On the Use of Machine Learning Techniques and Non-Invasive Indicators for Classifying and Predicting Cardiac Disorders
Abstract
:1. Introduction
2. Biological Indicators
2.1. Definition of Heart Disease
2.2. Description of Indicators
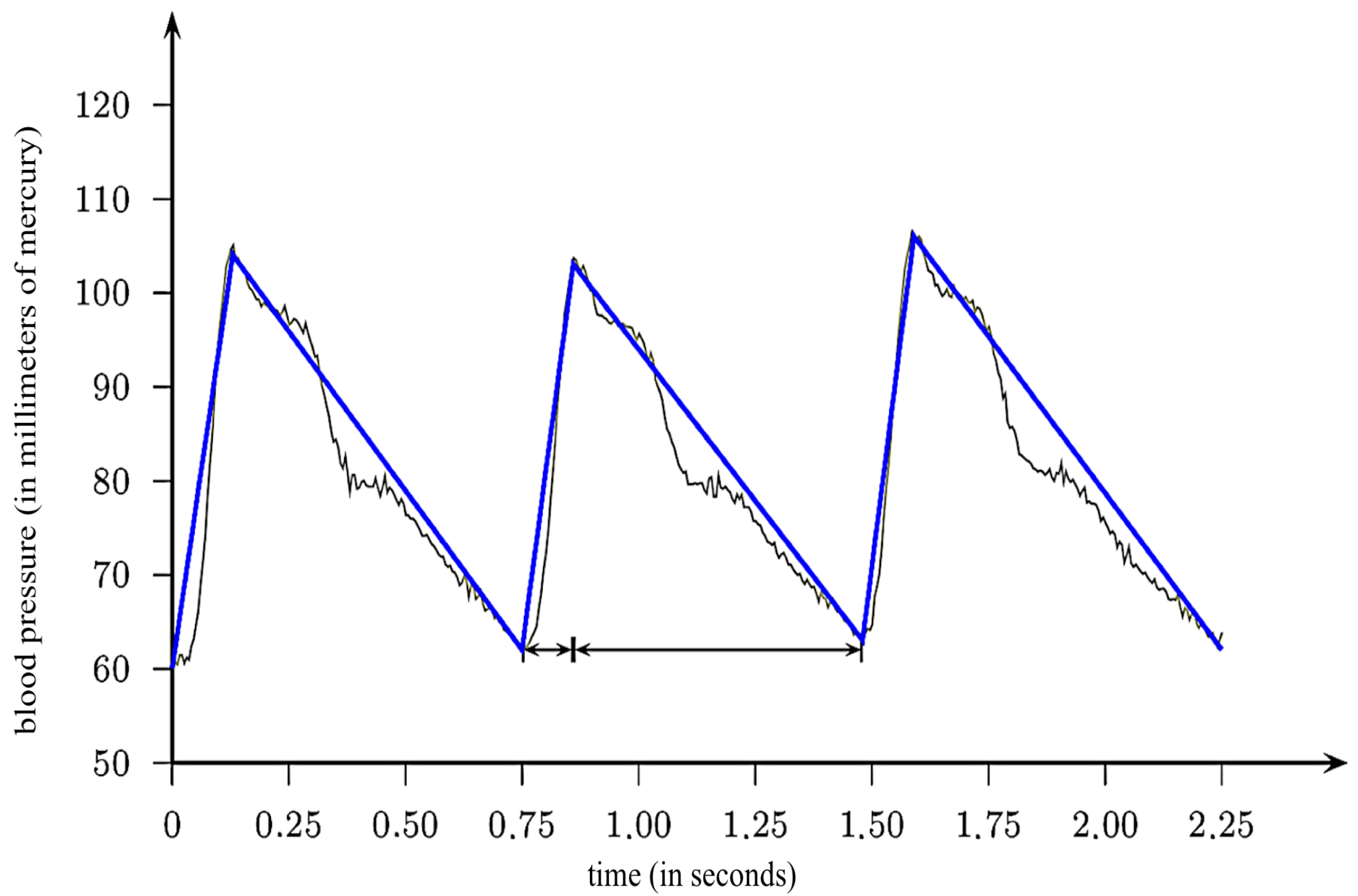
2.3. Mean Arterial Pressure
2.4. Product of Peripheral Resistance and Compliance
- Clinical relevance: The RC product is instrumental in evaluating the cardiovascular system’s resistance and elasticity. It is particularly relevant for patients with compromised vascular health, such as those with hypertension or atherosclerosis. Maintaining balanced RC values could be indicative of effective treatment strategies [11].
2.5. Pulsatile Blood Pressure Index
- Clinical relevance: The PBPI is a vital risk indicator for cardiovascular diseases, specifically for arterial hypertension. High PBPI levels can be a precursor to hypertension and other cardiovascular conditions, thus making it a tool for early diagnosis and prevention [13].
2.6. Harmony Measure
2.7. Modeling Ejection Time
3. ML Techniques
3.1. Datasets and Scenarios
- Scenario 1: It includes the variables V3 (age), V4 (gender), and V11 (history of hypertension); the indicators , , HM, MAP, PBPI, PBPIRC, and RC; as well as the response variable Y. The variables were selected to prioritize models that utilize low complexity in terms of data collection.
- Scenario 2: It includes the 75 variables in the heart disease directory; the indicators , , HM, MAP, PBPI, PBPIRC, and RC; as well as the response variable Y.
