The Ultraviolet Irradiation of Keratinocytes Induces Ectopic Expression of LINE-1 Retrotransposon Machinery and Leads to Cellular Senescence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Treatment Conditions
2.2. Immunofluorescence
2.3. Western Blotting
2.4. RT-qPCR
2.5. Statistical Analysis
3. Results
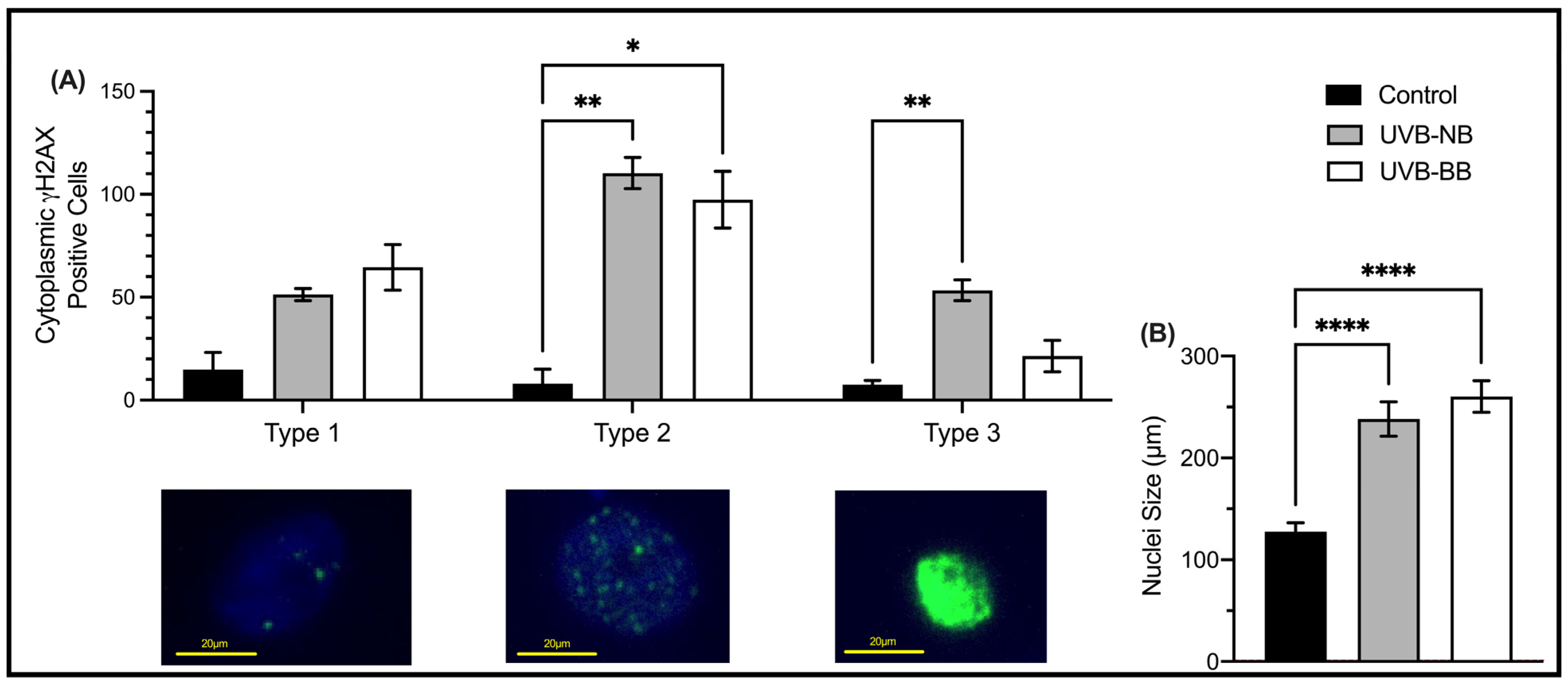
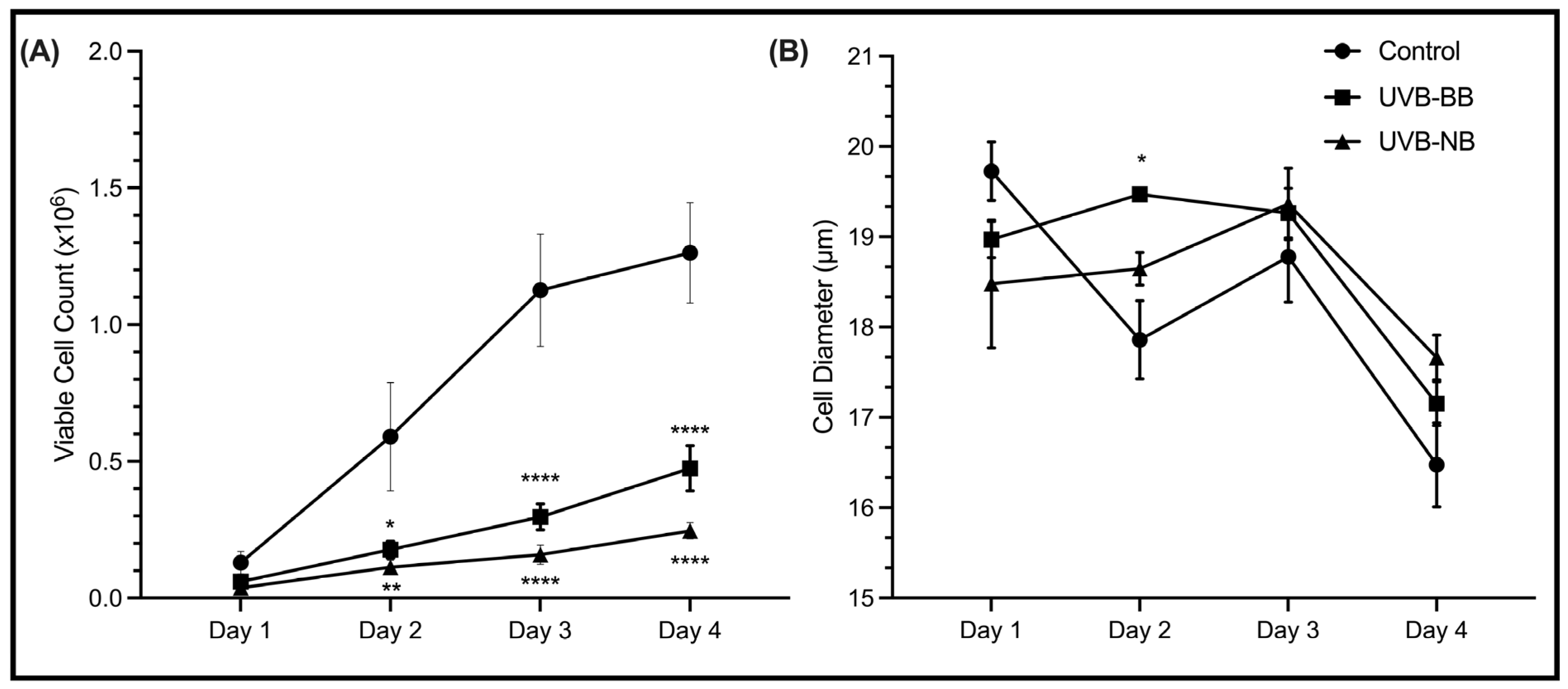
3.1. UV Irradiation Is Associated with DNA Damage and Impaired Proliferation
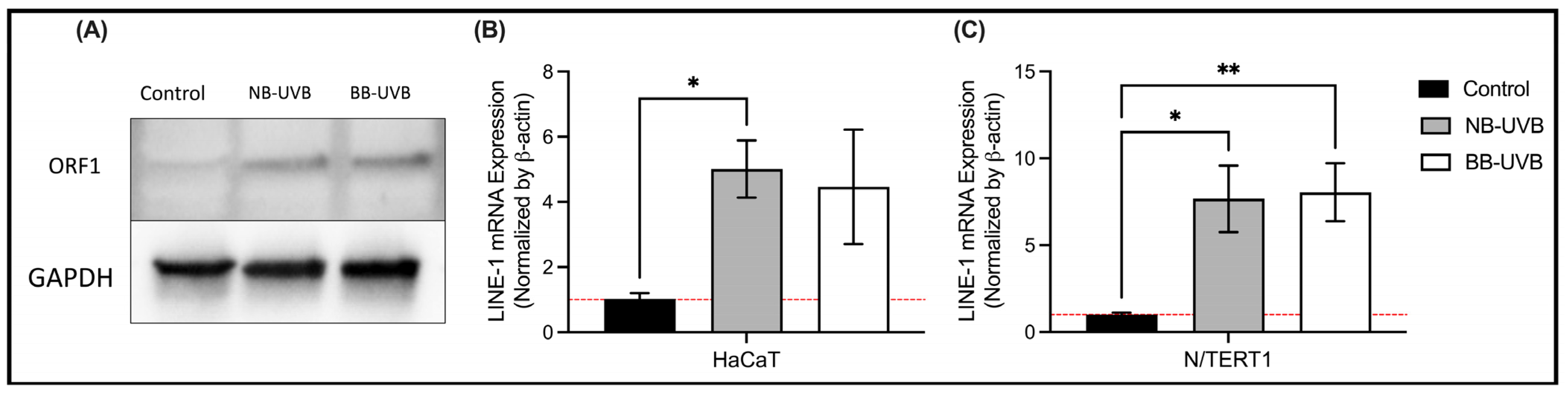
3.2. UV Irradiation Is Associated with LINE-1 Reactivation
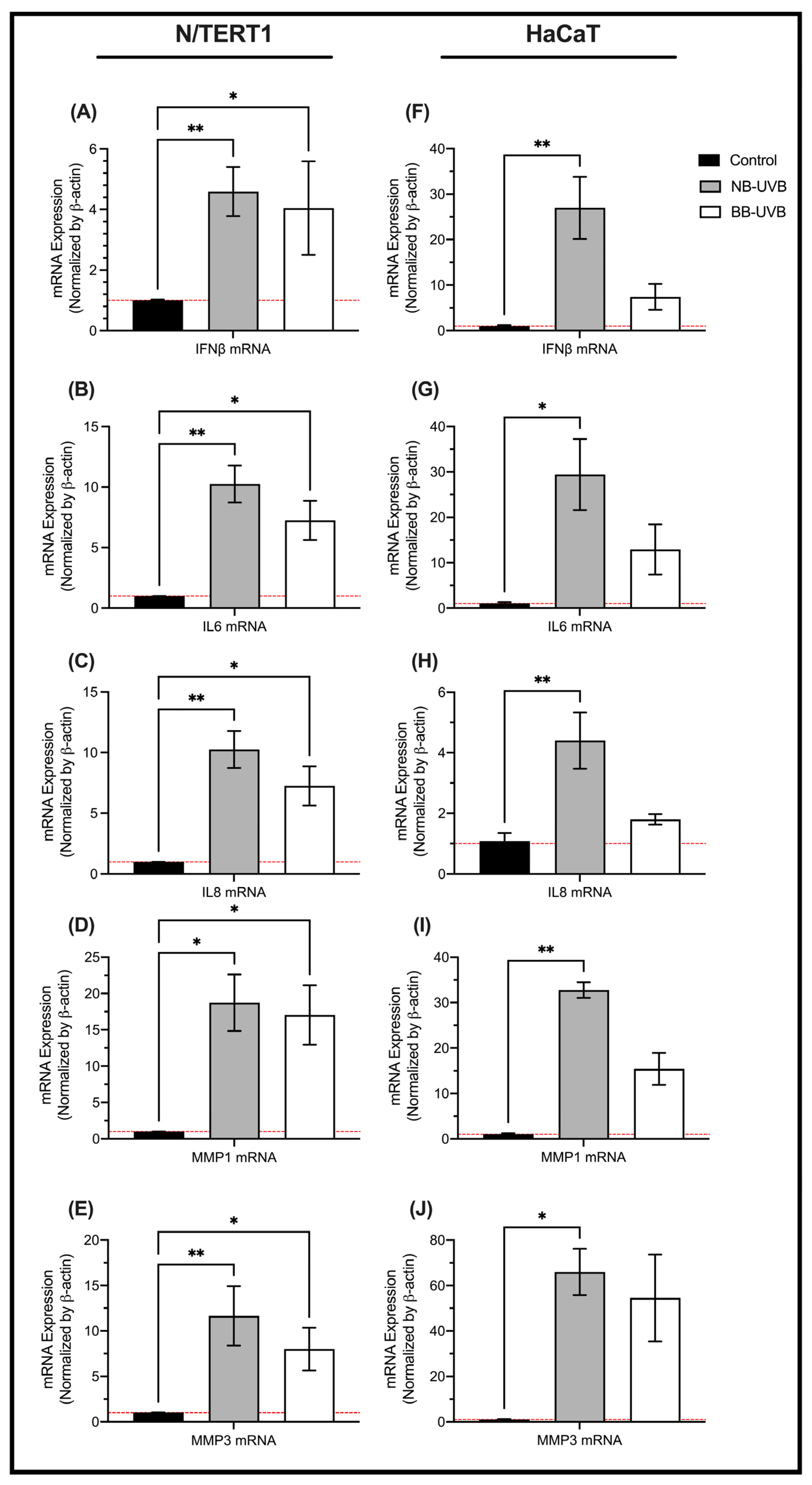
3.3. LINE-1 Reactivation and Cellular Senescence
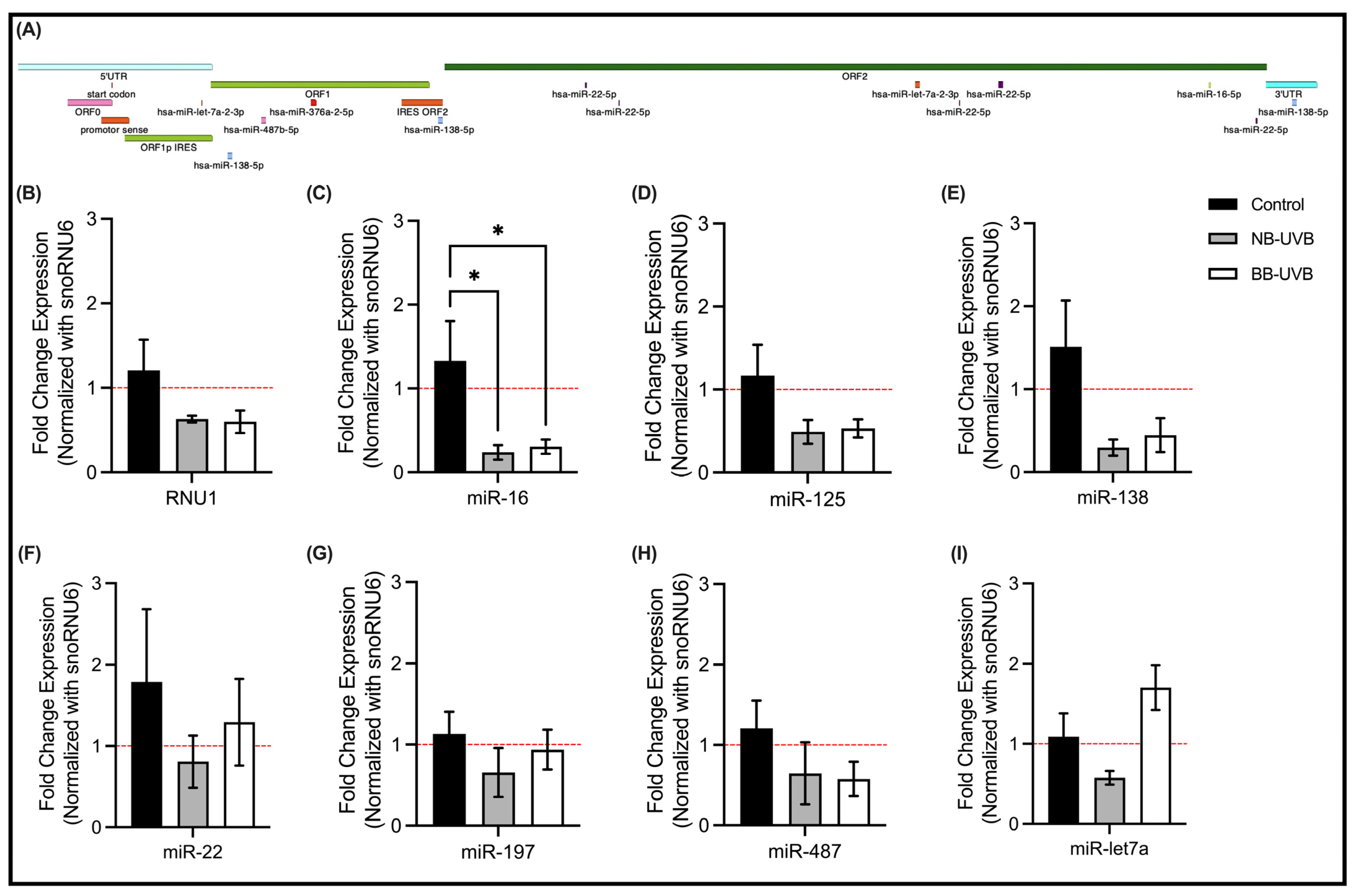
3.4. LINE-1 Reactivation and Associated miRNAs Expression Changes
4. Discussion
5. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

| Gene Name | Primer Sequence |
|---|---|
| ORF2 | FW-TCATAAAGCAAGTCCTCAGTGACC |
| RV-GGGGTGGAGAGTTCTGTAGATGTC | |
| IFN-B | FW-5′ACGCCGCATTGACCATCTAT-3′ |
| RV-5′GTCTCATTCCAGCCAGTGCT-3′ | |
| MMP1 | FW-5′AGCCTTCCAACTCTGGAGTAATGT-3′ |
| RV-5′CCGATGATCTCCCCTGACAA-3′ | |
| MMP3 | FW-5′CCCACCTTACATACAGGATTGTGA-3′ |
| RV-5′CCCAGACTTTCAGAGCTTTCTCA-3′ | |
| IL-6 | FW-5′CACTGGCAGAAAACAACCTGAA-3′ |
| RV 5′ACCAGGCAAGTCTCCTCATTGA-3′ | |
| IL-8 | FW-5′GTTTTTGAAGAGGGCTGAGAATTC-3′ |
| RV- 5′CCCTACAACAGACCCACACAATAC-3′ | |
| β-actin | FW- 5′CCAACCGCGAGAAGATGA-3′ |
| RV- 5′CCAGAGGCGTACAGGGATAG-3′ |
| miR | miRCURY LNA miRNA Custom PCR Assays |
|---|---|
| RNU1A1 | GeneGlobe ID- YP00203909 (Qiagen) |
| hsa-miR-16-5p | GeneGlobe ID- YP00205702 (Qiagen) |
| hsa-miR-125b-1-3p | GeneGlobe ID- YP00204400 (Qiagen) |
| hsa-miR-138-5p | GeneGlobe ID- YP00206078 (Qiagen) |
| hsa-miR-22-5p | GeneGlobe ID- YP00204255 (Qiagen) |
| hsa-miR-197-3p | GeneGlobe ID- YP00204380 (Qiagen) |
| hsa-miR-487b-5p | GeneGlobe ID- YP02115384 (Qiagen) |
| hsa-let-7a-5p | GeneGlobe ID- YP00205727 (Qiagen) |
| U6 snRNA | GeneGlobe ID- YP00203907 (Qiagen) |
References
- Solano, F. Photoprotection and Skin Pigmentation: Melanin-Related Molecules and Some Other New Agents Obtained from Natural Sources. Molecules 2020, 25, 1537. [Google Scholar] [CrossRef] [PubMed]
- De Gruijl, F.R. [33] Photocarcinogenesis: UVA vs. UVB. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2000; Volume 319, pp. 359–366. ISBN 978-0-12-182220-0. [Google Scholar]
- Myers, E.; Kheradmand, S.; Miller, R. An Update on Narrowband Ultraviolet B Therapy for the Treatment of Skin Diseases. Cureus 2021, 13, e19182. [Google Scholar] [CrossRef] [PubMed]
- Enninga, I.C.; Groenendijk, R.T.; Filon, A.R.; van Zeeland, A.A.; Simons, J.W. The Wavelength Dependence of u.v.-Induced Pyrimidine Dimer Formation, Cell Killing and Mutation Induction in Human Diploid Skin Fibroblasts. Carcinogenesis 1986, 7, 1829–1836. [Google Scholar] [CrossRef]
