Emerging Insights into Molecular Mechanisms of Inflammation in Myelodysplastic Syndromes
Abstract
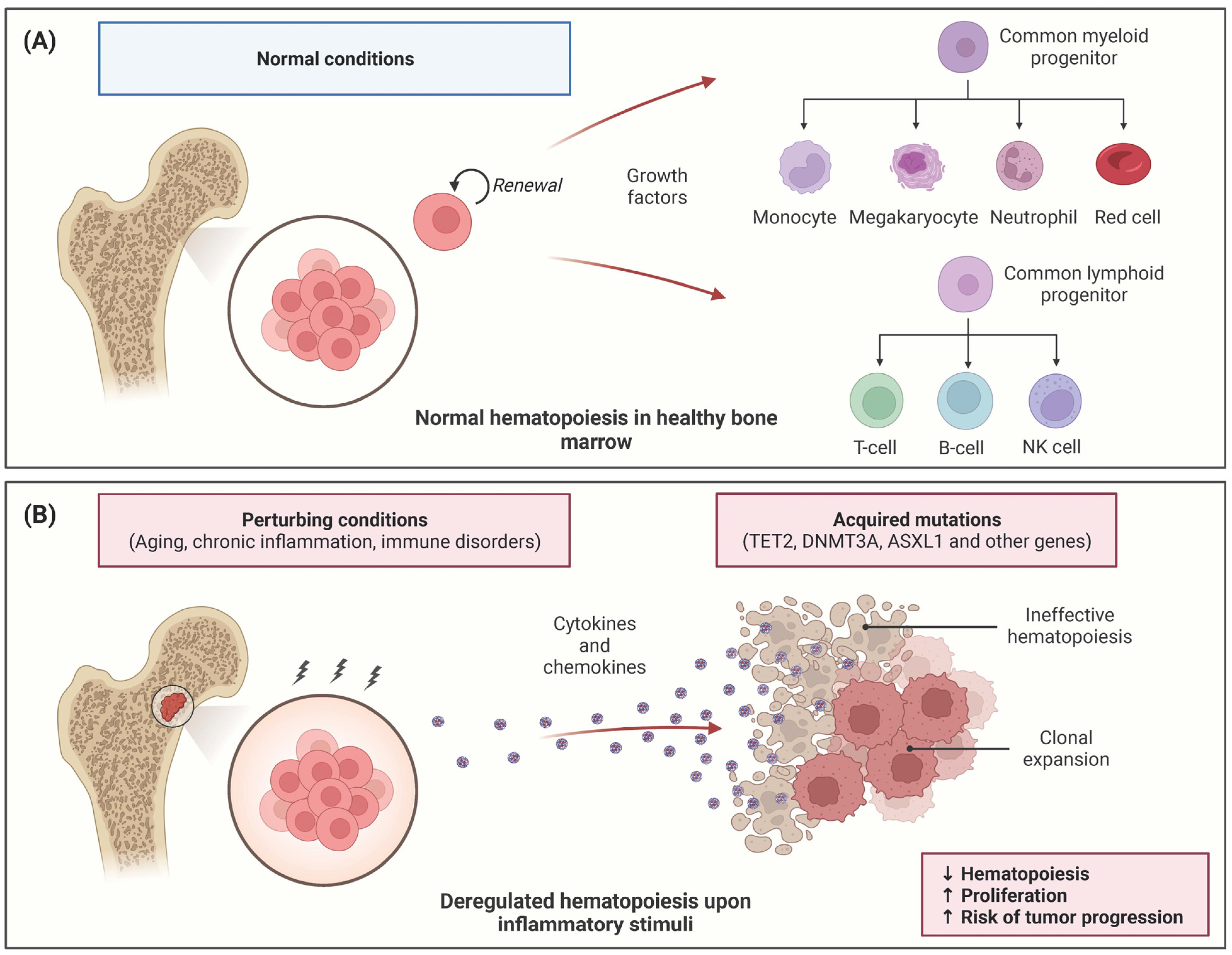
:1. Introduction
2. Inflammation Impact on MDS Hematopoiesis
2.1. Inflammation Promotes HSC Expansion in CHIP
2.2. MDS HSCs Show Altered Response to Chronic Inflammation
2.3. Heterogeneity of Inflammatory States in MDS
3. Multi-Omic Approaches to Dissect Immune Dysregulation in MDS
3.1. Single-Cell Sequencing Approaches to Characterize Clonal Evolution of MDS to sAML
3.2. sc-RNAseq Technology to Dissect Inflammatory Microenvironment
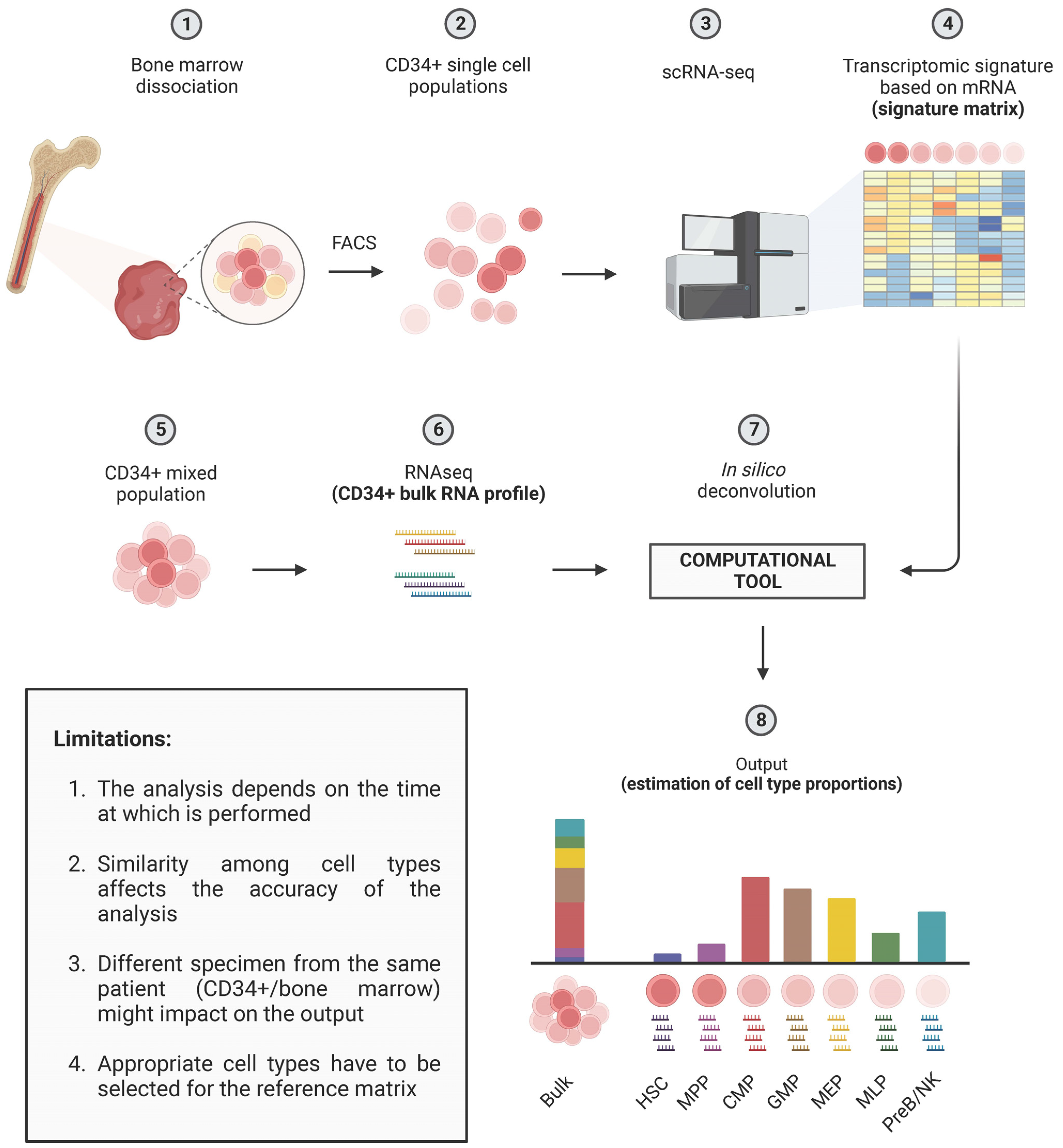
3.3. Deconvolution Methods to Dissect MDS Heterogeneity
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sezaki, M.; Hayashi, Y.; Wang, Y.; Johansson, A.; Umemoto, T.; Takizawa, H. Immuno-Modulation of Hematopoietic Stem and Progenitor Cells in Inflammation. Front. Immunol. 2020, 11, 585367. [Google Scholar] [CrossRef] [PubMed]
- Caiado, F.; Pietras, E.M.; Manz, M.G. Inflammation as a regulator of hematopoietic stem cell function in disease, aging, and clonal selection. J. Exp. Med. 2021, 218, e20201541. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S. Clonal hematopoiesis and non-hematologic disorders. Blood 2020, 136, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Barreyro, L.; Chlon, T.M.; Starczynowski, D.T. Chronic immune response dysregulation in MDS pathogenesis. Blood 2018, 132, 1553–1560. [Google Scholar] [CrossRef]
- Trowbridge, J.J.; Starczynowski, D.T. Innate immune pathways and inflammation in hematopoietic aging, clonal hematopoiesis, and MDS. J. Exp. Med. 2021, 218, e20201544. [Google Scholar] [CrossRef] [PubMed]
- Datar, G.K.; Goodell, M.A. Where There’s Smoke, There’s Fire: Inflammation Drives MDS. Trends Immunol. 2020, 41, 558–560. [Google Scholar] [CrossRef]
- Sallman, D.A.; List, A. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood 2019, 133, 1039–1048. [Google Scholar] [CrossRef]
- Bejar, R.; Steensma, D.P. Recent developments in myelodysplastic syndromes. Blood 2014, 124, 2793–2803. [Google Scholar] [CrossRef]
- Winter, S.; Shoaie, S.; Kordasti, S.; Platzbecker, U. Integrating the “Immunome” in the Stratification of Myelodysplastic Syndromes and Future Clinical Trial Design. J. Clin. Oncol. 2020, 38, 1723–1735. [Google Scholar] [CrossRef]