3.2. Adapted Consistency Measure
3.3. Features Selection
- Naive Bayes (NB): This classifier is based on the Bayes theorem, which estimates the probability that an event will occur considering prior information associated with this event [19,20]. The NB method is renowned for its remarkable simplicity and competitive performance compared to other classifiers. However, this method assumes independence between the explanatory variables [21].
- Random forests (RFs): This classifier is an extension of decision trees (DTs), formed by a collection of non-correlated trees. The classification or estimation is determined by a voting process among the trees. DTs are constructed through bootstrapping, where each tree is trained on a different subset of the data [21,22,23]. RFs offer several advantages, including robustness against outliers, low bias, and the ability to capture complex data interactions [21,22].
- Logistic regression (LR): This classifier corresponds to a linear regression in which the response variable is binary. A transformation is applied to ensure the response variable is continuous. Common transformations in the literature include logit, probit, and Cauchy [24,25,26,27]. In this study, the logit transformation was utilized.
- Adaboost: This is an ensemble learning method that combines the results of several weak learning algorithms to generate a more consistent joint response [28]. It iteratively adjusts the weights of misclassified instances to improve the classification performance, focusing especially on difficult instances. Initially, an adjustment takes place, where individuals who did not perform well in the current iteration have greater weight in the subsequent iteration. The classification error rate serves as a measure to evaluate whether this adjustment in weighting improves or worsens the classification. This method tries to enhance the classification performance, particularly for the most challenging individuals to categorize them correctly [29].
- Support vector machines (SVM): This classifier is based on finding an optimal hyperplane that maximally separates the response variable into two classes [30]. The categorization process of the response variable in this classifier utilizes the information from the matrix of explanatory variables to identify an optimal hyperplane. This hyperplane aims to achieve a maximum margin that separates the response into two classes, resulting in an improved classification performance [31]. It is also possible to perform a transformation in the original input space (explanatory matrix). While the maximum margin that distinguishes the classes may be linear in the transformed plane, it can exhibit non-linearity in the original space [32].
3.4. Performance Measures
- Variance inflation factor (VIF): This criterion uses the LR model and selects variables using the VIF [36]. The selection obtains a set of variables without collinearity (a strong correlation between two explanatory variables). VIF values greater than 10 indicate strong multicollinearity [37], which affects the estimates of the model [38]. Therefore, variables with a VIF are sequentially removed from the LR with all variables.
- Analysis of variance (ANOVA): This criterion employs ANOVA [39]. Variables that demonstrate statistical significance in ANOVA are chosen, indicating their influence on the response variable.
- ANOVA + VIF: This criterion utilizes ANOVA, followed by an analysis of the VIF. Variables with a VIF greater than 10 are removed from the model.
- Akaike information criterion (AIC): This criterion selects the best model that minimizes its value [40]. The AIC utilizes the model’s likelihood function and the number of explanatory variables in its calculation.
- AIC + VIF: This criterion selects the model by minimizing the AIC, followed by selection based on the VIF (the removal of variables with a VIF greater than 10).
- Average sensitivity (ASe): This is the average percentage of TPs, representing the cardiac patients who were correctly classified within the group of people with heart disease in each iteration of the multiple holdout. This measure is given by
