Role of Pericytes in Cardiomyopathy-Associated Myocardial Infarction Revealed by Multiple Single-Cell Sequencing Analysis
Abstract
:1. Introduction
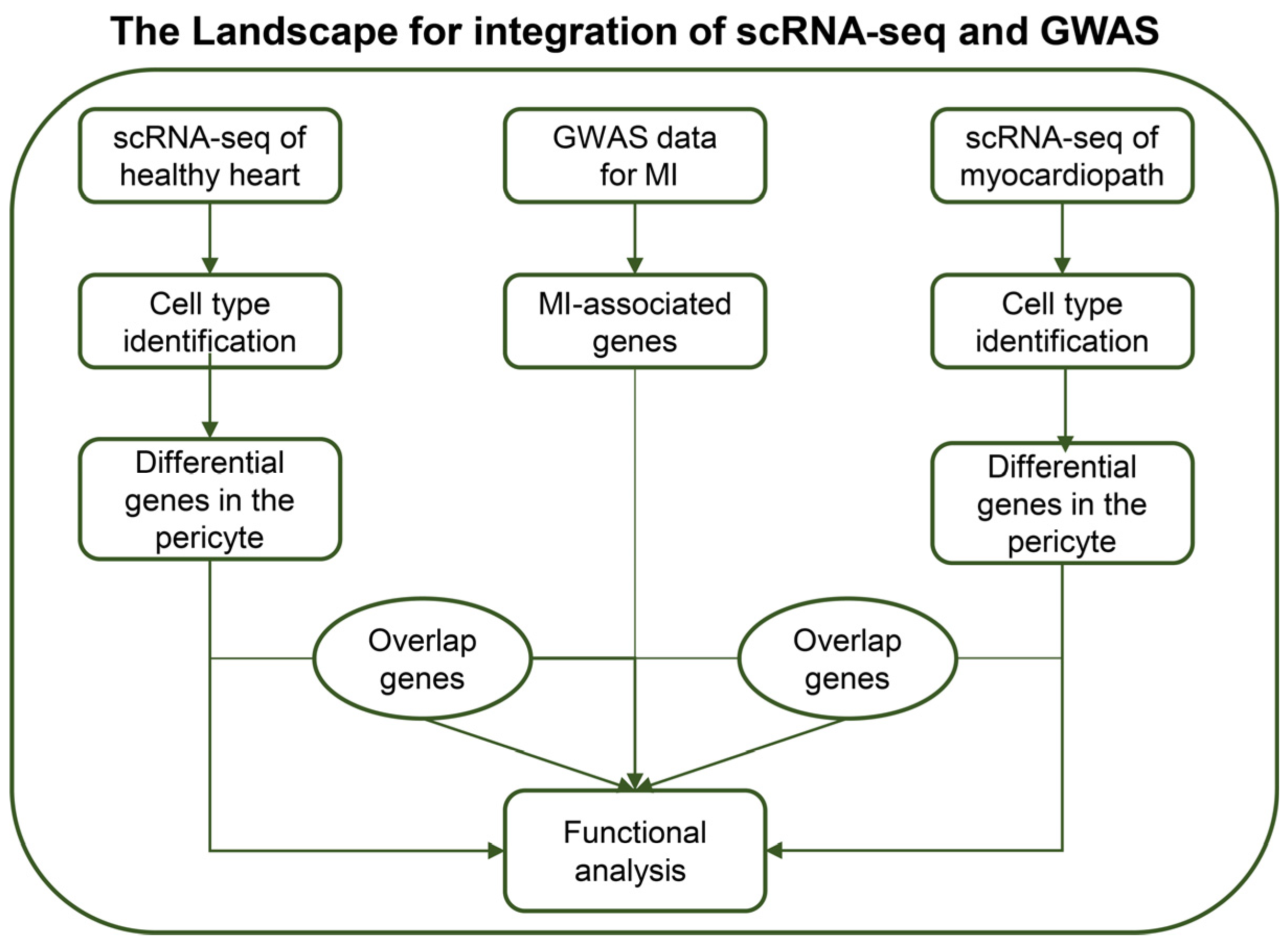
2. Materials and Methods
2.1. Dataset Download
2.2. Single-Cell RNA-Seq Analysis
2.3. Fine-Mapping
2.4. Pathway Enrichment Analysis
2.5. Statistics
3. Results
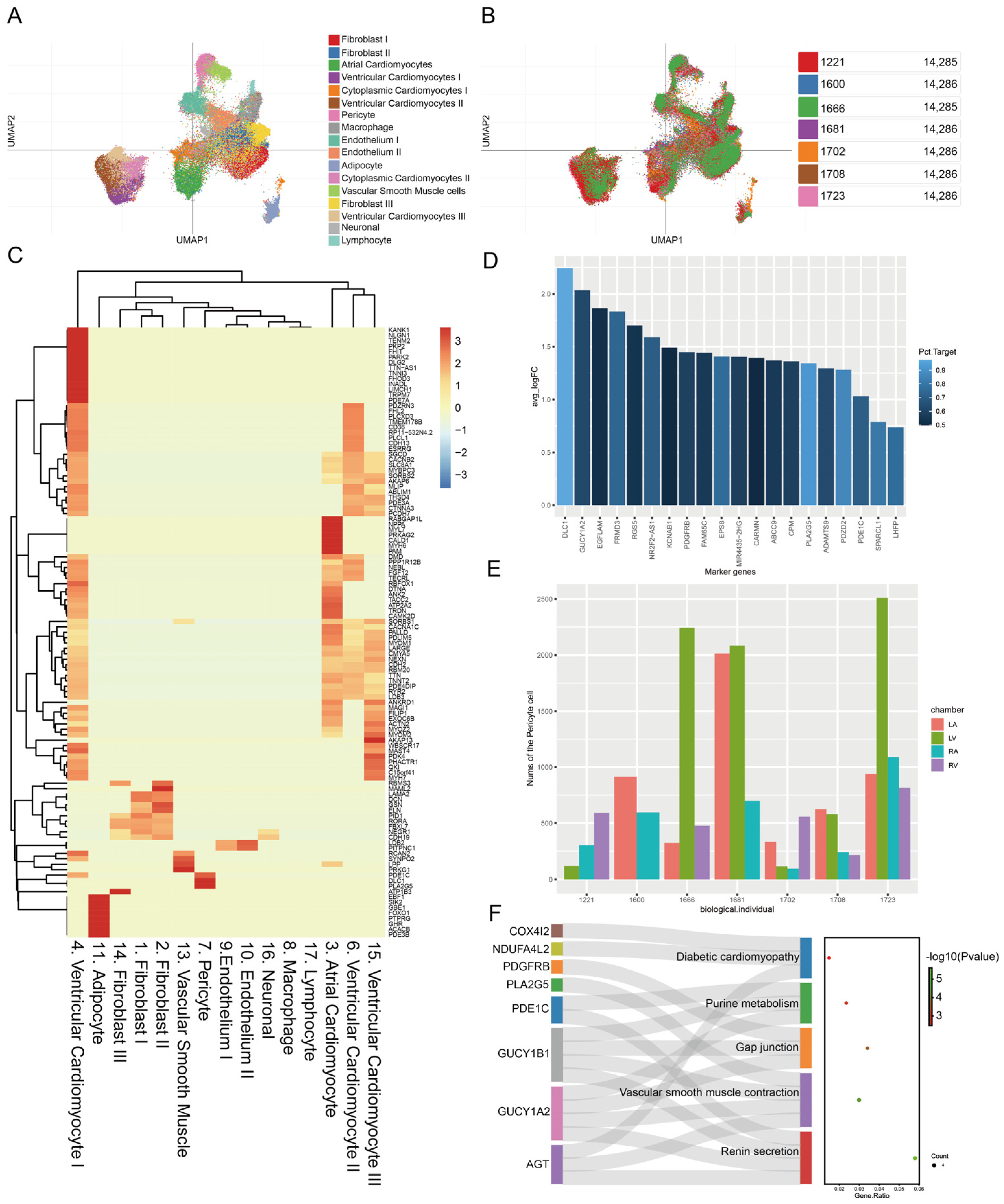
3.1. Identifying Unique Genes and Function Pathways of Pericytes in Healthy Donors
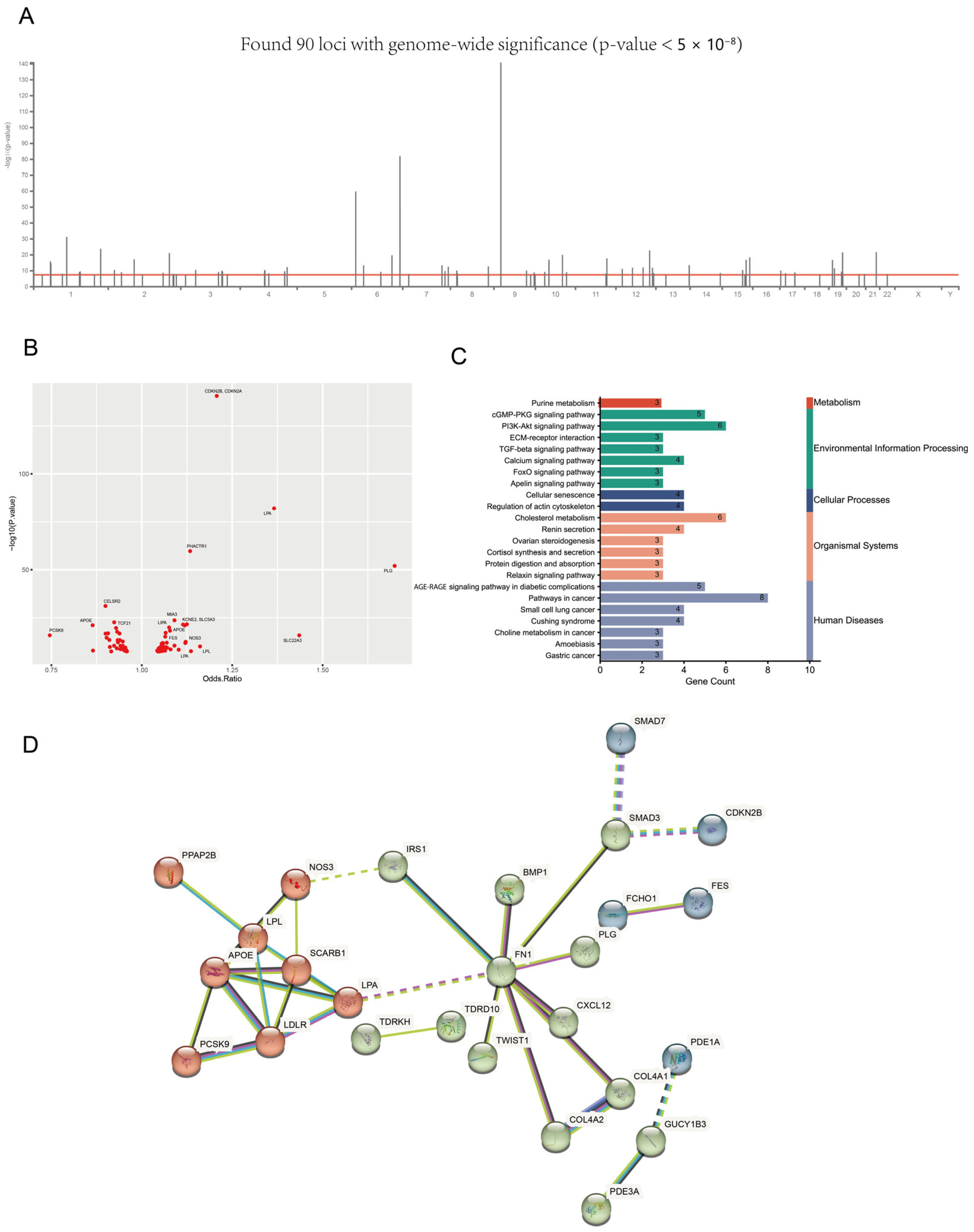
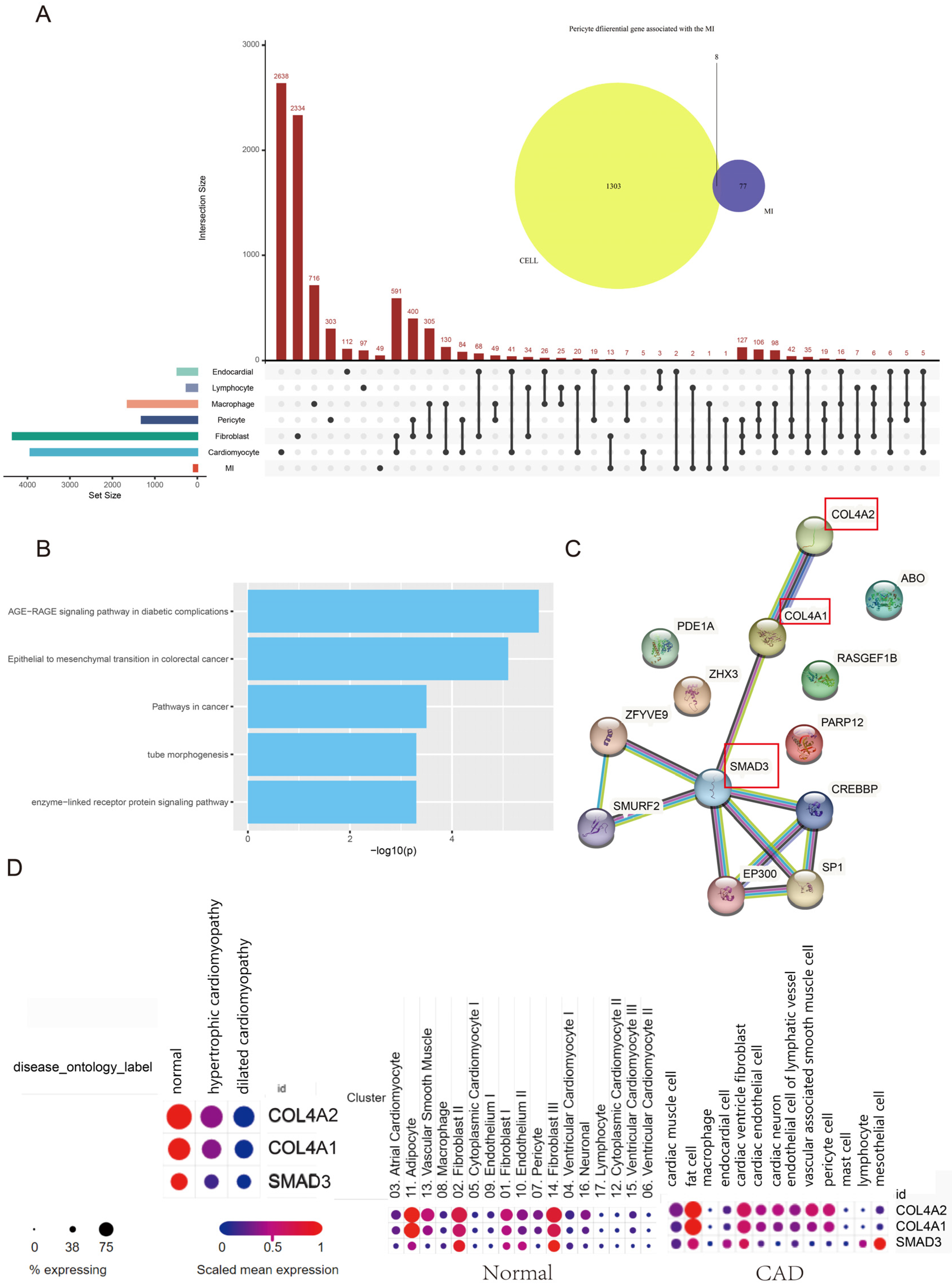
3.2. The Landscape of MI-Associated Genes
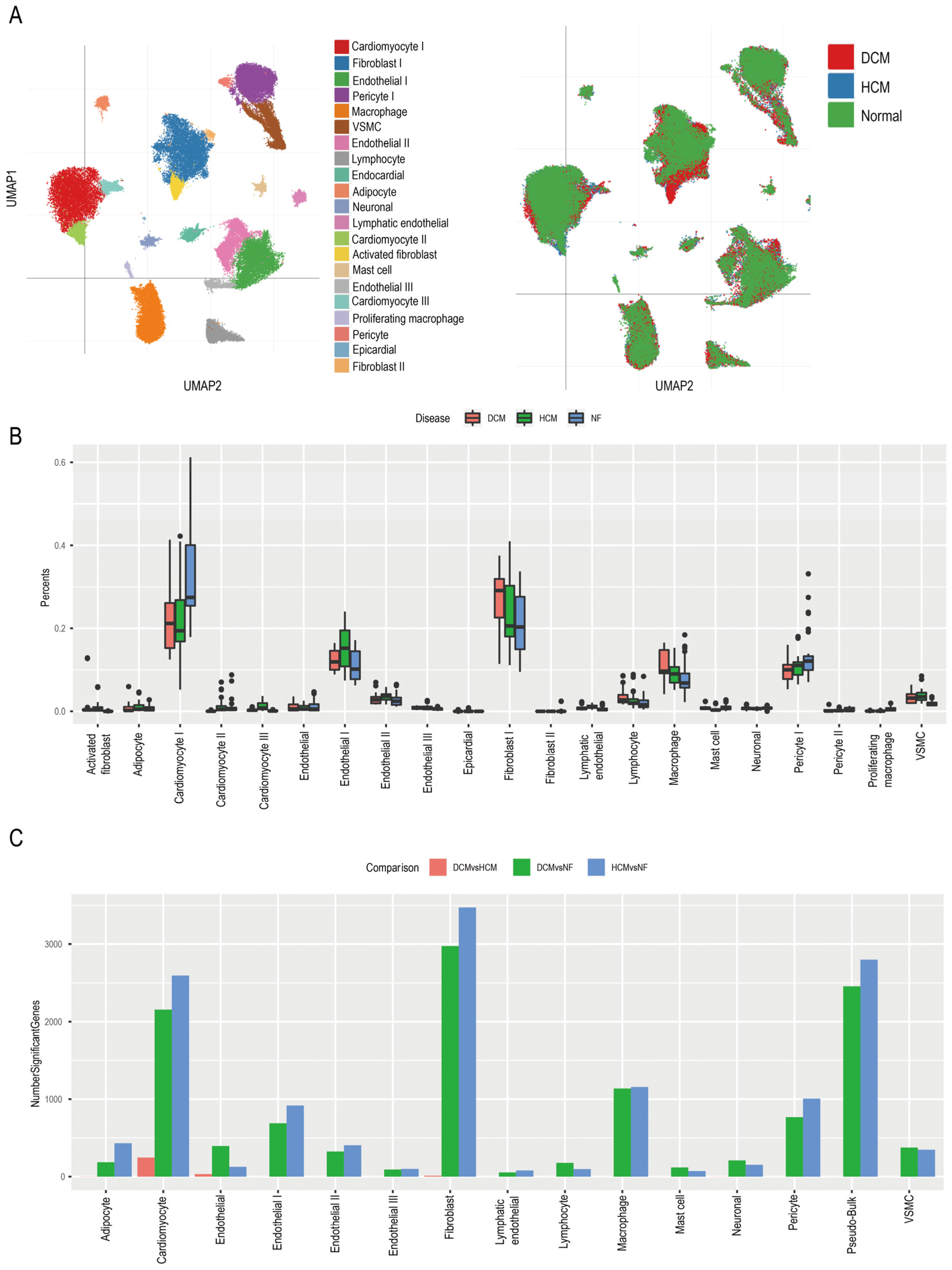
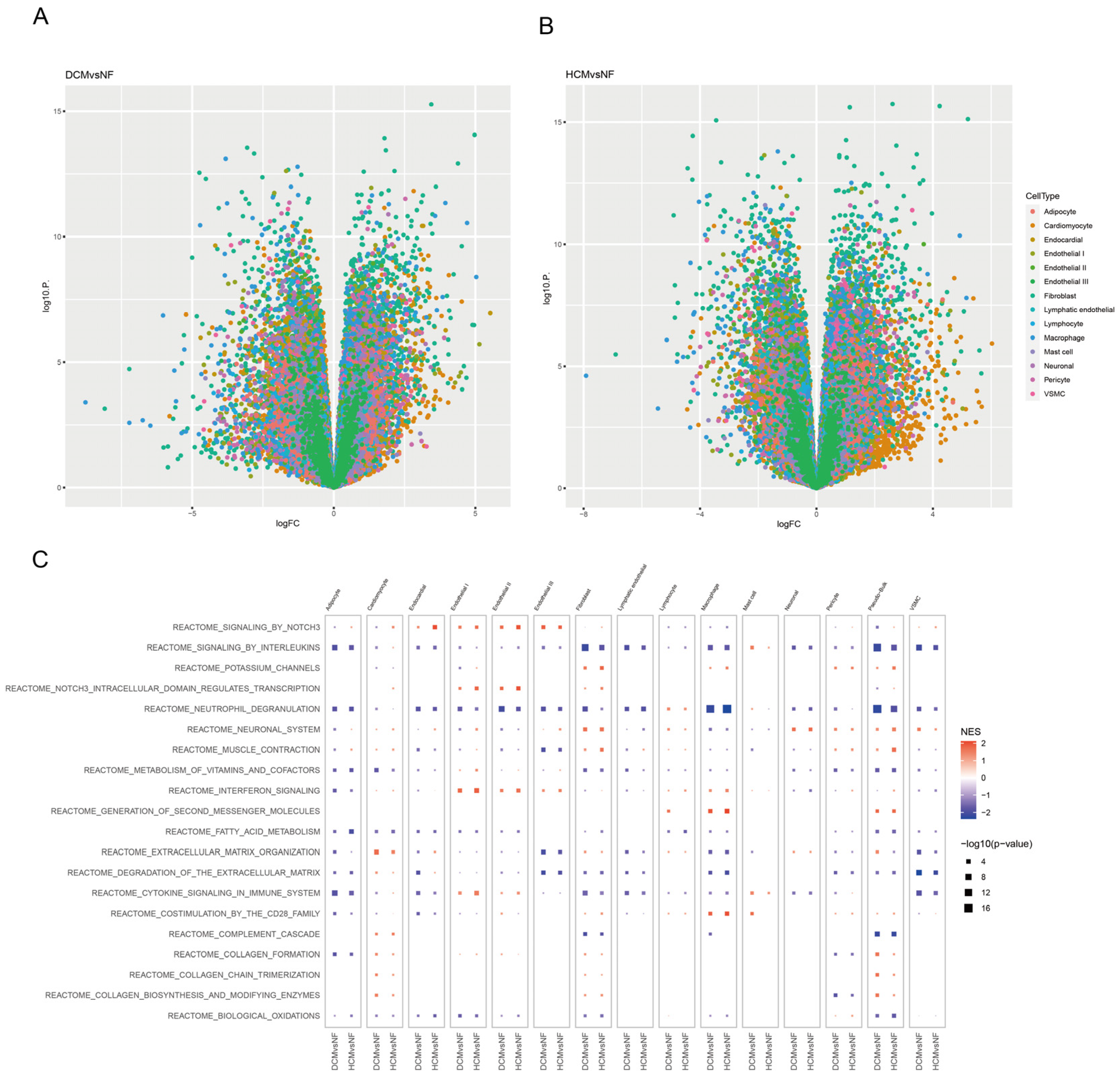
3.3. Identifying Function Pathways of Pericytes in Cardiomyopathy Patients
3.4. Hub Genes of Pericytes Involved in the MI Process
4. Discussion
5. Conclusions
6. Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CVD | cardiovascular disease |
| AMI | acute myocardial infarction |
| MI | myocardial infarction |
| PPI | protein–protein interaction |
| HCM | hypertrophic cardiomyopathy |
| DCM | dilated cardiomyopathy |
| EMT | epithelial–mesenchymal transition |
| LV | left ventricle |
| LA | left atrium |
| RA | right atrium |
| RV | right ventricle |
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.J.; Cheng, C.W.; Yang, N.I.; Hung, M.Y.; Cherng, W.J. Coronary vasospasm-induced acute coronary syndrome complicated by life-threatening cardiac arrhythmias in patients without hemodynamically significant coronary artery disease. Int. J. Cardiol. 2007, 117, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Tibaut, M.; Mekis, D.; Petrovic, D. Pathophysiology of Myocardial Infarction and Acute Management Strategies. Cardiovasc. Hematol. Agents Med. Chem. 2017, 14, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Westman, P.C.; Lipinski, M.J.; Luger, D.; Waksman, R.; Bonow, R.O.; Wu, E.; Epstein, S.E. Inflammation as a Driver of Adverse Left Ventricular Remodeling after Acute Myocardial Infarction. J. Am. Coll. Cardiol. 2016, 67, 2050–2060. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.H.; Arnaout, S.; Karrowni, W.; Dakik, H.A. The management of acute myocardial infarction in developing countries. Int. J. Cardiol. 2006, 111, 189–194. [Google Scholar] [CrossRef]