- Kitagawa, M.; Saito, I.; Kuwata, T.; Yoshida, S.; Yamaguchi, S.; Takahashi, M.; Hirokawa, K. Overexpression of tumor necrosis factor (TNF)-alpha and interferon (IFN)-gamma by bone marrow cells from patients with myelodysplastic syndromes. Leukemia 1997, 11, 2049–2054. [Google Scholar] [CrossRef]
- Raza, A.; Mundle, S.; Shetty, V.; Alvi, S.; Chopra, H.; Span, L.; Parcharidou, A.; Dar, S.; Venugopal, P.; Borok, R.; et al. Novel insights into the biology of myelodysplastic syndromes: Excessive apoptosis and the role of cytokines. Int. J. Hematol. 1996, 63, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, M.T.; King, K.Y.; Boles, N.C.; Weksberg, D.C.; Goodell, M.A. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 2010, 465, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Carey, A.; Edwards, D.K.; Eide, C.A.; Newell, L.; Traer, E.; Medeiros, B.C.; Pollyea, D.A.; Deininger, M.W.; Collins, R.H.; Tyner, J.W.; et al. Identification of Interleukin-1 by Functional Screening as a Key Mediator of Cellular Expansion and Disease Progression in Acute Myeloid Leukemia. Cell Rep. 2017, 18, 3204–3218. [Google Scholar] [CrossRef] [PubMed]
- Florez, M.A.; Tran, B.T.; Wathan, T.K.; DeGregori, J.; Pietras, E.M.; King, K.Y. Clonal hematopoiesis: Mutation-specific adaptation to environmental change. Cell Stem Cell 2022, 29, 882–904. [Google Scholar] [CrossRef]
- Bick, A.G.; Weinstock, J.S.; Nandakumar, S.K.; Fulco, C.P.; Bao, E.L.; Zekavat, S.M.; Szeto, M.D.; Liao, X.; Leventhal, M.J.; Nasser, J.; et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 2020, 586, 763–768. [Google Scholar] [CrossRef]
- Cook, E.K.; Izukawa, T.; Young, S.; Rosen, G.; Jamali, M.; Zhang, L.; Johnson, D.; Bain, E.; Hilland, J.; Ferrone, C.K.; et al. Comorbid and inflammatory characteristics of genetic subtypes of clonal hematopoiesis. Blood Adv. 2019, 3, 2482–2486. [Google Scholar] [CrossRef]
- Cai, Z.; Kotzin, J.J.; Ramdas, B.; Chen, S.; Nelanuthala, S.; Palam, L.R.; Pandey, R.; Mali, R.S.; Liu, Y.; Kelley, M.R.; et al. Inhibition of Inflammatory Signaling in Tet2 Mutant Preleukemic Cells Mitigates Stress-Induced Abnormalities and Clonal Hematopoiesis. Cell Stem Cell 2018, 23, 833–849. [Google Scholar] [CrossRef]
- Meisel, M.; Hinterleitner, R.; Pacis, A.; Chen, L.; Earley, Z.M.; Mayassi, T.; Pierre, J.F.; Ernest, J.D.; Galipeau, H.J.; Thuille, N.; et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 2018, 557, 580–584. [Google Scholar] [CrossRef]