- Kielbassa, C. Wavelength Dependence of Oxidative DNA Damage Induced by UV and Visible Light. Carcinogenesis 1997, 18, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.T.; Zmijewski, M.A.; Plonka, P.M.; Szaflarski, J.P.; Paus, R. How UV Light Touches the Brain and Endocrine System through Skin, and Why. Endocrinology 2018, 159, 1992–2007. [Google Scholar] [CrossRef]
- Hohenadl, C.; Germaier, H.; Walchner, M.; Hagenhofer, M.; Herrmann, M.; Stürzl, M.; Kind, P.; Hehlmann, R.; Erfle, V.; Leib-Mösch, C. Transcriptional Activation of Endogenous Retroviral Sequences in Human Epidermal Keratinocytes by UVB Irradiation. J. Investig. Dermatol. 1999, 113, 587–594. [Google Scholar] [CrossRef]
- Pelin, E.D.; Akay, B.N.; Seçil, V.; Canan, A.; Tuğçe, E.Y.; Hatice, Ş. Risk of Skin Cancers in Mycosis Fungoides Patients Receiving PUVA Therapy: A Real-Life Experience from a Single Tertiary Center. Photodermatol. Photoimmunol. Photomed. 2023, 39, 428–434. [Google Scholar] [CrossRef]
- Matsumura, Y.; Ananthaswamy, H.N. Toxic Effects of Ultraviolet Radiation on the Skin. Toxicol. Appl. Pharmacol. 2004, 195, 298–308. [Google Scholar] [CrossRef]
- Saladi, R.N.; Persaud, A.N. The Causes of Skin Cancer: A Comprehensive Review. Drugs Today 2005, 41, 37–53. [Google Scholar] [CrossRef]
- Butrón-Bris, B.; Daudén, E.; Rodríguez-Jiménez, P. Psoriasis Therapy and Skin Cancer: A Review. Life 2021, 11, 1109. [Google Scholar] [CrossRef]
- Archier, E.; Devaux, S.; Castela, E.; Gallini, A.; Aubin, F.; Le Maître, M.; Aractingi, S.; Bachelez, H.; Cribier, B.; Joly, P.; et al. Carcinogenic Risks of Psoralen UV-A Therapy and Narrowband UV-B Therapy in Chronic Plaque Psoriasis: A Systematic Literature Review: Carcinogenic Risks of PUV-A Therapy and NB-UVB. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Yu, Y.; Li, S.; Huang, Y.; Xu, D.; Tao, X.; Fan, Y. Ultraviolet Radiation and Basal Cell Carcinoma: An Environmental Perspective. Front. Public Health 2021, 9, 666528. [Google Scholar] [CrossRef] [PubMed]
- Stern, R.S. The Risk of Squamous Cell and Basal Cell Cancer Associated with Psoralen and Ultraviolet A Therapy: A 30-Year Prospective Study. J. Am. Acad. Dermatol. 2012, 66, 553–562. [Google Scholar] [CrossRef]
- Osmancevic, A.; Gillstedt, M.; Wennberg, A.-M.; Larkö, O. The Risk of Skin Cancer in Psoriasis Patients Treated with UVB Therapy. Acta Derm. Venereol. 2014, 94, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-L.; Wu, C.-Y.; Chang, Y.-T.; Juan, C.-K.; Chen, C.-C.; Yu, S.-H.; Chen, Y.-J. Risk of Skin Cancer in Psoriasis Patients Receiving Long-Term Narrowband Ultraviolet Phototherapy: Results from a Taiwanese Population-Based Cohort Study. Photodermatol. Photoimmunol. Photomed. 2019, 35, 164–171. [Google Scholar] [CrossRef]