- Average specificity (ASp): This is the average percentage of TNs, representing the non-cardiac patients correctly classified within the group of people without heart disease in each iteration of the multiple holdout. This measure is calculated as
- Average true positive predictive (ATPP): This is the average percentage of true positives in relation to all positive predictions, representing the cardiac patients who were correctly classified within the group of people who were estimated to have heart disease in each iteration of the multiple holdout. This measure is expressed as
3.5. Computational Environment and Conditions
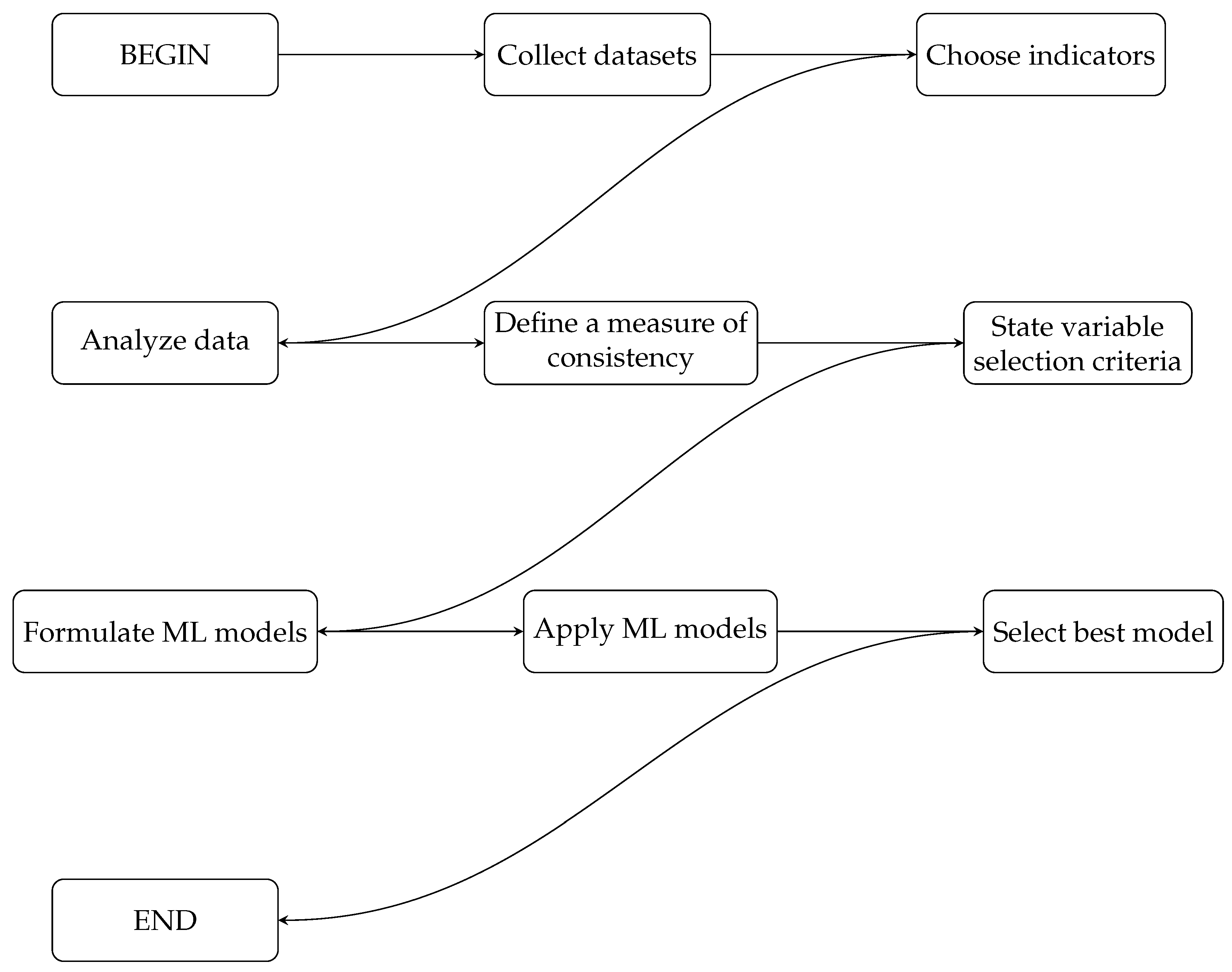
3.6. Summary of the Methodology
| Algorithm 1 Summary of the methodology using ML techniques to predict cardiac patients. |
|
4. Results and Discussions
4.1. Datasets
4.2. CS Indicators in the Datasets
4.3. CS Indicators in the Context of Selected Models
5. Conclusions, Limitations, and Future Research
5.1. Concluding Remarks
5.2. Limitations and Future Work
- Simplicity as a strength: A unique selling point of our approach is its resource efficiency, requiring only a tensiometer and a clock for data collection. This is not a limitation, but rather a strategic advantage, especially in resource-constrained environments where quick, yet effective, screenings are essential. In these contexts, the accessibility and ease of use of our model may outweigh the benefits of more complex and resource-intensive methods, offering a viable and immediate solution for diagnosing ischemic heart diseases. That said, we do acknowledge that the model’s simplicity may have boundaries when considering broader applicability. Nonetheless, the focus of this research is on maximizing diagnostic efficacy with minimal resources. Future work will aim to examine the impact of adding more variables and complexity, but the core benefit of our approach lies in its minimal resource demands.
- Hemodynamic indicators: The current study primarily employs hemodynamic indicators as described by Campello de Souza [4]. While effective in the context of ischemic heart diseases, these indicators may also be relevant in diagnosing other cardiovascular conditions, such as heart failure or valvular diseases. Future studies could extend the scope to evaluate such indicators in a wider range of cardiac conditions.
- Disease specificity: Our focus in this study has been primarily on coronary artery disease, which falls under the broader umbrella of ischemic heart disease. While our models have demonstrated effectiveness in this specific context, it is essential to note that heart disease is a broad category that includes various conditions such as heart failure, valvular diseases, and arrhythmias. Future research should explore the applicability of our machine learning models to these other types of heart disease, thereby enriching the diagnostic toolkit available to healthcare professionals.
- Model validation: Our approach already provides a robust baseline due to its simplicity and the diverse set of classifiers that we tested. Nevertheless, future work should engage in more rigorous validation techniques to confirm the generalizability of our models and to mitigate risks such as overfitting.
- Model performance heterogeneity: The variation in model performance across datasets underscores the model’s limitations but also suggests a path for future personalized medicine approaches. We see this not as a limitation but as an opportunity for tailored applications. In particular, the observed heterogeneity in model performance across different datasets raises critical questions about the need for population-specific models in the realm of personalized medicine. We recognize that different models may be more suitable for different populations, and understanding this interaction could lead to a more nuanced, individualized approach to diagnosing ischemic heart diseases. Future research should delve deeper into how our model can be fine-tuned to serve diverse populations effectively.
- Prospective and comparative studies: Our study sets a precedent for resource-efficient diagnostics, but it would benefit from prospective studies comparing its efficacy to that of more resource-intensive methods. This will help confirm its value as a standalone diagnostic tool.