- Heyde, A.; Rohde, D.; McAlpine, C.S.; Zhang, S.; Hoyer, F.F.; Gerold, J.M.; Cheek, D.; Iwamoto, Y.; Schloss, M.J.; Vandoorne, K.; et al. Faculty Opinions recommendation of Increased stem cell proliferation in atherosclerosis accelerates clonal hematopoiesis. Cell 2021, 184, 1348–1361. [Google Scholar] [CrossRef]
- Yeaton, A.; Cayanan, G.; Loghavi, S.; Dolgalev, I.; Leddin, E.M.; Loo, C.E.; Torabifard, H.; Nicolet, D.; Wang, J.; Corrigan, K.; et al. The Impact of Inflammation-Induced Tumor Plasticity during Myeloid Transformation. Cancer Discov. 2022, 12, 2392–2413. [Google Scholar] [CrossRef]
- Aivalioti, M.M.; Bartholdy, B.A.; Pradhan, K.; Bhagat, T.D.; Zintiridou, A.; Jeong, J.J.; Thiruthuvanathan, V.J.; Pujato, M.; Paranjpe, A.; Zhang, C.; et al. PU.1-Dependent Enhancer Inhibition Separates Tet2-Deficient Hematopoiesis from Malignant Transformation. Blood Cancer Discov. 2022, 3, 444–467. [Google Scholar] [CrossRef] [PubMed]
- Hormaechea-Agulla, D.; Matatall, K.A.; Le, D.T.; Kain, B.; Long, X.; Kus, P.; King, K.Y. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNgamma signaling. Cell Stem Cell 2021, 28, 1428–1442.e6. [Google Scholar] [CrossRef] [PubMed]
- Muto, T.; Walker, C.S.; Choi, K.; Hueneman, K.; Smith, M.A.; Gul, Z.; Garcia-Manero, G.; Ma, A.; Zheng, Y.; Starczynowski, D.T. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat. Immunol. 2020, 21, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Rolfs, C.; Trumpp, M.; Winter, S.; Fischer, L.; Richter, M.; Menger, V.; Nenoff, K.; Grieb, N.; Metzeler, K.H.; et al. Activation of distinct inflammatory pathways in subgroups of LR-MDS. Leukemia 2023, 37, 1709–1718. [Google Scholar] [CrossRef] [PubMed]
- van Galen, P.; Hovestadt, V.; Wadsworth, M.H., II; Hughes, T.K.; Griffin, G.K.; Battaglia, S.; Bernstein, B.E. Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell 2019, 176, 1265–1281.e24. [Google Scholar] [CrossRef] [PubMed]
- Guess, T.; Potts, C.R.; Bhat, P.; Cartailler, J.A.; Brooks, A.; Holt, C.; Yenamandra, A.; Wheeler, F.C.; Savona, M.R.; Cartailler, J.-P.; et al. Distinct Patterns of Clonal Evolution Drive Myelodysplastic Syndrome Progression to Secondary Acute Myeloid Leukemia. Blood Cancer Discov. 2022, 3, 316–329. [Google Scholar] [CrossRef]