- Ozawa, M.; Ferenczi, K.; Kikuchi, T.; Cardinale, I.; Austin, L.M.; Coven, T.R.; Burack, L.H.; Krueger, J.G. 312-Nanometer Ultraviolet B Light (Narrow-Band UVB) Induces Apoptosis of T Cells within Psoriatic Lesions. J. Exp. Med. 1999, 189, 711–718. [Google Scholar] [CrossRef]
- Bulat, V.; Situm, M.; Dediol, I.; Ljubicić, I.; Bradić, L. The Mechanisms of Action of Phototherapy in the Treatment of the Most Common Dermatoses. Coll. Antropol. 2011, 35 (Suppl. S2), 147–151. [Google Scholar]
- Situm, M.; Bulat, V.; Majcen, K.; Dzapo, A.; Jezovita, J. Benefits of Controlled Ultraviolet Radiation in the Treatment of Dermatological Diseases. Coll. Antropol. 2014, 38, 1249–1253. [Google Scholar]
- Pfeifer, G.P. Mechanisms of UV-Induced Mutations and Skin Cancer. Genome Instab. Dis. 2020, 1, 99–113. [Google Scholar] [CrossRef]
- Mullenders, L.H.F. Solar UV Damage to Cellular DNA: From Mechanisms to Biological Effects. Photochem. Photobiol. Sci. 2018, 17, 1842–1852. [Google Scholar] [CrossRef]
- Cadet, J.; Douki, T. Formation of UV-Induced DNA Damage Contributing to Skin Cancer Development. Photochem. Photobiol. Sci. 2018, 17, 1816–1841. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, G.; Gupta, N.; Tiwari, J.; Raman, G. Ultraviolet-Induced Transformation of Keratinocytes: Possible Involvement of Long Interspersed Element-1 Reverse Transcriptase. Photoderm. Photoimm. Photomed. 2005, 21, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Pace, J.K.; Feschotte, C. The Evolutionary History of Human DNA Transposons: Evidence for Intense Activity in the Primate Lineage. Genome Res. 2007, 17, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Craig, N.L. (Ed.) Mobile DNA II; ASM Press: Washington, DC, USA, 2002; ISBN 978-1-55581-209-6. [Google Scholar]
- Cordaux, R.; Batzer, M.A. The Impact of Retrotransposons on Human Genome Evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H.; Wong, C.; Youssoufian, H.; Scott, A.F.; Phillips, D.G.; Antonarakis, S.E. Haemophilia A Resulting from de Novo Insertion of L1 Sequences Represents a Novel Mechanism for Mutation in Man. Nature 1988, 332, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P.L.; Batzer, M.A. Alu Repeats and Human Disease. Mol. Genet. Metab. 1999, 67, 183–193. [Google Scholar] [CrossRef]
- Chen, J.-M.; Stenson, P.D.; Cooper, D.N.; Férec, C. A Systematic Analysis of LINE-1 Endonuclease-Dependent Retrotranspositional Events Causing Human Genetic Disease. Hum. Genet. 2005, 117, 411–427. [Google Scholar] [CrossRef]
- Callinan, P.A.; Batzer, M.A. Retrotransposable Elements and Human Disease. Genome Dyn. 2006, 1, 104–115. [Google Scholar] [CrossRef]