- Future methodologies: To advance in this area, further research employing diverse and larger datasets is required. Studies involving more diverse sample sizes could offer additional insights into the generalizability and reliability of our models.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cofiño-Fabres, C.; Passier, R.; Schwach, V. Towards improved human in vitro models for cardiac arrhythmia: Disease mechanisms, treatment, and models of atrial fibrillation. Biomedicines 2023, 11, 2355. [Google Scholar] [CrossRef]
- World Health Organization. The Top 10 Causes of Death; Technical Report; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Carvalho, A.C.C.; Sousa, J.M.A. Ischemic heart disease. Rev. Bras. Hipertens. 2001, 8, 297–305. (In Portuguese) [Google Scholar]
- de Souza, F.M.C. Support for Medical Diagnosis: What Can Be Done with a Blood Pressure Monitor and a Watch; Vade Mecum: Recife, Brazil, 2010. (In Portuguese) [Google Scholar]
- Cingolani, H.E.; Perez, N.G.; Cingolani, O.H.; Ennis, I.L. The importance of mean arterial pressure in cardiovascular physiology. J. Hypertens. 2013, 31, 16–25. [Google Scholar]
- Hoppensteadt, F.C.; Peskin, C.S. Mathematics in Medicine and the Life Sciences; Springer: New York, NY, USA, 2013. [Google Scholar]
- McQueen, D.M.; Peskin, C.S. Heart simulation by an immersed boundary method with formal second-order accuracy and reduced numerical viscosity. In Mechanics for a New Millennium; Aref, H., Phillips, J.W., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 429–444. [Google Scholar]
- Franz, M.; Chin, M.; Wang, D.; Stern, R.; Scheinman, M.M. Monitoring of radiofrequency ablation effect by simultaneous monophasic action potential recording. Pacing Clin. Electrophysiol. 1991, 14, 703. [Google Scholar]
- Rego, L.; Campello de Souza, F. Improved estimation of left ventricular hypertrophy. IEEE Eng. Med. Biol. Mag. 2002, 21, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Maximiano, J. A chronobiological look at the heart and depression. Psilogos 2008, 5, 54–62. (In Portuguese) [Google Scholar]
- Mohrman, D.E.; Heller, L.J. Cardiovascular Physiology; McGraw-Hill: New York, NY, USA, 2018. [Google Scholar]
- Jan, M.Y.; Hsiu, H.; Hsu, T.L.; Wang, Y.Y.L.; Wang, W.K. The importance of pulsatile microcirculation in relation to hypertension. IEEE Eng. Med. Biol. Mag. 2000, 19, 106–111. [Google Scholar]
- Franklin, S.S.; Gustin, W.; Wong, N.D.; Larson, M.G.; Weber, M.A.; Kannel, W.B.; Levy, D. Hemodynamic patterns of age-related changes in blood pressure: The Framingham heart study. Circulation 1997, 96, 308–315. [Google Scholar] [CrossRef]
- Ronan, C.A. (Ed.) Illustrated History of Science in the University of Cambridge; Jorge Zahar Editorial: Rio de Janeiro, Brazil, 1994. [Google Scholar]
- Leite, W.A.; Sa, D. (Eds.) Cardiovascular; Jorge Zahar Editorial: Rio de Janeiro, Brazil, 2005. [Google Scholar]
- Tortora, G.J.; Derrickson, B. Principles of Anatomy and Physiology; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Lee, L.; Berger, T.; Aviczer, E. Reliable online human signature verification systems. IEEE Trans. Pattern Anal. Mach. Intell. 1996, 18, 643–647. [Google Scholar] [CrossRef]
- Antal, M.; Szabó, L.Z. Some remarks on a set of information theory features used for on-line signature verification. In Proceedings of the 5th International Symposium on Digital Forensic and Security, Tirgu Mures, Romania, 26–28 April 2017; pp. 1–5. [Google Scholar]
- Dos Santos Machado, L.; da Mota Resende Machado, D. Assessment systems for training based on virtual reality: A comparison study. J. Interact. Syst. 2012, 3, 9–17. [Google Scholar]
- D’Agostini, G. A multidimensional unfolding method based on Bayes’ theorem. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 1995, 362, 487–498. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. The Elements of Statistical Learning; Springer: New York, NY, USA, 2009. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Davison, A.C.; Hinkley, D.V. Bootstrap Methods and Their Application; Cambridge University Press: Cambridge, MA, USA, 1997. [Google Scholar]