- Shi, X.; Zhang, X.; Zhuang, F.; Lu, Y.; Liang, F.; Zhao, N.; Wang, X.; Li, Y.; Cai, Z.; Wu, Z.; et al. Congestive heart failure detection based on attention mechanism-enabled bi-directional long short-term memory model in the internet of medical things. J. Ind. Inf. Integr. 2022, 30, 100402. [Google Scholar] [CrossRef]
- Reddy, K.; Khaliq, A.; Henning, R.J. Recent advances in the diagnosis and treatment of acute myocardial infarction. World J. Cardiol. 2015, 7, 243–276. [Google Scholar] [CrossRef]
- Liu, F.; Ma, Y.; Ge, H.; Zhao, Y.; Shen, H.; Zhang, D.; Sun, Y.; Ma, X.; Cheng, Y.; Zhou, Y. Long-Term Outcomes of Acute Myocardial Infarction in Patients with Hypertrophic Cardiomyopathy. Angiology 2018, 69, 900–908. [Google Scholar] [CrossRef]
- Jia, D.; Chen, S.; Bai, P.; Luo, C.; Liu, J.; Sun, A.; Ge, J. Cardiac Resident Macrophage-Derived Legumain Improves Cardiac Repair by Promoting Clearance and Degradation of Apoptotic Cardiomyocytes After Myocardial Infarction. Circulation 2022, 145, 1542–1556. [Google Scholar] [CrossRef]
- Shi, X.; Cao, Y.; Zhang, X.; Gu, C.; Liang, F.; Xue, J.; Ni, H.W.; Wang, Z.; Li, Y.; Wang, X.; et al. Comprehensive Analysis of N6-Methyladenosine RNA Methylation Regulators Expression Identify Distinct Molecular Subtypes of Myocardial Infarction. Front. Cell Dev. Biol. 2021, 9, 756483. [Google Scholar] [CrossRef]
- Zhao, M.; Nakada, Y.; Wei, Y.; Bian, W.; Chu, Y.; Borovjagin, A.V.; Xie, M.; Zhu, W.; Nguyen, T.; Zhou, Y.; et al. Cyclin D2 Overexpression Enhances the Efficacy of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Myocardial Repair in a Swine Model of Myocardial Infarction. Circulation 2021, 144, 210–228. [Google Scholar] [CrossRef] [PubMed]