- Belancio, V.P.; Hedges, D.J.; Deininger, P. Mammalian Non-LTR Retrotransposons: For Better or Worse, in Sickness and in Health. Genome Res. 2008, 18, 343–358. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Swergold, G.D. Identification, Characterization, and Cell Specificity of a Human LINE-1 Promoter. Mol. Cell. Biol. 1990, 10, 6718–6729. [Google Scholar] [CrossRef] [PubMed]
- Babushok, D.V.; Kazazian, H.H. Progress in Understanding the Biology of the Human Mutagen LINE-1. Hum. Mutat. 2007, 28, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Boeke, J.D. LINE-1 Retrotransposons: Modulators of Quantity and Quality of Mammalian Gene Expression? Bioessays 2005, 27, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.T.; Sokolowski, M.; Roy-Engel, A.M.; Smither, M.E.; Belancio, V.P. Identification of Charged Amino Acids Required for Nuclear Localization of Human L1 ORF1 Protein. Mob. DNA 2019, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Mathias, S.L.; Scott, A.F.; Kazazian, H.H.; Boeke, J.D.; Gabriel, A. Reverse Transcriptase Encoded by a Human Transposable Element. Science 1991, 254, 1808–1810. [Google Scholar] [CrossRef]
- Naufer, M.N.; Furano, A.V.; Williams, M.C. Protein-Nucleic Acid Interactions of LINE-1 ORF1p. Semin. Cell Dev. Biol. 2019, 86, 140–149. [Google Scholar] [CrossRef]
- Symer, D.E.; Connelly, C.; Szak, S.T.; Caputo, E.M.; Cost, G.J.; Parmigiani, G.; Boeke, J.D. Human L1 Retrotransposition Is Associated with Genetic Instability in Vivo. Cell 2002, 110, 327–338. [Google Scholar] [CrossRef]
- Gilbert, N.; Lutz-Prigge, S.; Moran, J.V. Genomic Deletions Created upon LINE-1 Retrotransposition. Cell 2002, 110, 315–325. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Burhans, W.C.; Curcio, M.J. Retrotransposition Is Associated with Genome Instability during Chronological Aging. Proc. Natl. Acad. Sci. USA 2011, 108, 20376–20381. [Google Scholar] [CrossRef]
- Laurent, G.S.; Hammell, N.; McCaffrey, T.A. A LINE-1 Component to Human Aging: Do LINE Elements Exact a Longevity Cost for Evolutionary Advantage? Mech. Ageing Dev. 2010, 131, 299–305. [Google Scholar] [CrossRef]
- Ichiyanagi, T.; Ichiyanagi, K.; Ogawa, A.; Kuramochi-Miyagawa, S.; Nakano, T.; Chuma, S.; Sasaki, H.; Udono, H. HSP90α Plays an Important Role in piRNA Biogenesis and Retrotransposon Repression in Mouse. Nucleic Acids Res. 2014, 42, 11903–11911. [Google Scholar] [CrossRef]
- Specchia, V.; Piacentini, L.; Tritto, P.; Fanti, L.; D’Alessandro, R.; Palumbo, G.; Pimpinelli, S.; Bozzetti, M.P. Hsp90 Prevents Phenotypic Variation by Suppressing the Mutagenic Activity of Transposons. Nature 2010, 463, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Protasova, M.S.; Andreeva, T.V.; Rogaev, E.I. Factors Regulating the Activity of LINE1 Retrotransposons. Genes 2021, 12, 1562. [Google Scholar] [CrossRef] [PubMed]