- Shi, M.; Rendell, M.M. Modelling mortality of a stored grain insect pest with fumigation: Probit, logistic or cauchy model? Math. Biosci. 2013, 243, 137–146. [Google Scholar] [CrossRef]
- de Oliveira, J.S.C.; Ospina, R.; Leiva, V.; Figueroa-Zúñiga, J.; Castro, C. Quasi-Cauchy regression modeling for fractiles based on data supported in the unit interval. Fractal Fract. 2023, 7, 667. [Google Scholar] [CrossRef]
- Jiang, J. Linear and Generalized Linear Mixed Models and Their Applications; Springer: New York, NY, USA, 2006. [Google Scholar]
- Lindsey, J.K. Applying Generalized Linear Models; Springer: New York, NY, USA, 2000. [Google Scholar]
- Freund, Y.; Schapire, R.E. Experiments with a new boosting algorithm. In Proceedings of the Thirteenth International Conference on International Conference on Machine Learning, San Francisco, CA, USA, 3–6 July 1996; pp. 148–156. [Google Scholar]
- Freund, Y.; Schapire, R.E. A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 1999, 14, 1612. [Google Scholar]
- de Carvalho, J.B.; Silva, M.C.; von Borries, G.F.; de Pinho, A.L.S.; von Borries, R.F. A combined Fourier analysis and support vector machine for EEG classification. Chil. J. Stat. 2019, 10, 3–20. [Google Scholar]
- Hussain, M.; Wajid, S.K.; Elzaart, A.; Berbar, M. A comparison of SVM kernel functions for breast cancer detection. In Proceedings of the Eighth International Conference Computer Graphics, Imaging and Visualization, Singapore, 17–19 August 2011; pp. 145–150. [Google Scholar]
- Shawe-Taylor, J.; Cristianini, N. Kernel Methods for Pattern Analysis; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. ACM Sigmobile Mob. Comput. Commun. Rev. 2001, 5, 3–55. [Google Scholar] [CrossRef]
- de Oliveira, H.M.; Ospina, R. Leiva, V.; Martin-Barreiro, C.; Chesneau, C. On the use of variability measures to analyze source coding data based on the Shannon entropy. Mathematics 2023, 11, 293. [Google Scholar] [CrossRef]
- Borland, L.; Plastino, A.R.; Tsallis, C. Information gain within nonextensive thermostatistics. J. Math. Phys. 1998, 39, 6490–6501. [Google Scholar] [CrossRef]
- Mason, O.R. A caution regarding rules of thumb for variance inflation factors. Qual. Quant. 2007, 41, 673–690. [Google Scholar]
- Menard, S. Applied Logistic Regression Analysis; Sage University Series: New York, NY, USA, 1995. [Google Scholar]
- Neter, J.; Kutner, M.H.; Nachtsheim, C.J.; Wasserman, W. Applied Linear Statistical Models; McGraw-Hill: New York, NY, USA, 1996. [Google Scholar]
- Hair, J.F.; Black, W.C.; Babin, B.J.; Anderson, R.E.; Tatham, R.L. Multivariate Data Analysis; Pearson Education: Essex, UK, 2014. [Google Scholar]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef]
- Glaros, A.G.; Kline, R.B. Understanding the accuracy of tests with cutting scores: The sensitivity, specificity, and predictive value model. J. Clin. Psychol. 1988, 44, 1013–1023. [Google Scholar] [CrossRef]
- Tu, M.C.; Shin, D.; Shin, D. Effective diagnosis of heart disease through bagging approach. In Proceedings of the 2nd International Conference on Biomedical Engineering and Informatics, Tianjin, China, 17–19 October 2009; pp. 1–4. [Google Scholar]
- Kahramanli, H.; Allahverdi, N. Design of a hybrid system for the diabetes and heart diseases. Expert Syst. Appl. 2008, 35, 82–89. [Google Scholar] [CrossRef]
- Baldi, P.; Brunak, S.; Chauvin, Y.; Andersen, C.A.F.; Nielsen, H. Assessing the accuracy of prediction algorithms for classification: An overview. Bioinformatics 2000, 16, 412–424. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: www.r-project.org (accessed on 13 September 2023).
- Korosteleva, O. Advanced Regression Models with SAS and R; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Etaati, L. Machine Learning with Microsoft Technologies; Springer: New York, NY, USA, 2019. [Google Scholar]
- Taylan, O.; Alkabaa, A.S.; Alqabbaa, H.S.; Pamukçu, E.; Leiva, V. Early prediction in classification of cardiovascular diseases with machine learning, neuro-fuzzy and statistical methods. Biology 2023, 12, 117. [Google Scholar] [CrossRef]
- Detrano, R.; Janosi, A.; Steinbrunn, W.; Pfisterer, M.; Schmid, J.J.; Sandhu, S.; Guppy, K.H.; Lee, S.; Froelicher, V. International application of a new probability algorithm for the diagnosis of coronary artery disease. Am. J. Cardiol. 1989, 64, 304–310. [Google Scholar] [CrossRef]