- Menssen, A.J.; Khanna, A.; Miller, C.A.; Srivatsan, S.N.; Chang, G.S.; Shao, J.; Robinson, J.; O’Laughlin, M.; Fronick, C.C.; Fulton, R.S.; et al. Convergent Clonal Evolution of Signaling Gene Mutations Is a Hallmark of Myelodysplastic Syndrome Progression. Blood Cancer Discov. 2022, 3, 330–345. [Google Scholar] [CrossRef]
- Ganan-Gomez, I.; Yang, H.; Ma, F.; Montalban-Bravo, G.; Thongon, N.; Marchica, V.; Richard-Carpentier, G.; Chien, K.; Manyam, G.; Wang, F.; et al. Stem cell architecture drives myelodysplastic syndrome progression and predicts response to venetoclax-based therapy. Nat. Med. 2022, 28, 557–567. [Google Scholar] [CrossRef]
- Lasry, A.; Nadorp, B.; Fornerod, M.; Nicolet, D.; Wu, H.; Walker, C.J.; Aifantis, I. An inflammatory state remodels the immune microenvironment and improves risk stratification in acute myeloid leukemia. Nat Cancer 2023, 4, 27–42. [Google Scholar] [CrossRef]
- Dai, C.; Chen, M.; Wang, C.; Hao, X. Deconvolution of Bulk Gene Expression Profiles with Single-Cell Transcriptomics to Develop a Cell Type Composition-Based Prognostic Model for Acute Myeloid Leukemia. Front. Cell Dev. Biol. 2021, 9, 762260. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Hou, H.-A.; Lin, C.-C.; Kuo, Y.-Y.; Yao, C.-Y.; Hsu, C.-L.; Tseng, M.-H.; Tsai, C.-H.; Peng, Y.-L.; Kao, C.-J.; et al. A CIBERSORTx-based immune cell scoring system could independently predict the prognosis of patients with myelodysplastic syndromes. Blood Adv. 2021, 5, 4535–4548. [Google Scholar] [CrossRef] [PubMed]
- Paracatu, L.C.; Schuettpelz, L.G. Contribution of Aberrant Toll Like Receptor Signaling to the Pathogenesis of Myelodysplastic Syndromes. Front. Immunol. 2020, 11, 1236. [Google Scholar] [CrossRef] [PubMed]
- Maratheftis, C.I.; Andreakos, E.; Moutsopoulos, H.M.; Voulgarelis, M. Toll-like Receptor-4 Is Up-Regulated in Hematopoietic Progenitor Cells and Contributes to Increased Apoptosis in Myelodysplastic Syndromes. Clin. Cancer Res. 2007, 13, 1154–1160. [Google Scholar] [CrossRef] [PubMed]
- Pellagatti, A.; Cazzola, M.; Giagounidis, A.; Perry, J.; Malcovati, L.; Della Porta, M.G.; Jädersten, M.; Killick, S.; Verma, A.; Norbury, C.J.; et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia 2010, 24, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Dimicoli, S.; Wei, Y.; Bueso-Ramos, C.; Yang, H.; DiNardo, C.; Jia, Y.; Zheng, H.; Fang, Z.; Nguyen, M.; Pierce, S.; et al. Overexpression of the Toll-Like Receptor (TLR) Signaling Adaptor MYD88, but Lack of Genetic Mutation, in Myelodysplastic Syndromes. PLoS ONE 2013, 8, e71120. [Google Scholar] [CrossRef]