- Miousse, I.R.; Chalbot, M.-C.G.; Lumen, A.; Ferguson, A.; Kavouras, I.G.; Koturbash, I. Response of Transposable Elements to Environmental Stressors. Mutat. Res. Mol. Mech. Mutagen. 2015, 765, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Gluud, M.; Willerslev-Olsen, A.; Gjerdrum, L.M.R.; Lindahl, L.M.; Buus, T.B.; Andersen, M.H.; Bonefeld, C.M.; Krejsgaard, T.; Litvinov, I.V.; Iversen, L.; et al. MicroRNAs in the Pathogenesis, Diagnosis, Prognosis and Targeted Treatment of Cutaneous T-Cell Lymphomas. Cancers 2020, 12, 1229. [Google Scholar] [CrossRef] [PubMed]
- Querfeld, C.; Foss, F.M.; Kim, Y.H.; Pinter-Brown, L.; William, B.M.; Porcu, P.; Pacheco, T.; Haverkos, B.M.; DeSimone, J.; Guitart, J.; et al. Phase 1 Trial of Cobomarsen, an Inhibitor of Mir-155, in Cutaneous T Cell Lymphoma. Blood 2018, 132, 2903. [Google Scholar] [CrossRef]
- Litvinov, I.V.; Pehr, K.; Sasseville, D. Connecting the Dots in Cutaneous T Cell Lymphoma (CTCL): STAT5 Regulates Malignant T Cell Proliferation via miR-155. Cell Cycle 2013, 12, 2172. [Google Scholar] [CrossRef]
- Houwing, S.; Kamminga, L.M.; Berezikov, E.; Cronembold, D.; Girard, A.; van den Elst, H.; Filippov, D.V.; Blaser, H.; Raz, E.; Moens, C.B.; et al. A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell 2007, 129, 69–82. [Google Scholar] [CrossRef]
- Bernard, J.J.; Cowing-Zitron, C.; Nakatsuji, T.; Muehleisen, B.; Muto, J.; Borkowski, A.W.; Martinez, L.; Greidinger, E.L.; Yu, B.D.; Gallo, R.L. Ultraviolet Radiation Damages Self Noncoding RNA and Is Detected by TLR3. Nat. Med. 2012, 18, 1286–1290. [Google Scholar] [CrossRef]
- Smits, J.P.H.; Niehues, H.; Rikken, G.; van Vlijmen-Willems, I.M.J.J.; van de Zande, G.W.H.J.F.; Zeeuwen, P.L.J.M.; Schalkwijk, J.; van den Bogaard, E.H. Immortalized N/TERT Keratinocytes as an Alternative Cell Source in 3D Human Epidermal Models. Sci. Rep. 2017, 7, 11838. [Google Scholar] [CrossRef]
- Dickson, M.A.; Hahn, W.C.; Ino, Y.; Ronfard, V.; Wu, J.Y.; Weinberg, R.A.; Louis, D.N.; Li, F.P.; Rheinwald, J.G. Human Keratinocytes That Express hTERT and Also Bypass a P16(INK4a)-Enforced Mechanism That Limits Life Span Become Immortal yet Retain Normal Growth and Differentiation Characteristics. Mol. Cell. Biol. 2000, 20, 1436–1447. [Google Scholar] [CrossRef] [PubMed]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal Keratinization in a Spontaneously Immortalized Aneuploid Human Keratinocyte Cell Line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Gantchev, J.; Ramchatesingh, B.; Berman-Rosa, M.; Sikorski, D.; Raveendra, K.; Amar, L.; Xu, H.H.; Villarreal, A.M.; Ordaz, D.J.G.; Litvinov, I.V. Tools Used to Assay Genomic Instability in Cancers and Cancer Meiomitosis. J. Cell Commun. Signal. 