- Dua, D.; Graff, C. UCI Machine Learning Repository. 2017. Available online: http://archive.ics.uci.edu/ml (accessed on 13 September 2023).
- Veerabaku, M.G.; Nithiyanantham, J.; Urooj, S.; Md, A.Q.; Sivaraman, A.K.; Tee, K.F. Intelligent Bi-LSTM with architecture optimization for heart disease prediction in WBAN through optimal channel selection and feature selection. Biomedicines 2023, 11, 1167. [Google Scholar] [CrossRef]
- Cavalcante, T.; Ospina, R.; Leiva, V.; Cabezas, X.; Martin-Barreiro, C. Weibull regression and machine learning survival models: Methodology, comparison, and application to biomedical data related to cardiac surgery. Biology 2023, 12, 442. [Google Scholar] [CrossRef] [PubMed]
- Alkadya, W.; ElBahnasy, K.; Leiva, V.; Gad, W. Classifying COVID-19 based on amino acids encoding with machine learning algorithms. Chemom. Intell. Lab. Syst. 2022, 224, 104535. [Google Scholar] [CrossRef]
- Feng, H.; Wang, F.; Li, N.; Xu, Q.; Zheng, G.; Sun, X.; Zhang, G. A random forest model for peptide classification based on virtual docking data. Int. J. Mol. Sci. 2023, 24, 11409. [Google Scholar] [CrossRef]
- Sardar, I.; Akbar, M.A.; Leiva, V.; Alsanad, A.; Mishra, P. Machine learning and automatic ARIMA/Prophet models-based forecasting of COVID-19: Methodology, evaluation, and case study in SAARC countries. Stoch. Environ. Res. Risk Assess. 2023, 37, 345–359. [Google Scholar] [CrossRef]
- Leiva, V.; Alcudia, E.; Montano, A.; Castro, C. An epidemiological analysis for assessing and evaluating COVID-19 based on data analytics in Latin American countries. Biology 2023, 12, 887. [Google Scholar] [CrossRef]
- Martin-Barreiro, C.; Cabezas, X.; Leiva, V.; Ramos de Santis, P.; Ramirez-Figueroa, J.A.; Delgado, E. Statistical characterization of vaccinated cases and deaths due to COVID-19: Methodology and case study in South America. AIMS Math. 2023, 8, 22693–22713. [Google Scholar] [CrossRef]
- Ospina, R.; Gondim, J.A.M.; Leiva, V.; Castro, C. An overview of forecast analysis with ARIMA models during the COVID-19 pandemic: Methodology and case study in Brazil. Mathematics 2023, 11, 3069. [Google Scholar] [CrossRef]
- Rahman, M.Z.U.; Akbar, M.A.; Leiva, V.; Tahir, A.; Riaz, M.T.; Martin-Barreiro, C. An intelligent health monitoring and diagnosis system based on the internet of things and fuzzy logic for cardiac arrhythmia COVID-19 patients. Comput. Biol. Med. 2023, 154, 106583. [Google Scholar] [CrossRef]
- Jerez-Lillo, N.; Lagos Alvarez, B.; Munoz Gutierrez, J.; Figueroa-Zúñiga, J.; Leiva, V. A statistical analysis for the epidemiological surveillance of COVID-19 in Chile. Signa Vitae 2022, 18, 19–30. [Google Scholar]
- Ospina, R.; Leite, A.; Ferraz, C.; Magalhaes, A.; Leiva, V. Data-driven tools for assessing and combating COVID-19 out-breaks based on analytics and statistical methods in Brazil. Signa Vitae 2022, 18, 18–32. [Google Scholar]
- Dominic, V.; Gupta, D.; Khare, S. An effective performance analysis of machine learning techniques for cardiovascular disease. Appl. Med. Inform. 2015, 36, 23–32. [Google Scholar]
- Bustos, N.; Tello, M.; Droppelmann, G.; Garcia, N.; Feijoo, F.; Leiva, V. Machine learning techniques as an efficient alternative diagnostic tool for COVID-19 cases. Signa Vitae 2022, 18, 23–33. [Google Scholar]
- Cox, D.R.; Hinkley, D.V. Theoretical Statistics; CRC Press: Boca-Raton, FL, USA, 1979. [Google Scholar]
- Das, R.; Turkoglu, I.; Sengur, A. Effective diagnosis of heart disease through neural networks ensembles. Expert Syst. Appl. 2009, 36, 7675–7680. [Google Scholar] [CrossRef]
- Sayad, A.; Harb, H. Diagnosis of heart disease using neural network approach. Int. J. Adv. Sci. Eng. Technol. 2014, 2, 88–92. [Google Scholar]
- Johnson, N.L.; Kotz, S.; Balakrishnan, N. Continuous Univariate Distributions; Wiley: New York, NY, USA, 1994; Volume 1. [Google Scholar]
- Kotz, S.; Leiva, V.; Sanhueza, A. Two new mixture models related to the inverse Gaussian distribution. Methodol. Comput. Appl. Probab. 2010, 12, 199–212. [Google Scholar] [CrossRef]
- Johnson, N.L.; Kotz, S.; Balakrishnan, N. Continuous Univariate Distributions; Wiley: New York, NY, USA, 1995; Volume 2. [Google Scholar]
