mRNA Metabolism and Hypertension
Abstract
1. Introduction
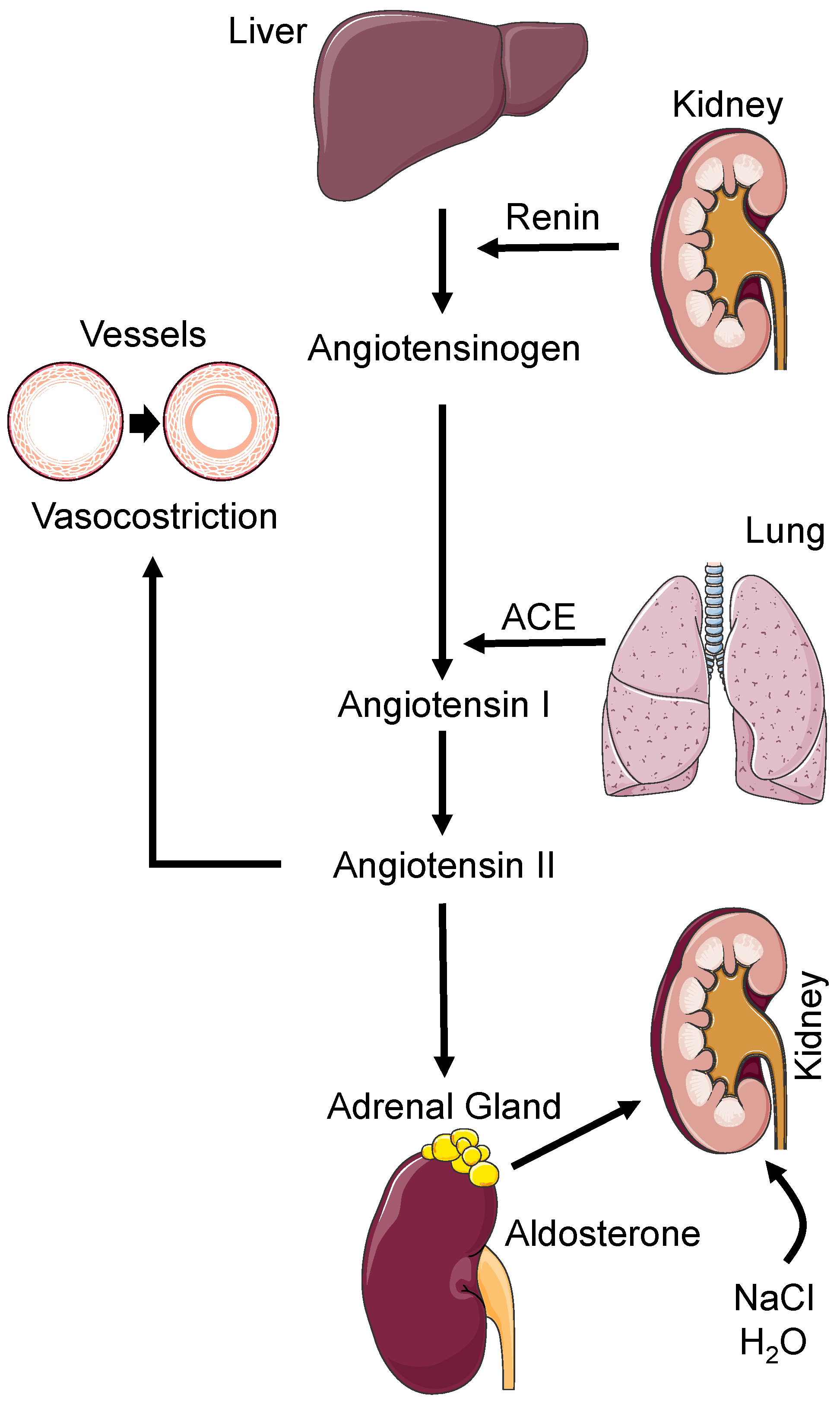
2. Pathophysiology of Hypertension
3. Genetics of Hypertension
4. The Role of RNA
5. mRNA Metabolism
| Author | Modification | Function |
|---|---|---|
| Marcadenti, A et al. [54] | FTO gene rs9939609 and MC4R gene rs17782313 | m6 A-SNP loci are associated with hypertension |
| Bengtsson, K. et al. [57] | Arg389 variant of the beta(1)-adrenergic receptor gene | Homozygous for the Arg389 allele of the beta(1)-adrenergic receptor gene is at increased risk to develop hypertension |
| Lombari, P. et al. [64] | miR-23a | Expression is downregulated in the mTAL of HSD rats whereas NHE1 is upregulated |
| Fang, Z. et al. [68] | Arntl rs6486121 | mRNA expression of Arntl was downregulated in hypertension cases compared with controls in women |
| Liu, C. et al. [71] | FLNA rs2070816 (CT + TT vs. CC) and rs2070829 (CG + GG vs. CC) | They were significantly associated with hypertension in <55 years group |
| FLNB rs839240 (AG + GG vs. AA) | It was significantly associated with hypertension in females |
6. Clinical Trials
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Angeli, F.; Reboldi, G.; Verdecchia, P. Hypertension, inflammation and atrial fibrillation. J. Hypertens. 2014, 32, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Mills, K.T.; Bundy, J.D.; Kelly, T.N.; Reed, J.E.; Kearney, P.M.; Reynolds, K.; Chen, J.; He, J. Global Disparities of Hypertension Prevalence and Control: A Systematic Analysis of Population-Based Studies From 90 Countries. Circulation 2016, 134, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Reboldi, G.; Angeli, F.; de Simone, G.; Staessen, J.A.; Verdecchia, P.; Cardio-Sis, I. Tight versus standard blood pressure control in patients with hypertension with and without cardiovascular disease. Hypertension 2014, 63, 475–482. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Verdecchia, P.; Reboldi, G.; Angeli, F.; Trimarco, B.; Mancia, G.; Pogue, J.; Gao, P.; Sleight, P.; Teo, K.; Yusuf, S. Systolic and diastolic blood pressure changes in relation with myocardial infarction and stroke in patients with coronary artery disease. Hypertension 2015, 65, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Verdecchia, P.; Reboldi, G.; Angeli, F. The 2020 International Society of Hypertension global hypertension practice guidelines—Key messages and clinical considerations. Eur. J. Intern. Med. 2020, 82, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Worldwide trends in blood pressure from 1975 to 2015: A pooled analysis of 1479 population-based measurement studies with 19·1 million participants. Lancet 2017, 389, 37–55. [CrossRef]
- Lim, S.S.; Vos, T.; Flaxman, A.D.; Danaei, G.; Shibuya, K.; Adair-Rohani, H.; Amann, M.; Anderson, H.R.; Andrews, K.G.; Aryee, M.; et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2224–2260. [Google Scholar] [CrossRef]
- Lloyd-Jones, D.; Adams, R.; Carnethon, M.; De Simone, G.; Ferguson, T.B.; Flegal, K.; Ford, E.; Furie, K.; Go, A.; Greenlund, K.; et al. Heart disease and stroke statistics—2009 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2009, 119, e21–e181. [Google Scholar] [CrossRef]