2022, 16, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open Source Software for Digital Pathology Image Analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Dickey, J.S.; Redon, C.E.; Nakamura, A.J.; Baird, B.J.; Sedelnikova, O.A.; Bonner, W.M. H2AX: Functional Roles and Potential Applications. Chromosoma 2009, 118, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Solier, S.; Pommier, Y. The Apoptotic Ring: A Novel Entity with Phosphorylated Histones H2AX and H2B and Activated DNA Damage Response Kinases. Cell Cycle 2009, 8, 1853–1859. [Google Scholar] [CrossRef]
- Marti, T.M.; Hefner, E.; Feeney, L.; Natale, V.; Cleaver, J.E. H2AX Phosphorylation within the G1 Phase after UV Irradiation Depends on Nucleotide Excision Repair and Not DNA Double-Strand Breaks. Proc. Natl. Acad. Sci. USA 2006, 103, 9891–9896. [Google Scholar] [CrossRef]
- Lu, T.; Zhang, Y.; Kidane, Y.; Feiveson, A.; Stodieck, L.; Karouia, F.; Ramesh, G.; Rohde, L.; Wu, H. Cellular Responses and Gene Expression Profile Changes Due to Bleomycin-Induced DNA Damage in Human Fibroblasts in Space. PLoS ONE 2017, 12, e0170358. [Google Scholar] [CrossRef]
- Halicka, H.D.; Huang, X.; Traganos, F.; King, M.A.; Dai, W.; Darzynkiewicz, Z. Histone H2AX Phosphorylation after Cell Irradiation with UV-B: Relationship to Cell Cycle Phase and Induction of Apoptosis. Cell Cycle 2005, 4, 339–345. [Google Scholar] [CrossRef]
- Ding, D.; Zhang, Y.; Wang, J.; Zhang, X.; Gao, Y.; Yin, L.; Li, Q.; Li, J.; Chen, H. Induction and Inhibition of the Pan-Nuclear Gamma-H2AX Response in Resting Human Peripheral Blood Lymphocytes after X-Ray Irradiation. Cell Death Discov. 2016, 2, 16011. [Google Scholar] [CrossRef]
- Fischer, E.G. Nuclear Morphology and the Biology of Cancer Cells. Acta Cytol. 2020, 64, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Barrandon, Y.; Green, H. Cell Size as a Determinant of the Clone-Forming Ability of Human Keratinocytes. Proc. Natl. Acad. Sci. USA 1985, 82, 5390–5394. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, Y.; Permatasari, F.; Liu, W.; Li, W.; Guo, X.; Huang, Q.; Guo, Z.; Luo, D. Characterization of the miRNA Profile in UVB-Irradiated Normal Human Keratinocytes. Exp. Dermatol. 2012, 21, 317–319. [Google Scholar] [CrossRef]
- Pothof, J.; Verkaik, N.S.; van IJcken, W.; Wiemer, E.A.C.; Ta, V.T.B.; van der Horst, G.T.J.; Jaspers, N.G.J.; van Gent, D.C.; Hoeijmakers, J.H.J.; Persengiev, S.P. MicroRNA-Mediated Gene Silencing Modulates the UV-Induced DNA-Damage Response. EMBO J. 2009, 28, 2090–2099. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.J.; Yang, L.-X. Gamma-H2AX—A Novel Biomarker for DNA Double-Strand Breaks. In Vivo 2008, 22, 305–309. [Google Scholar] [PubMed]