- Craig, D.J.; James, A.W.; Wang, Y.; Tavian, M.; Crisan, M.; Péault, B.M. Blood Vessel Resident Human Stem Cells in Health and Disease. Stem Cells Transl. Med. 2022, 11, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Litviňuková, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Worth, C.L.; Lindberg, E.L.; Kanda, M.; Polanski, K.; Heinig, M.; Lee, M.; et al. Cells of the adult human heart. Nature 2020, 588, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Cantrell, A.C.; Zeng, H.; Zhu, S.H.; Chen, J.X. Emerging Role of Pericytes and Their Secretome in the Heart. Cells 2021, 10, 548. [Google Scholar] [CrossRef] [PubMed]
- Spencer, H.L.; Jover, E.; Cathery, W.; Avolio, E.; Rodriguez-Arabaolaza, I.; Thomas, A.C.; Alvino, V.V.; Sala-Newby, G.; Dang, Z.; Fagnano, M.; et al. Role of TPBG (Trophoblast Glycoprotein) Antigen in Human Pericyte Migratory and Angiogenic Activity. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1113–1124. [Google Scholar] [CrossRef]
- Yoshida, Y.; Kabara, M.; Kano, K.; Horiuchi, K.; Hayasaka, T.; Tomita, Y.; Takehara, N.; Minoshima, A.; Aonuma, T.; Maruyama, K.; et al. Capillary-resident EphA7(+) pericytes are multipotent cells with anti-ischemic effects through capillary formation. Stem Cells Transl. Med. 2020, 9, 120–130. [Google Scholar] [CrossRef]
- Quijada, P.; Park, S.; Zhao, P.; Kolluri, K.S.; Wong, D.; Shih, K.D.; Fang, K.; Pezhouman, A.; Wang, L.; Daraei, A.; et al. Cardiac pericytes mediate the remodeling response to myocardial infarction. J. Clin. Investig. 2023, 133, e162188. [Google Scholar] [CrossRef]
- Alex, L.; Tuleta, I.; Hernandez, S.C.; Hanna, A.; Venugopal, H.; Astorkia, M.; Humeres, C.; Kubota, A.; Su, K.; Zheng, D.; et al. Cardiac Pericytes Acquire a Fibrogenic Phenotype and Contribute to Vascular Maturation After Myocardial Infarction. Circulation 2023, 148, 882–898. [Google Scholar] [CrossRef]
- Tucker, N.R.; Chaffin, M.; Fleming, S.J.; Hall, A.W.; Parsons, V.A.; Bedi, K.C., Jr.; Akkad, A.D.; Herndon, C.N.; Arduini, A.; Papangeli, I.; et al. Transcriptional and Cellular Diversity of the Human Heart. Circulation 2020, 142, 466–482. [Google Scholar] [CrossRef]