- DeGoma, E.M.; Knowles, J.W.; Angeli, F.; Budoff, M.J.; Rader, D.J. The evolution and refinement of traditional risk factors for cardiovascular disease. Cardiol. Rev. 2012, 20, 118–129. [Google Scholar] [CrossRef]
- Forouzanfar, M.H.; Liu, P.; Roth, G.A.; Ng, M.; Biryukov, S.; Marczak, L.; Alexander, L.; Estep, K.; Hassen Abate, K.; Akinyemiju, T.F.; et al. Global Burden of Hypertension and Systolic Blood Pressure of at Least 110 to 115 mm Hg, 1990–2015. JAMA 2017, 317, 165–182. [Google Scholar] [CrossRef]
- Verdecchia, P.; Angeli, F.; Mazzotta, G.; Garofoli, M.; Reboldi, G. Aggressive blood pressure lowering is dangerous: The J-curve: Con side of the arguement. Hypertension 2014, 63, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Angeli, F.; Verdecchia, P.; Gattobigio, R.; Sardone, M.; Reboldi, G. White-coat hypertension in adults. Blood Press. Monit. 2005, 10, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Vasan, R.S. Epidemiology of uncontrolled hypertension in the United States. Circulation 2005, 112, 1651–1662. [Google Scholar] [CrossRef] [PubMed]
- Verdecchia, P.; Cavallini, C.; Angeli, F. Advances in the Treatment Strategies in Hypertension: Present and Future. J. Cardiovasc. Dev. Dis. 2022, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Saxena, T.; Ali, A.O.; Saxena, M. Pathophysiology of essential hypertension: An update. Expert Rev. Cardiovasc. Ther. 2018, 16, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Di Giosia, P.; Giorgini, P.; Stamerra, C.A.; Petrarca, M.; Ferri, C.; Sahebkar, A. Gender Differences in Epidemiology, Pathophysiology, and Treatment of Hypertension. Curr. Atheroscler. Rep. 2018, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Rauf, A.; Khan, H.; Abu-Izneid, T. Renin-angiotensin-aldosterone (RAAS): The ubiquitous system for homeostasis and pathologies. Biomed. Pharmacother. 2017, 94, 317–325. [Google Scholar] [CrossRef]
- Verano-Braga, T.; Martins, A.L.V.; Motta-Santos, D.; Campagnole-Santos, M.J.; Santos, R.A.S. ACE2 in the renin-angiotensin system. Clin. Sci. 2020, 134, 3063–3078. [Google Scholar] [CrossRef]
- Fournier, D.; Luft, F.C.; Bader, M.; Ganten, D.; Andrade-Navarro, M.A. Emergence and evolution of the renin-angiotensin-aldosterone system. J. Mol. Med. 2012, 90, 495–508. [Google Scholar] [CrossRef]
- Angeli, F.; Verdecchia, P.; Balestrino, A.; Bruschi, C.; Ceriana, P.; Chiovato, L.; Dalla Vecchia, L.A.; Fanfulla, F.; La Rovere, M.T.; Perego, F.; et al. Renin Angiotensin System Blockers and Risk of Mortality in Hypertensive Patients Hospitalized for COVID-19: An Italian Registry. J. Cardiovasc. Dev. Dis. 2022, 9, 15. [Google Scholar] [CrossRef]