- Beukers, R.; Eker, A.P.M.; Lohman, P.H.M. 50 Years Thymine Dimer. DNA Repair 2008, 7, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Markovitsi, D. UV-Induced DNA Damage: The Role of Electronic Excited States. Photochem. Photobiol. 2016, 92, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, G.P. Formation and Processing of UV Photoproducts: Effects of DNA Sequence and Chromatin Environment. Photochem. Photobiol. 1997, 65, 270–283. [Google Scholar] [CrossRef]
- Sinha, R.P.; Häder, D.P. UV-Induced DNA Damage and Repair: A Review. Photochem. Photobiol. Sci. 2002, 1, 225–236. [Google Scholar] [CrossRef]
- Sharma, R.; Rodić, N.; Burns, K.H.; Taylor, M.S. Immuno-Detection of Human LINE-1 Expression. Methods Mol. Biol. 2016, 1400, 261–280. [Google Scholar] [CrossRef]
- Pavez Lorie, E.; Stricker, N.; Plitta-Michalak, B.; Chen, I.-P.; Volkmer, B.; Greinert, R.; Jauch, A.; Boukamp, P.; Rapp, A. Characterisation of the Novel Spontaneously Immortalized and Invasively Growing Human Skin Keratinocyte Line HaSKpw. Sci. Rep. 2020, 10, 15196. [Google Scholar] [CrossRef] [PubMed]
- Lehman, T.A.; Modali, R.; Boukamp, P.; Stanek, J.; Bennett, W.P.; Welsh, J.A.; Metcalf, R.A.; Stampfer, M.R.; Fusenig, N.; Rogan, E.M. P53 Mutations in Human Immortalized Epithelial Cell Lines. Carcinogenesis 1993, 14, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Miyata, Y.; Okada, K.; Fujimoto, A.; Hata, K.-I.; Kagami, H.; Tomita, Y.; Ueda, M. The Effect of the Long-Term Cultivation on Telomere Length and Morphology of Cultured Epidermis. J. Dermatol. Sci. 2004, 34, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Härle-Bachor, C.; Boukamp, P. Telomerase Activity in the Regenerative Basal Layer of the Epidermis Inhuman Skin and in Immortal and Carcinoma-Derived Skin Keratinocytes. Proc. Natl. Acad. Sci. USA 1996, 93, 6476–6481. [Google Scholar] [CrossRef]
- Gorbunova, V.; Seluanov, A.; Pereira-Smith, O.M. Expression of Human Telomerase (hTERT) Does Not Prevent Stress-Induced Senescence in Normal Human Fibroblasts but Protects the Cells from Stress-Induced Apoptosis and Necrosis. J. Biol. Chem. 2002, 277, 38540–38549. [Google Scholar] [CrossRef]
- Muñoz-Espín, D.; Serrano, M. Cellular Senescence: From Physiology to Pathology. Nat. Rev. Mol. Cell Biol. 2014, 15, 482–496. [Google Scholar] [CrossRef]
- Shmulevich, R.; Krizhanovsky, V. Cell Senescence, DNA Damage, and Metabolism. Antioxid. Redox Signal. 2021, 34, 324–334. [Google Scholar] [CrossRef]
- Coppé, J.-P.; Desprez, P.-Y.; Krtolica, A.; Campisi, J. The Senescence-Associated Secretory Phenotype: The Dark Side of Tumor Suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef]
- Tang, L.; Wu, W.; Fu, W.; Hu, Y. The Effects of Phototherapy and Melanocytes on Keratinocytes. Exp. Ther. Med. 2018, 15, 3459–3466. [Google Scholar] [CrossRef]