- Hartiala, J.A.; Han, Y.; Jia, Q.; Hilser, J.R.; Huang, P.; Gukasyan, J.; Schwartzman, W.S.; Cai, Z.; Biswas, S.; Trégouët, D.A.; et al. Genome-wide analysis identifies novel susceptibility loci for myocardial infarction. Eur. Heart J. 2021, 42, 919–933. [Google Scholar] [CrossRef]
- Chaffin, M.; Papangeli, I.; Simonson, B.; Akkad, A.D.; Hill, M.C.; Arduini, A.; Fleming, S.J.; Melanson, M.; Hayat, S.; Kost-Alimova, M.; et al. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy. Nature 2022, 608, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Dobie, R.; Wilson-Kanamori, J.R.; Dora, E.F.; Henderson, B.E.P.; Luu, N.T.; Portman, J.R.; Matchett, K.P.; Brice, M.; Marwick, J.A.; et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019, 575, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Lun, A.T.; McCarthy, D.J.; Marioni, J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research 2016, 5, 2122. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Campbell, K.R.; Lun, A.T.; Wills, Q.F. Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 2017, 33, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.J.; Fan, C.M.; Yuan, J.Q.; Zhang, H.B.; Duan, F.J.; Wang, Z.M.; Guo, X.Y.; Zhai, S.S.; An, S.Y.; Hang, F.; et al. Long-term survival after acute myocardial infarction in patients with hypertrophic cardiomyopathy. Clin. Cardiol. 2017, 40, 26–31. [Google Scholar] [CrossRef]
- Hwang, B.; Lee, J.H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018, 50, 96. [Google Scholar] [CrossRef]
- Iqbal, F.; Lupieri, A.; Aikawa, M.; Aikawa, E. Harnessing Single-Cell RNA Sequencing to Better Understand How Diseased Cells Behave the Way They Do in Cardiovascular Disease. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 585–600. [Google Scholar] [CrossRef]
- Paik, D.T.; Cho, S.; Tian, L.; Chang, H.Y.; Wu, J.C. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat. Rev. Cardiol. 2020, 17, 457–473. [Google Scholar] [CrossRef]