- Verdecchia, P.; Angeli, F.; Cavallini, C.; Gattobigio, R.; Gentile, G.; Staessen, J.A.; Reboldi, G. Blood pressure reduction and renin-angiotensin system inhibition for prevention of congestive heart failure: A meta-analysis. Eur. Heart J. 2009, 30, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Verdecchia, P.; Angeli, F.; Mazzotta, G.; Gentile, G.; Reboldi, G. The renin angiotensin system in the development of cardiovascular disease: Role of aliskiren in risk reduction. Vasc. Health Risk Manag. 2008, 4, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Verdecchia, P.; Gentile, G.; Angeli, F.; Reboldi, G. Beyond blood pressure: Evidence for cardiovascular, cerebrovascular, and renal protective effects of renin-angiotensin system blockers. Ther. Adv. Cardiovasc. Dis. 2012, 6, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Verdecchia, P.; Reboldi, G.; Angeli, F.; Avanzini, F.; de Simone, G.; Pede, S.; Perticone, F.; Schillaci, G.; Vanuzzo, D.; Maggioni, A.P.; et al. Prognostic value of serial electrocardiographic voltage and repolarization changes in essential hypertension: The HEART Survey study. Am. J. Hypertens. 2007, 20, 997–1004. [Google Scholar] [CrossRef][Green Version]
- Verdecchia, P.; Angeli, F. The Seventh Report of the Joint National Committee on the Prevention, Detection, Evaluation and Treatment of High Blood Pressure: The weapons are ready. Rev. Esp. Cardiol. 2003, 56, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Grossman, E.; Verdecchia, P.; Shamiss, A.; Angeli, F.; Reboldi, G. Diuretic treatment of hypertension. Diabetes Care 2011, 34 (Suppl 2), S313–S319. [Google Scholar] [CrossRef] [PubMed]
- Reboldi, G.; Gentile, G.; Angeli, F.; Verdecchia, P. Choice of ACE inhibitor combinations in hypertensive patients with type 2 diabetes: Update after recent clinical trials. Vasc. Health Risk Manag. 2009, 5, 411–427. [Google Scholar] [CrossRef] [PubMed]
- Evangelou, E.; Warren, H.R.; Mosen-Ansorena, D.; Mifsud, B.; Pazoki, R.; Gao, H.; Ntritsos, G.; Dimou, N.; Cabrera, C.P.; Karaman, I.; et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 2018, 50, 1412–1425. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Ehret, G.B.; Rice, K.; Verwoert, G.C.; Launer, L.J.; Dehghan, A.; Glazer, N.L.; Morrison, A.C.; Johnson, A.D.; Aspelund, T.; et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009, 41, 677–687. [Google Scholar] [CrossRef]
- Newton-Cheh, C.; Johnson, T.; Gateva, V.; Tobin, M.D.; Bochud, M.; Coin, L.; Najjar, S.S.; Zhao, J.H.; Heath, S.C.; Eyheramendy, S.; et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009, 41, 666–676. [Google Scholar] [CrossRef]
- Ehret, G.B.; Munroe, P.B.; Rice, K.M.; Bochud, M.; Johnson, A.D.; Chasman, D.I.; Smith, A.V.; Tobin, M.D.; Verwoert, G.C.; Hwang, S.J.; et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011, 478, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Ehret, G.B.; Ferreira, T.; Chasman, D.I.; Jackson, A.U.; Schmidt, E.M.; Johnson, T.; Thorleifsson, G.; Luan, J.; Donnelly, L.A.; Kanoni, S.; et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat. Genet. 2016, 48, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.J.; Ehret, G.B.; Nandakumar, P.; Ranatunga, D.; Schaefer, C.; Kwok, P.Y.; Iribarren, C.; Chakravarti, A.; Risch, N. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat. Genet. 2017, 49, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661–678. [CrossRef]