| Observed Value: Y | Estimated Value: | |
|---|---|---|
| 0 | 1 | |
| 0 | TN | FP |
| 1 | FN | TP |
| Method | Package | Function | Argument |
|---|---|---|---|
| NB | e1071 | naiveBayes | laplace = 0, na.action = na.pass |
| RF | randomForest | randomForest | ntree = 500, na.action = na.omit |
| SVM | e1071 | svm | scale = F, kernel = “poly”, cost = 100, epsilon = 1.0 × , na.action = na.omit |
| LR | stats | glm | family = binomial (link = “logit”), na.action = na.omit |
| Adaboost | fastAdaboost | adaboost | nIter = 10 |
| Dataset | n | Y | |
|---|---|---|---|
| 0 | 1 | ||
| Cleveland | 282 | 157 | 125 |
| Hungarian | 294 | 188 | 106 |
| Long Beach | 200 | 51 | 149 |
| Switzerland | 123 | 8 | 115 |
| Indicator | p-Value | ||||||
|---|---|---|---|---|---|---|---|
| 0.0038 | 0.0040 | 0.0029 | 0.0033 | 0.0029 | 0.0033 | 0.4837 | |
| 5.8280 | 5.7834 | 5.8284 | 5.7121 | 0.7035 | 0.7473 | 0.4837 | |
| HM | 88.2808 | 79.0296 | 51.6722 | 42.4812 | 118.2997 | 94.5672 | 0.3004 |
| MAP | 105.1306 | 108.0998 | 105.9115 | 107.8869 | 11.6754 | 11.6864 | |
| PBPI | 0.5439 | 0.5692 | 0.5294 | 0.5556 | 0.1824 | 0.1760 | 0.2434 |
| PBPIRC | 19.3680 | 19.8608 | 15.7457 | 17.3187 | 12.5346 | 11.7759 | 0.5273 |
| RC | 0.0339 | 0.0340 | 0.0329 | 0.0325 | 0.0115 | 0.0113 | 0.9683 |
| Indicator | p-Value | ||||||
|---|---|---|---|---|---|---|---|
| 0.0048 | 0.0047 | 0.0039 | 0.0038 | 0.0043 | 0.0035 | 0.7747 | |
| 5.5739 | 5.5588 | 5.5552 | 5.5628 | 0.6453 | 0.6335 | 0.7747 | |
| HM | 53.8264 | 55.4877 | 38.9873 | 37.2640 | 51.6480 | 77.3438 | 0.2751 |
| MAP | 105.4446 | 108.6070 | 102.9850 | 108.0512 | 11.4091 | 12.3450 | |
| PBPI | 0.5639 | 0.5971 | 0.5500 | 0.5556 | 0.1502 | 0.2195 | 0.1196 |
| PBPIRC | 21.7992 | 22.9755 | 18.6552 | 19.2456 | 13.1868 | 18.9023 | 0.6145 |
| RC | 0.0295 | 0.0300 | 0.0280 | 0.0291 | 0.0081 | 0.0080 | 0.5268 |
| Indicator | p-Value | ||||||
|---|---|---|---|---|---|---|---|
| 0.0038 | 0.0044 | 0.0029 | 0.0039 | 0.0030 | 0.0028 | 0.1033 | |
| 5.8083 | 5.8785 | 5.8440 | 5.5574 | 0.6754 | 3.0681 | 0.1033 | |
| HM | 67.7813 | 47.4181 | 44.0492 | 34.3783 | 69.1907 | 49.5149 | |
| MAP | 102.7137 | 106.1425 | 99.0182 | 104.4601 | 13.1111 | 11.7342 | 0.1067 |
| PBPI | 0.6404 | 0.6842 | 0.6085 | 0.6500 | 0.2085 | 0.2005 | 0.2863 |
| PBPIRC | 23.5717 | 25.9667 | 20.7905 | 21.8479 | 15.9530 | 13.7833 | 0.2079 |
| RC | 0.0327 | 0.0299 | 0.0307 | 0.0288 | 0.0106 | 0.0083 | 0.1720 |
| Indicator | p-Value | ||||||
|---|---|---|---|---|---|---|---|
| 0.0039 | 0.0038 | 0.0042 | 0.0026 | 0.0031 | 0.0036 | 0.9293 | |
| 5.9888 | 5.9288 | 5.4949 | 5.9362 | 1.1305 | 0.8388 | 0.9293 | |
| HM | 197.6817 | 112.8445 | 33.3719 | 56.5868 | 285.4129 | 166.4005 | 0.9293 |
| MAP | 98.9302 | 104.4039 | 102.2406 | 102.0779 | 16.6840 | 14.5477 | 0.5347 |
| PBPI | 0.5972 | 0.5976 | 0.6587 | 0.5714 | 0.2409 | 0.2411 | 0.7942 |
| PBPIRC | 22.1667 | 21.8520 | 26.9187 | 15.5101 | 13.9135 | 19.4161 | 0.6276 |
| RC | 0.0355 | 0.0358 | 0.0247 | 0.0341 | 0.0161 | 0.0134 | 0.5839 |
| Dataset | HM | MAP | PBPI | PBPIRC | RC | ||
|---|---|---|---|---|---|---|---|
| Cleveland | 0.0648 | 0.0435 | 0.0611 | 0.1797 | 0.0996 | 0.0286 | 0.0083 |
| Hungarian | 0.0091 | 0.0167 | 0.0179 | 0.1881 | 0.1249 | 0.0510 | 0.0473 |
| Long Beach | 0.1522 | 0.0223 | 0.2393 | 0.1949 | 0.1514 | 0.1136 | 0.2037 |
| Switzerland | 0.0204 | 0.0426 | 0.2568 | 0.2473 | 0.0011 | 0.0132 | 0.0163 |
| MAP | PBPI | RC | PBPIRC | HM | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | % | % | % | % | % | % | ||||||||
| 22 | 52.38 | 13 | 30.95 | 11 | 26.19 | 9 | 21.43 | 9 | 21.43 | 9 | 21.43 | 8 | 19.05 | 42 |
| Model | Variables |
|---|---|
| Cleveland | |
| A | V4, V9, V11, V16, V18, V23, V24, V25, V26, V27, V29, V30, V31, V32, V38, V39, V40, V41, V44, V51, V60, V61, V63, V65, V67, V68, V72 |
| B | V3, V4, V9, V11, V12, V14, V15, V16, V18, V19, V23, V24, V25, V26, V27, V29, V31, V32, V33, V34, V35, V38, V40, V41, V43, V44, V51, V59, V60, V61, V63, V65, V67, V68, V71, V72, V73, MAP, PBPI, HM, |
| C | V3, V4, V9, V10, V23, V24, V32, V34, V38, V40, V44, V51, V60, V61 |
| D | V4, V10, V34, V40, V44, V51 |