- Qian, J.; Gao, Y.; Lai, Y.; Ye, Z.; Yao, Y.; Ding, K.; Tong, J.; Lin, H.; Zhu, G.; Yu, Y.; et al. Single-Cell RNA Sequencing of Peripheral Blood Mononuclear Cells from Acute Myocardial Infarction. Front. Immunol. 2022, 13, 908815. [Google Scholar] [CrossRef]
- Shi, X.; Zhang, L.; Li, Y.; Xue, J.; Liang, F.; Ni, H.W.; Wang, X.; Cai, Z.; Shen, L.H.; Huang, T.; et al. Integrative Analysis of Bulk and Single-Cell RNA Sequencing Data Reveals Cell Types Involved in Heart Failure. Front. Bioeng. Biotechnol. 2021, 9, 779225. [Google Scholar] [CrossRef]
- Jin, K.; Gao, S.; Yang, P.; Guo, R.; Li, D.; Zhang, Y.; Lu, X.; Fan, G.; Fan, X. Single-Cell RNA Sequencing Reveals the Temporal Diversity and Dynamics of Cardiac Immunity after Myocardial Infarction. Small Methods 2022, 6, e2100752. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Gao, P.; Zhong, X.; Li, M.; Wang, M.; Song, X. Identification of Five Hub Genes Based on Single-Cell RNA Sequencing Data and Network Pharmacology in Patients with Acute Myocardial Infarction. Front. Public Health 2022, 10, 894129. [Google Scholar] [CrossRef]
- Longden, T.A.; Zhao, G.; Hariharan, A.; Lederer, W.J. Pericytes and the Control of Blood Flow in Brain and Heart. Annu. Rev. Physiol. 2023, 85, 137–164. [Google Scholar] [CrossRef] [PubMed]
- Teichert, M.; Milde, L.; Holm, A.; Stanicek, L.; Gengenbacher, N.; Savant, S.; Ruckdeschel, T.; Hasanov, Z.; Srivastava, K.; Hu, J.; et al. Pericyte-expressed Tie2 controls angiogenesis and vessel maturation. Nat. Commun. 2017, 8, 16106. [Google Scholar] [CrossRef] [PubMed]
- Korte, N.; Ilkan, Z.; Pearson, C.L.; Pfeiffer, T.; Singhal, P.; Rock, J.R.; Sethi, H.; Gill, D.; Attwell, D.; Tammaro, P. The Ca2+-gated channel TMEM16A amplifies capillary pericyte contraction and reduces cerebral blood flow after ischemia. J. Clin. Investig. 2022, 132, e154118. [Google Scholar] [CrossRef]