- Kasprowicz-Maluśki, A.; Kwiatkowski, W.; Starosta, A.; Wojtaszek, P. Journey from the Center of the Cell—The intra- and intercellular transport of mRNA. Acta Biochim. Pol. 2016, 63, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Nachtergaele, S.; He, C. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 2017, 14, 156–163. [Google Scholar] [CrossRef]
- Ogorodnikov, A.; Kargapolova, Y.; Danckwardt, S. Processing and transcriptome expansion at the mRNA 3′ end in health and disease: Finding the right end. Pflug. Arch. 2016, 468, 993–1012. [Google Scholar] [CrossRef]
- Gebauer, F.; Schwarzl, T.; Valcárcel, J.; Hentze, M.W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2021, 22, 185–198. [Google Scholar] [CrossRef]
- Chen, Q.; Hu, G. Post-transcriptional regulation of the pluripotent state. Curr. Opin. Genet. Dev. 2017, 46, 15–23. [Google Scholar] [CrossRef]
- Corbett, A.H. Post-transcriptional regulation of gene expression and human disease. Curr. Opin. Cell Biol. 2018, 52, 96–104. [Google Scholar] [CrossRef]
- Odenbach, J.; Wang, X.; Cooper, S.; Chow, F.L.; Oka, T.; Lopaschuk, G.; Kassiri, Z.; Fernandez-Patron, C. MMP-2 mediates angiotensin II-induced hypertension under the transcriptional control of MMP-7 and TACE. Hypertension 2011, 57, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Garlick, P.J.; McNurlan, M.A.; Preedy, V.R. A rapid and convenient technique for measuring the rate of protein synthesis in tissues by injection of [3H]phenylalanine. Biochem. J. 1980, 192, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.E.; Kelly, F.J.; Goldspink, D.F. Pre- and post-natal growth and protein turnover in smooth muscle, heart and slow- and fast-twitch skeletal muscles of the rat. Biochem. J. 1984, 217, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Wiener, D.; Schwartz, S. The epitranscriptome beyond m(6)A. Nat. Rev. Genet. 2021, 22, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell. Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2011, 2, 611–631. [Google Scholar] [CrossRef]
- Glaich, O.; Parikh, S.; Bell, R.E.; Mekahel, K.; Donyo, M.; Leader, Y.; Shayevitch, R.; Sheinboim, D.; Yannai, S.; Hollander, D.; et al. DNA methylation directs microRNA biogenesis in mammalian cells. Nat. Commun. 2019, 10, 5657. [Google Scholar] [CrossRef]
- He, R.Z.; Jiang, J.; Luo, D.X. The functions of N6-methyladenosine modification in lncRNAs. Genes Dis. 2020, 7, 598–605. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Wu, Y.; Zhan, S.; Xu, Y.; Gao, X. RNA modifications in cardiovascular diseases, the potential therapeutic targets. Life Sci. 2021, 278, 119565. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell. Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yuan, X.; Han, R.; Zhang, H.; Xiu, R. Epitranscriptomic mechanisms of N6-methyladenosine methylation regulating mammalian hypertension development by determined spontaneously hypertensive rats pericytes. Epigenomics 2019, 11, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Marcadenti, A.; Fuchs, F.D.; Matte, U.; Sperb, F.; Moreira, L.B.; Fuchs, S.C. Effects of FTO RS9939906 and MC4R RS17782313 on obesity, type 2 diabetes mellitus and blood pressure in patients with hypertension. Cardiovasc. Diabetol. 2013, 12, 103. [Google Scholar] [CrossRef]