| E | V3, V4, V10, V15, V16, V18, V19, V23, V25, V27, V29, V31, V33, V37, V38, V40, V43, V44, V51, V59, V60, V61, V63, V65, V67, V68, V71, V72, V73, MAP, PBPI, RC, |
| F | V3, V4, V15, V16, V18, V19, V23, V25, V27, V29, V38, V40, V43, V44, V51, V60, V61, V63, V65, V67, V68, V71, V72, V73, MAP, PBPI, RC |
| Hungarian | |
| A | V4, V5, V6, V7, V9, V11, V16, V24, V25, V26, V27, V32, V38, V39, V40, V41, V72, V73 |
| B | V3, V4, V5, V6, V11, V12, V16, V19, V24, V25, V27, V32, V35, V38, V40, V41, V43, V72 V73, MAP, PBPI, HM |
| C | V4, V6, V11, V28, V29 |
| D | V4, V6, V11, V28 |
| E | V3, V4, V5, V6, V7, V9, V10, V11, V12, V16, V19, V24, V25, V26, V27, V28, V29, V34, V30, V31, V32, V33, V35, V37, V38, V40, V41, V42, V43, V72, V73, MAP, PBPI, RC, PBPIRC, HM, , |
| F | V4, V5, V6, V12, V16, V19, V24, V27, V31, V32, V34, V35, V38, V40, V41, V42, V72, V73, MAP, RC |
| Long Beach | |
| A | V4, V5, V6, V7, V9, V11, V13, V16, V18, V23, V24, V25, V26, V27, V38, V39, V41, V60, V61, V63, V65, V67, V75 |
| B | V4, V5, V6, V7, V11, V12, V14, V15, V19, V28, V59, V60, V62, V63, V64, V65, V68, V70, V71, |
| C | V4, V6, V43, V60, V61 |
| D | V3, V4, V5, V6, V7, V10, V11, V12, V13, V14, V15, V16, V18, V19, V28, V29, V31, V32, V33, V37, V38, V39, V40, V42, V43, V59, V60,V61, V62, V63, V65, V66, V67, V68, V70, V71, V72, V73, V74, MAP, PBPI, RC, PBPIRC, HM, , |
| E | V4, V5, V6, V7, V11, V14, V18, V28, V33, V42, V59, V61, V63, V65, V66, V67, V71, V73, MAP, PBPI |
| Switzerland | |
| A | V4, V5, V6, V7, V9, V11, V24, V25, V26, V27, V38, V39, V41 |
| B | V4, V7, V25, V27, V33, V38, V39, V40, V59, V61, V62, V65, V67, MAP |
| C | V7, V61, V67, MAP |
| D | V4, V6, V7, V19, V24, V25, V27, V32, V33, V36, V38, V39, V40, V60, V61, V62, V64, MAP, PBPI, RC, PBPIRC, HM |
| E | V4, V7, V19, V27, V33, V38, V39, V40, V61, V64, MAP, HM |
| Dataset | Model | # Features | Classifier | Accuracy | ASe | ASp | ATPP | Indicator |
|---|---|---|---|---|---|---|---|---|
| Cleveland | B | 41 | Adaboost | 98.58 (1.80) | 96.82 (3.93) | 99.98 (0.20) | 99.97 (0.32) | , HM, MAP, PBPI |
| D | 6 | LR | 81.32 (3.83) | 75.76 (7.15) | 85.94 (4.53) | 81.00 (6.40) | - | |
| F | 27 | LR | 99.20 (1.17) | 98.23 (2.46) | 100.00 (0.00) | 100.00 (0.00) | MAP, PBPI, RC | |
| Hungarian | B | 20 | NB | 83.10 (4.25) | 64.71 (10.51) | 93.17 (3.95) | 84.39 (7.72) | HM, MAP, PBPI |
| D | 4 | LR | 80.84 (3.52) | 74.60 (7.16) | 84.31 (4.83) | 72.15 (7.42) | - | |
| F | 18 | NB | 83.56 (4.03) | 64.16 (10.09) | 94.17 (3.52) | 86.20 (6.75) | MAP, RC | |
| Long Beach | B | 19 | Adaboost | 79.52 (4.90) | 88.00 (4.83) | 55.05 (14.21) | 85.33 (5.30) | |
| C | 5 | Adaboost | 78.77 (4.54) | 88.65 (5.60) | 50.55 (16.23) | 84.19 (5.76) | - | |
| E | 19 | LR | 85.47 (6.14) | 88.78 (6.16) | 74.23 (16.85) | 92.22 (5.13) | MAP, PBPI | |
| Switzerland | B | 14 | RF | 93.12 (3.06) | 99.33 (1.35) | 0.00 (0.00) | 93.38 (3.03) | MAP |
| C | 4 | Adaboost | 92.52 (3.25) | 98.44 (2.18) | 5.96 (16.78) | 93.74 (3.32) | MAP | |
| E | 12 | RF | 93.51 (3.25) | 99.74 (0.86) | 0.00 (0.00) | 93.33 (3.08) | HM, MAP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ospina, R.; Ferreira, A.G.O.; de Oliveira, H.M.; Leiva, V.; Castro, C. On the Use of Machine Learning Techniques and Non-Invasive Indicators for Classifying and Predicting Cardiac Disorders. Biomedicines 2023, 11, 2604. https://doi.org/10.3390/biomedicines11102604
Ospina R, Ferreira AGO, de Oliveira HM, Leiva V, Castro C. On the Use of Machine Learning Techniques and Non-Invasive Indicators for Classifying and Predicting Cardiac Disorders. Biomedicines. 2023; 11(10):2604. https://doi.org/10.3390/biomedicines11102604
Chicago/Turabian StyleOspina, Raydonal, Adenice G. O. Ferreira, Hélio M. de Oliveira, Víctor Leiva, and Cecilia Castro. 2023. "On the Use of Machine Learning Techniques and Non-Invasive Indicators for Classifying and Predicting Cardiac Disorders" Biomedicines 11, no. 10: 2604. https://doi.org/10.3390/biomedicines11102604
APA StyleOspina, R., Ferreira, A. G. O., de Oliveira, H. M., Leiva, V., & Castro, C. (2023). On the Use of Machine Learning Techniques and Non-Invasive Indicators for Classifying and Predicting Cardiac Disorders. Biomedicines, 11(10), 2604. https://doi.org/10.3390/biomedicines11102604