- Aufiero, B.M.; Talwar, H.; Young, C.; Krishnan, M.; Hatfield, J.S.; Lee, H.K.; Wong, H.K.; Hamzavi, I.; Murakawa, G.J. Narrow-Band UVB Induces Apoptosis in Human Keratinocytes. J. Photochem. Photobiol. B Biol. 2006, 82, 132–139. [Google Scholar] [CrossRef]
- Sapam, R.; Agrawal, S.; Dhali, T.K. Systemic PUVA vs. Narrowband UVB in the Treatment of Vitiligo: A Randomized Controlled Study. Int. J. Dermatol. 2012, 51, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and Consequences of microRNA Dysregulation in Cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA Genes Are Frequently Located at Fragile Sites and Genomic Regions Involved in Cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA Expression Profiles Classify Human Cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Döhner, H.; Stilgenbauer, S.; Benner, A.; Leupolt, E.; Kröber, A.; Bullinger, L.; Döhner, K.; Bentz, M.; Lichter, P. Genomic Aberrations and Survival in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2000, 343, 1910–1916. [Google Scholar] [CrossRef]
- Guo, S.; Guo, W.; Li, S.; Dai, W.; Zhang, N.; Zhao, T.; Wang, H.; Ma, J.; Yi, X.; Ge, R.; et al. Serum miR-16: A Potential Biomarker for Predicting Melanoma Prognosis. J. Investig. Dermatol. 2016, 136, 985–993. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Cretoiu, S.M.; Zurac, S. miRNAs in the Diagnosis and Prognosis of Skin Cancer. Front. Cell Dev. Biol. 2020, 8, 71. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Touma, F.; Lambert, M.; Martínez Villarreal, A.; Gantchev, J.; Ramchatesingh, B.; Litvinov, I.V. The Ultraviolet Irradiation of Keratinocytes Induces Ectopic Expression of LINE-1 Retrotransposon Machinery and Leads to Cellular Senescence. Biomedicines 2023, 11, 3017. https://doi.org/10.3390/biomedicines11113017
Touma F, Lambert M, Martínez Villarreal A, Gantchev J, Ramchatesingh B, Litvinov IV. The Ultraviolet Irradiation of Keratinocytes Induces Ectopic Expression of LINE-1 Retrotransposon Machinery and Leads to Cellular Senescence. Biomedicines. 2023; 11(11):3017. https://doi.org/10.3390/biomedicines11113017
Chicago/Turabian StyleTouma, Fadi, Marine Lambert, Amelia Martínez Villarreal, Jennifer Gantchev, Brandon Ramchatesingh, and Ivan V. Litvinov. 2023. "The Ultraviolet Irradiation of Keratinocytes Induces Ectopic Expression of LINE-1 Retrotransposon Machinery and Leads to Cellular Senescence" Biomedicines 11, no. 11: 3017. https://doi.org/10.3390/biomedicines11113017
APA StyleTouma, F., Lambert, M., Martínez Villarreal, A., Gantchev, J., Ramchatesingh, B., & Litvinov, I. V. (2023). The Ultraviolet Irradiation of Keratinocytes Induces Ectopic Expression of LINE-1 Retrotransposon Machinery and Leads to Cellular Senescence. Biomedicines, 11(11), 3017. https://doi.org/10.3390/biomedicines11113017