- Fang, J.; Bolanos, L.C.; Choi, K.; Liu, X.; Christie, S.; Akunuru, S.; Kumar, R.; Wang, D.; Chen, X.; Greis, K.D.; et al. Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia. Nat. Immunol. 2016, 18, 236–245. [Google Scholar] [CrossRef]
- Varney, M.E.; Niederkorn, M.; Konno, H.; Matsumura, T.; Gohda, J.; Yoshida, N.; Starczynowski, D.T. Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling. J. Exp. Med. 2015, 212, 1967–1985. [Google Scholar] [CrossRef]
- Fang, J.; Barker, B.; Bolanos, L.; Liu, X.; Jerez, A.; Makishima, H.; Starczynowski, D.T. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-kappaB gene network. Cell Rep. 2014, 8, 1328–1338. [Google Scholar] [CrossRef]
- Sallman, D.A.; Cluzeau, T.; Basiorka, A.A.; List, A. Unraveling the Pathogenesis of MDS: The NLRP3 Inflammasome and Pyroptosis Drive the MDS Phenotype. Front. Oncol. 2016, 6, 151. [Google Scholar] [CrossRef]
- Basiorka, A.A.; McGraw, K.L.; Eksioglu, E.A.; Chen, X.; Johnson, J.; Zhang, L.; Zhang, Q.; Irvine, B.A.; Cluzeau, T.; Sallman, D.A.; et al. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood 2016, 128, 2960–2975. [Google Scholar] [CrossRef]
- Levine, J.H.; Simonds, E.F.; Bendall, S.C.; Davis, K.L.; El-ad, D.A.; Tadmor, M.D.; Litvin, O.; Fienberg, H.G.; Jager, A.; Zunder, E.R. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 2015, 162, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Notta, F.; Zandi, S.; Takayama, N.; Dobson, S.; Gan, O.I.; Wilson, G.; Kaufmann, K.B.; McLeod, J.; Laurenti, E.; Dunant, C.F.; et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 2016, 351, aab2116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Grimes, H.L. Why Single-Cell Sequencing Has Promise in MDS. Front. Oncol. 2021, 11, 769753. [Google Scholar] [CrossRef] [PubMed]
- Tikhonova, A.N.; Dolgalev, I.; Hu, H.; Sivaraj, K.K.; Hoxha, E.; Cuesta-Domínguez, Á.; Pinho, S.; Akhmetzyanova, I.; Gao, J.; Witkowski, M.; et al. The bone marrow microenvironment at single-cell resolution. Nature 2019, 569, 222–228. [Google Scholar] [CrossRef]