- Mo, X.B.; Lei, S.F.; Zhang, Y.H.; Zhang, H. Examination of the associations between m(6)A-associated single-nucleotide polymorphisms and blood pressure. Hypertens. Res. 2019, 42, 1582–1589. [Google Scholar] [CrossRef]
- Visvanathan, A.; Somasundaram, K. mRNA Traffic Control Reviewed: N6-Methyladenosine (m(6) A) Takes the Driver’s Seat. Bioessays 2018, 40, 1700093. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, K.; Melander, O.; Orho-Melander, M.; Lindblad, U.; Ranstam, J.; Råstam, L.; Groop, L. Polymorphism in the beta(1)-adrenergic receptor gene and hypertension. Circulation 2001, 104, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Li, X.; Zhang, S.; Guo, S.; Niu, W. The β1-adrenoreceptor gene Arg389Gly and Ser49Gly polymorphisms and hypertension: A meta-analysis. Mol. Biol. Rep. 2013, 40, 4047–4053. [Google Scholar] [CrossRef]
- Wang, H.; Liu, J.; Liu, K.; Liu, Y.; Wang, Z.; Lou, Y.; Niu, Q.; Gu, W.; Wang, L.; Li, M.; et al. β1-adrenoceptor gene Arg389Gly polymorphism and essential hypertension risk in general population: A meta-analysis. Mol. Biol. Rep. 2013, 40, 4055–4063. [Google Scholar] [CrossRef]
- Krüger, N.; Biwer, L.A.; Good, M.E.; Ruddiman, C.A.; Wolpe, A.G.; DeLalio, L.J.; Murphy, S.; Macal, E.H., Jr.; Ragolia, L.; Serbulea, V.; et al. Loss of Endothelial FTO Antagonizes Obesity-Induced Metabolic and Vascular Dysfunction. Circ. Res. 2020, 126, 232–242. [Google Scholar] [CrossRef]
- Félétou, M.; Huang, Y.; Vanhoutte, P.M. Endothelium-mediated control of vascular tone: COX-1 and COX-2 products. Br. J. Pharmacol. 2011, 164, 894–912. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Liu, X.; Zhang, J.J.; Zhao, J.J.; Xiong, Y.J.; Chang, Q.; Wang, H.Y.; Su, P.; Meng, J.; Zhao, Y.B. Vascular Smooth Muscle FTO Promotes Aortic Dissecting Aneurysms via m6A Modification of Klf5. Front. Cardiovasc. Med. 2020, 7, 592550. [Google Scholar] [CrossRef]
- Zhu, B.; Gong, Y.; Shen, L.; Li, J.; Han, J.; Song, B.; Hu, L.; Wang, Q.; Wang, Z. Total Panax notoginseng saponin inhibits vascular smooth muscle cell proliferation and migration and intimal hyperplasia by regulating WTAP/p16 signals via m(6)A modulation. Biomed. Pharmacother. 2020, 124, 109935. [Google Scholar] [CrossRef] [PubMed]
- Lombari, P.; Mallardo, M.; Petrazzuolo, O.; Nagoth, J.A.; Fiume, G.; Scanni, R.; Iervolino, A.; Damiano, S.; Coppola, A.; Borriello, M.; et al. miRNA-23a modulates sodium-hydrogen exchanger 1 expression: Studies in medullary thick ascending limb of salt induced hypertensive rats. Nephrol. Dial. Transplant. 2022, gfac232. [Google Scholar] [CrossRef] [PubMed]
- Kolomeĭchuk, S.N.; Makeeva, I.V.; Topchieva, L.V.; Korneva, V.A.; Nemova, N.N. Association of T3111C polymorphism in 3′-untranslated region of the CLOCK gene with the risk of essential arterial hypertension and coronary artery disease in the Russian population Karelia. Genetika 2011, 47, 1411–1415. [Google Scholar] [CrossRef]