- Zhang, G.; Li, J.; Sun, H.; Yang, G. Screening for the Biomarkers Associated with Myocardial Infarction by Bioinformatics Analysis. J. Comput. Biol. 2020, 27, 779–785. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, J.; Hong, L.; Zhou, Y. EGFLAM correlates with cell proliferation, migration, invasion and poor prognosis in glioblastoma. Cancer Biomark. 2019, 24, 343–350. [Google Scholar] [CrossRef]
- Yamada, Y.; Sakuma, J.; Takeuchi, I.; Yasukochi, Y.; Kato, K.; Oguri, M.; Fujimaki, T.; Horibe, H.; Muramatsu, M.; Sawabe, M.; et al. Identification of EGFLAM, SPATC1L and RNASE13 as novel susceptibility loci for aortic aneurysm in Japanese individuals by exome-wide association studies. Int. J. Mol. Med. 2017, 39, 1091–1100. [Google Scholar] [CrossRef]
- Chen, L.; Li, X.; Chen, M.; Feng, Y.; Xiong, C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020, 116, 1097–1100. [Google Scholar] [CrossRef]
- Peuhkurinen, K.; Risteli, L.; Jounela, A.; Risteli, J. Changes in interstitial collagen metabolism during acute myocardial infarction treated with streptokinase or tissue plasminogen activator. Am. Heart J. 1996, 131, 7–13. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. The extracellular matrix in myocardial injury, repair, and remodeling. J. Clin. Investig. 2017, 127, 1600–1612. [Google Scholar] [CrossRef] [PubMed]
- Basso, C.; Thiene, G.; Della Barbera, M.; Angelini, A.; Kirchengast, M.; Iliceto, S. Endothelin A-receptor antagonist administration immediately after experimental myocardial infarction with reperfusion does not affect scar healing in dogs. Cardiovasc. Res. 2002, 55, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Aharonov, A.; Shakked, A.; Umansky, K.B.; Savidor, A.; Genzelinakh, A.; Kain, D.; Lendengolts, D.; Revach, O.Y.; Morikawa, Y.; Dong, J.; et al. ERBB2 drives YAP activation and EMT-like processes during cardiac regeneration. Nat. Cell Biol. 2020, 22, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Blom, J.N.; Feng, Q. Cardiac repair by epicardial EMT: Current targets and a potential role for the primary cilium. Pharmacol. Ther. 2018, 186, 114–129. [Google Scholar] [CrossRef]