- Wolock, S.L.; Krishnan, I.; Tenen, D.E.; Matkins, V.; Camacho, V.; Patel, S.; Agarwal, P.; Bhatia, R.; Tenen, D.G.; Klein, A.M.; et al. Mapping Distinct Bone Marrow Niche Populations and Their Differentiation Paths. Cell Rep. 2019, 28, 302–311. [Google Scholar] [CrossRef]
- Baccin, C.; Al-Sabah, J.; Velten, L.; Helbling, P.M.; Grünschläger, F.; Hernández-Malmierca, P.; Nombela-Arrieta, C.; Steinmetz, L.M.; Trumpp, A.; Haas, S. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 2020, 22, 38–48. [Google Scholar] [CrossRef]
- Baryawno, N.; Przybylski, D.; Kowalczyk, M.S.; Kfoury, Y.; Severe, N.; Gustafsson, K.; Scadden, D.T. A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell 2019, 177, 1915–1932.e16. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Q.; Johnson, C.B.; Pham, G.; Kinder, J.M.; Olsson, A.; Slaughter, A.; May, M.; Weinhaus, B.; D’alessandro, A.; et al. In situ mapping identifies distinct vascular niches for myelopoiesis. Nature 2021, 590, 457–462. [Google Scholar] [CrossRef]
- Pei, W.; Feyerabend, T.B.; Rössler, J.; Wang, X.; Postrach, D.; Busch, K.; Rode, I.; Klapproth, K.; Dietlein, N.; Quedenau, C.; et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 2017, 548, 456–460. [Google Scholar] [CrossRef]
- Velten, L.; Haas, S.F.; Raffel, S.; Blaszkiewicz, S.; Islam, S.; Hennig, B.P.; Hirche, C.; Lutz, C.; Buss, E.C.; Nowak, D.; et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 2017, 19, 271–281. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Corces, M.R.; Lareau, C.A.; Wu, B.; Schep, A.N.; Aryee, M.J.; Majeti, R.; Chang, H.Y.; Greenleaf, W.J. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 2018, 173, 1535–1548. [Google Scholar] [CrossRef] [PubMed]
- Pellin, D.; Loperfido, M.; Baricordi, C.; Wolock, S.L.; Montepeloso, A.; Weinberg, O.K.; Biffi, A.; Klein, A.M.; Biasco, L. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat. Commun. 2019, 10, 2395. [Google Scholar] [CrossRef] [PubMed]
- Avila Cobos, F.; Alquicira-Hernandez, J.; Powell, J.E.; Mestdagh, P.; De Preter, K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat. Commun. 2020, 11, 5650. [Google Scholar] [CrossRef] [PubMed]