- Woon, P.Y.; Kaisaki, P.J.; Bragança, J.; Bihoreau, M.T.; Levy, J.C.; Farrall, M.; Gauguier, D. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc. Natl. Acad. Sci. USA 2007, 104, 14412–14417. [Google Scholar] [CrossRef]
- Kurbatova, I.V.; Topchieva, L.V.; Korneva, V.A.; Kolomeichuk, S.N.; Nemova, N.N. Expression of circadian rhythm genes CLOCK, BMAL1, and PER1 in buccal epithelial cells of patients with essential arterial hypertension in dependence on polymorphic variants of CLOCK and BMAL1 genes. Bull. Exp. Biol. Med. 2014, 157, 360–363. [Google Scholar] [CrossRef]
- Fang, Z.; Zhu, L.; Jin, Y.; Chen, Y.; Chang, W.; Yao, Y. Downregulation of Arntl mRNA Expression in Women with Hypertension: A Case-Control Study. Kidney Blood Press. Res. 2021, 46, 741–748. [Google Scholar] [CrossRef]
- Cunningham, C.C.; Gorlin, J.B.; Kwiatkowski, D.J.; Hartwig, J.H.; Janmey, P.A.; Byers, H.R.; Stossel, T.P. Actin-binding protein requirement for cortical stability and efficient locomotion. Science 1992, 255, 325–327. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, M.H.; Moskowitz, I.P.; Mendonza, A.M.; Vidali, L.; Nakamura, F.; Kwiatkowski, D.J.; Walsh, C.A. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19836–19841. [Google Scholar] [CrossRef]
- Liu, C.; Tang, W.; Zhao, H.; Yang, S.; Ren, Z.; Li, J.; Chen, Y.; Zhao, X.; Xu, D.; Zhao, Y.; et al. The variants at FLNA and FLNB contribute to the susceptibility of hypertension and stroke with differentially expressed mRNA. Pharmacogenom. J. 2021, 21, 458–466. [Google Scholar] [CrossRef]
- Jain, M.; Mann, T.D.; Stulić, M.; Rao, S.P.; Kirsch, A.; Pullirsch, D.; Strobl, X.; Rath, C.; Reissig, L.; Moreth, K.; et al. RNA editing of Filamin A pre-mRNA regulates vascular contraction and diastolic blood pressure. EMBO J. 2018, 37, e94813. [Google Scholar] [CrossRef]
- Huang, S.; Taubel, J.; Fiore, G.; Dewland, P.; Bakris, G.; Desai, A.; Cheng, Y.; Agarwal, S.; Harrop, J.; Nguyen, H. Dose-related reductions in blood pressure with a rna interference (rnai) therapeutic targeting angiotensinogen in hypertensive patients: Interim results from a first-in-human phase 1 study of aln-agt01. Circulation 2020, 142, A14387. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zappa, M.; Verdecchia, P.; Spanevello, A.; Golino, M.; Angeli, F. mRNA Metabolism and Hypertension. Biomedicines 2023, 11, 118. https://doi.org/10.3390/biomedicines11010118
Zappa M, Verdecchia P, Spanevello A, Golino M, Angeli F. mRNA Metabolism and Hypertension. Biomedicines. 2023; 11(1):118. https://doi.org/10.3390/biomedicines11010118
Chicago/Turabian StyleZappa, Martina, Paolo Verdecchia, Antonio Spanevello, Michele Golino, and Fabio Angeli. 2023. "mRNA Metabolism and Hypertension" Biomedicines 11, no. 1: 118. https://doi.org/10.3390/biomedicines11010118
APA StyleZappa, M., Verdecchia, P., Spanevello, A., Golino, M., & Angeli, F. (2023). mRNA Metabolism and Hypertension. Biomedicines, 11(1), 118. https://doi.org/10.3390/biomedicines11010118








