| TX | 2 | 1.7% | 0 | 0.0% | |
| N Stage | 0.839 | ||||
| N-negative | 35 | 29.2% | 15 | 25.0% | |
| N-positive | 81 | 67.5% | 43 | 71.7% | |
| NX | 4 | 3.3% | 2 | 3.3% | |
| Grade | 0.373 | ||||
| G1 | 24 | 20.0% | 7 | 11.7% | |
| G2 | 61 | 50.8% | 35 | 58.3% | |
| G3 | 33 | 27.5% | 18 | 30.0% | |
| GX | 2 | 1.7% | 0 | 0.0% | |
| SMAD4 Wild-Type | SMAD4 Mutant | p | |||
|---|---|---|---|---|---|
| (n = 115) | (n = 26) | ||||
| # | % | # | % | ||
| Age | 0.147 | ||||
| Median (IQR) | 64 (59–71) | 66 (63–72) | |||
| Sex | 0.556 | ||||
| Female | 56 | 48.7% | 11 | 42.3% | |
| Male | 59 | 51.3% | 15 | 57.7% | |
| AJCC Stage a | 0.491 | ||||
| I | 18 | 16.4% | 5 | 20.8% | |
| II | 48 | 43.6% | 12 | 50.0% | |
| III | 35 | 31.8% | 7 | 29.2% | |
| IV | 9 | 8.2% | 0 | 0.0% | |
| T Stage | 0.799 | ||||
| pT1 | 7 | 6.1% | 3 | 11.5% | |
| pT2 | 72 | 62.6% | 17.2 | 57.7% | |
| pT3 | 33 | 28.7% | 19.5 | 30.8% | |
| pT4 | 1 | 0.9% | 0 | 0.0% | |
| pTX | 2 | 1.7% | 0 | 0.0% | |
| N Stage | 0.842 | ||||
| pN0 | 24 | 20.9% | 7 | 26.9% | |
| pN1 | 44 | 38.2% | 10 | 38.5% | |
| pN2 | 40 | 34.8% | 7 | 26.9% | |
| pNX | 7 | 6.1% | 2 | 7.7% | |
| LVI b | 0.429 | ||||
| Absent | 31 | 29.2% | 9 | 37.5% | |
| Present | 75 | 70.8% | 15 | 62.5% | |
| PNI c | 0.471 | ||||
| Absent | 15 | 13.8% | 2 | 8.3% | |
| Present | 94 | 86.2% | 22 | 91.7% | |
| SMAD4 Wild-Type | SMAD4 Mutant | p | |||
|---|---|---|---|---|---|
| # | % | # | % | ||
| TCGA | |||||
| GATA6 Tertile | 0.928 | ||||
| Low | 41 | 34.2% | 19 | 31.7% | |
| Medium | 40 | 33.3% | 20 | 33.3% | |
| High | 39 | 32.5% | 21 | 35.0% | |
| Moffitt Subtype a | 0.783 | ||||
| Classical | 54 | 57.4% | 32 | 55.2% | |
| Basal-like | 40 | 42.6% | 26 | 44.8% | |
| CPTAC | |||||
| GATA6 Tertile | 0.828 | ||||
| Low | 39 | 31.5% | 8 | 30.8% | |
| Medium | 39 | 31.5% | 8 | 30.8% | |
| High | 46 | 37.0% | 10 | 38.4% | |
| Moffitt Subtype b | 0.416 | ||||
| Classical | 55 | 55.0% | 16 | 64.0% | |
| Basal-like | 45 | 45.0% | 9 | 36.0% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greendyk, J.D.; Allen, W.E.; Alexander, H.R.; Beninato, T.; Eskander, M.F.; Grandhi, M.S.; Kennedy, T.J.; Langan, R.C.; Maggi, J.C.; De, S.; et al. Association between SMAD4 Mutations and GATA6 Expression in Paired Pancreatic Ductal Adenocarcinoma Tumor Specimens: Data from Two Independent Molecularly-Characterized Cohorts. Biomedicines 2023, 11, 3058. https://doi.org/10.3390/biomedicines11113058
Greendyk JD, Allen WE, Alexander HR, Beninato T, Eskander MF, Grandhi MS, Kennedy TJ, Langan RC, Maggi JC, De S, et al. Association between SMAD4 Mutations and GATA6 Expression in Paired Pancreatic Ductal Adenocarcinoma Tumor Specimens: Data from Two Independent Molecularly-Characterized Cohorts. Biomedicines. 2023; 11(11):3058. https://doi.org/10.3390/biomedicines11113058
Chicago/Turabian StyleGreendyk, Joshua D., William E. Allen, H. Richard Alexander, Toni Beninato, Mariam F. Eskander, Miral S. Grandhi, Timothy J. Kennedy, Russell C. Langan, Jason C. Maggi, Subhajyoti De, and et al. 2023. "Association between SMAD4 Mutations and GATA6 Expression in Paired Pancreatic Ductal Adenocarcinoma Tumor Specimens: Data from Two Independent Molecularly-Characterized Cohorts" Biomedicines 11, no. 11: 3058. https://doi.org/10.3390/biomedicines11113058
APA StyleGreendyk, J. D., Allen, W. E., Alexander, H. R., Beninato, T., Eskander, M. F., Grandhi, M. S., Kennedy, T. J., Langan, R. C., Maggi, J. C., De, S., Court, C. M., & Ecker, B. L. (2023). Association between SMAD4 Mutations and GATA6 Expression in Paired Pancreatic Ductal Adenocarcinoma Tumor Specimens: Data from Two Independent Molecularly-Characterized Cohorts. Biomedicines, 11(11), 3058. https://doi.org/10.3390/biomedicines11113058