- Vallania, F.; Tam, A.; Lofgren, S.; Schaffert, S.; Azad, T.D.; Bongen, E.; Haynes, W.; Alsup, M.; Alonso, M.; Davis, M.; et al. Leveraging heterogeneity across multiple datasets increases cell-mixture deconvolution accuracy and reduces biological and technical biases. Nat. Commun. 2018, 9, 4735. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Wang, X.; Park, J.; Susztak, K.; Zhang, N.R.; Li, M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 2019, 10, 380. [Google Scholar] [CrossRef]
- Menden, K.; Marouf, M.; Oller, S.; Dalmia, A.; Magruder, D.S.; Kloiber, K.; Heutink, P.; Bonn, S. Deep learning–based cell composition analysis from tissue expression profiles. Sci. Adv. 2020, 6, eaba2619. [Google Scholar] [CrossRef]
- Gong, T.; Szustakowski, J.D. DeconRNASeq: A statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics 2013, 29, 1083–1085. [Google Scholar] [CrossRef]
- Dong, M.; Thennavan, A.; Urrutia, E.; Li, Y.; Perou, C.M.; Zou, F.; Jiang, Y. SCDC: Bulk gene expression deconvolution by multiple single-cell RNA sequencing references. Brief. Bioinform. 2020, 22, 416–427. [Google Scholar] [CrossRef]
- Charytonowicz, D.; Brody, R.; Sebra, R. Interpretable and context-free deconvolution of multi-scale whole transcriptomic data with UniCell deconvolve. Nat. Commun. 2023, 14, 1350. [Google Scholar] [CrossRef] [PubMed]
- Ostendorf, B.N.; Flenner, E.; Flörcken, A.; Westermann, J. Phenotypic characterization of aberrant stem and progenitor cell populations in myelodysplastic syndromes. PLoS ONE 2018, 13, e0197823. [Google Scholar] [CrossRef] [PubMed]
| Reference | Publication | Relevance |
|---|---|---|
| [20] | Yeaton et al., Cancer Discov (2022) | By sc-RNAseq of bone marrow from mice carrying Tet2 mutations, MDS/AML progression in aged animals has been shown to correlate with an enhanced inflammatory response and with the emergence of a novel population of inflammatory monocytes. |
| [22] | Hormaechea-Agulla et al., Cell Stem Cell (2021) | IFNγ signaling induced by chronic infection promoted the expansion of mouse Dnmt3aloss-of-function HSCs. |
| [23] | Muto et al., Nat Immunol (2020) | In a mouse model with increased TRAF6 expression in HSCs, an inflammatory stimulus has been shown to induce sustained myelopoiesis. |
| [24] | Schneider at al., Leukemia (2023) | Bulk RNA-seq profiling in a large cohort of MDS patients revealed distinct inflammatory pathways in subgroups of LR-MDS. |
| [25] | van Galen et al., Cell (2019) | sc-RNAseq and genotyping profiling of bone marrow from AML patients were used to characterize the heterogeneity of this disease. |