- Poschl, E.; Pollner, R.; Kuhn, K. The genes for the alpha 1(IV) and alpha 2(IV) chains of human basement membrane collagen type IV are arranged head-to-head and separated by a bidirectional promoter of unique structure. EMBO J. 1988, 7, 2687–2695. [Google Scholar] [CrossRef]
- Yang, W.; Ng, F.L.; Chan, K.; Pu, X.; Poston, R.N.; Ren, M.; An, W.; Zhang, R.; Wu, J.; Yan, S.; et al. Coronary-Heart-Disease-Associated Genetic Variant at the COL4A1/COL4A2 Locus Affects COL4A1/COL4A2 Expression, Vascular Cell Survival, Atherosclerotic Plaque Stability and Risk of Myocardial Infarction. PLoS Genet. 2016, 12, e1006127. [Google Scholar] [CrossRef]
- Yurchenco, P.D. Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 2011, 3, a004911. [Google Scholar] [CrossRef]
- Verbeek, E.; Meuwissen, M.E.; Verheijen, F.W.; Govaert, P.P.; Licht, D.J.; Kuo, D.S.; Poulton, C.J.; Schot, R.; Lequin, M.H.; Dudink, J.; et al. COL4A2 mutation associated with familial porencephaly and small-vessel disease. Eur. J. Hum. Genet. 2012, 20, 844–851. [Google Scholar] [CrossRef]
- Hao, J.; Ju, H.; Zhao, S.; Junaid, A.; Scammell-La Fleur, T.; Dixon, I.M. Elevation of expression of Smads 2, 3, and 4, decorin and TGF-beta in the chronic phase of myocardial infarct scar healing. J. Mol. Cell. Cardiol. 1999, 31, 667–678. [Google Scholar] [CrossRef]
- Dobaczewski, M.; Bujak, M.; Li, N.; Gonzalez-Quesada, C.; Mendoza, L.H.; Wang, X.F.; Frangogiannis, N.G. Smad3 signaling critically regulates fibroblast phenotype and function in healing myocardial infarction. Circ. Res. 2010, 107, 418–428. [Google Scholar] [CrossRef]
- Kong, P.; Shinde, A.V.; Su, Y.; Russo, I.; Chen, B.; Saxena, A.; Conway, S.J.; Graff, J.M.; Frangogiannis, N.G. Opposing Actions of Fibroblast and Cardiomyocyte Smad3 Signaling in the Infarcted Myocardium. Circulation 2018, 137, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Bujak, M.; Ren, G.; Kweon, H.J.; Dobaczewski, M.; Reddy, A.; Taffet, G.; Wang, X.F.; Frangogiannis, N.G. Essential role of Smad3 in infarct healing and in the pathogenesis of cardiac remodeling. Circulation 2007, 116, 2127–2138. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Huang, S.; Su, Y.; Wu, Y.J.; Hanna, A.; Brickshawana, A.; Graff, J.; Frangogiannis, N.G. Macrophage Smad3 Protects the Infarcted Heart, Stimulating Phagocytosis and Regulating Inflammation. Circ. Res. 2019, 125, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, F.; Zhang, M.; Xue, H.; Fan, L.; Weng, Y. The role of SMAD signaling in hypertrophic obstructive cardiomyopathy: An immunohistopathological study in pediatric and adult patients. Sci. Rep. 2023, 13, 3706. [Google Scholar] [CrossRef]
- Su, S.A.; Yang, D.; Wu, Y.; Xie, Y.; Zhu, W.; Cai, Z.; Shen, J.; Fu, Z.; Wang, Y.; Jia, L.; et al. EphrinB2 Regulates Cardiac Fibrosis Through Modulating the Interaction of Stat3 and TGF-β/Smad3 Signaling. Circ. Res. 2017, 121, 617–627. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Huo, H.; Liang, F.; Xue, J.; Fang, L.; Miao, Y.; Shen, L.; He, B. Role of Pericytes in Cardiomyopathy-Associated Myocardial Infarction Revealed by Multiple Single-Cell Sequencing Analysis. Biomedicines 2023, 11, 2896. https://doi.org/10.3390/biomedicines11112896
Lu Y, Huo H, Liang F, Xue J, Fang L, Miao Y, Shen L, He B. Role of Pericytes in Cardiomyopathy-Associated Myocardial Infarction Revealed by Multiple Single-Cell Sequencing Analysis. Biomedicines. 2023; 11(11):2896. https://doi.org/10.3390/biomedicines11112896
Chicago/Turabian StyleLu, Yanqiao, Huanhuan Huo, Feng Liang, Jieyuan Xue, Liang Fang, Yutong Miao, Lan Shen, and Ben He. 2023. "Role of Pericytes in Cardiomyopathy-Associated Myocardial Infarction Revealed by Multiple Single-Cell Sequencing Analysis" Biomedicines 11, no. 11: 2896. https://doi.org/10.3390/biomedicines11112896
APA StyleLu, Y., Huo, H., Liang, F., Xue, J., Fang, L., Miao, Y., Shen, L., & He, B. (2023). Role of Pericytes in Cardiomyopathy-Associated Myocardial Infarction Revealed by Multiple Single-Cell Sequencing Analysis. Biomedicines, 11(11), 2896. https://doi.org/10.3390/biomedicines11112896