| [26] | Guess et al., Blood Cancer Discov (2022) | Single-cell sequencing of MDS and sAML bone marrow patients revealed patient-specific clonal evolution patterns. |
| [27] | Menssen et al., Blood Cancer Discov (2022) | Single-cell sequencing of MDS and sAML indicated that subclone expansion is a hallmark of MDS progression. |
| [28] | Ganan-Gomez et al., Nat Med (2022) | In this study, by sc-RNAseq of HSCs from MDS patients combined with immunophenotypic characterization, they found MDS HSCs in two distinct differentiation states. |
| [29] | Lasry et al., Nat Cancer (2023) | Single-cell CITE-seq profiling was used to characterize the effects of inflammation on the composition of bone marrow immune microenvironment in adult and pediatric patients with AML. |
| [30] | Dai et al., Front Cell Dev Biol (2021) | In this study, a novel prognostic model based on cell type composition deconvolution of bulk RNA-seq datasets of AML bone marrow or peripheral blood patients was developed. |
| [31] | Wang et al., Front Blood Adv (2021) | CIBERSORTx was used to estimate immune cell type compositions in bone marrow of MDS patients. A CIBERSORTx-based scoring system could predict the patients’ prognosis. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vallelonga, V.; Gandolfi, F.; Ficara, F.; Della Porta, M.G.; Ghisletti, S. Emerging Insights into Molecular Mechanisms of Inflammation in Myelodysplastic Syndromes. Biomedicines 2023, 11, 2613. https://doi.org/10.3390/biomedicines11102613
Vallelonga V, Gandolfi F, Ficara F, Della Porta MG, Ghisletti S. Emerging Insights into Molecular Mechanisms of Inflammation in Myelodysplastic Syndromes. Biomedicines. 2023; 11(10):2613. https://doi.org/10.3390/biomedicines11102613
Chicago/Turabian StyleVallelonga, Veronica, Francesco Gandolfi, Francesca Ficara, Matteo Giovanni Della Porta, and Serena Ghisletti. 2023. "Emerging Insights into Molecular Mechanisms of Inflammation in Myelodysplastic Syndromes" Biomedicines 11, no. 10: 2613. https://doi.org/10.3390/biomedicines11102613
APA StyleVallelonga, V., Gandolfi, F., Ficara, F., Della Porta, M. G., & Ghisletti, S. (2023). Emerging Insights into Molecular Mechanisms of Inflammation in Myelodysplastic Syndromes. Biomedicines, 11(10), 2613. https://doi.org/10.3390/biomedicines11102613





