GJA1/CX43 High Expression Levels in the Cervical Spinal Cord of ALS Patients Correlate to Microglia-Mediated Neuroinflammatory Profile
Abstract
1. Introduction
2. Materials and Methods
2.1. Daset Selection
2.2. Sample Stratification
2.3. Clinical and Pathological Criteria
2.4. Data Processing and Experimental Design
2.5. Drugs Analysis Prediction (DAP)
2.6. Statistical Analysis
3. Results
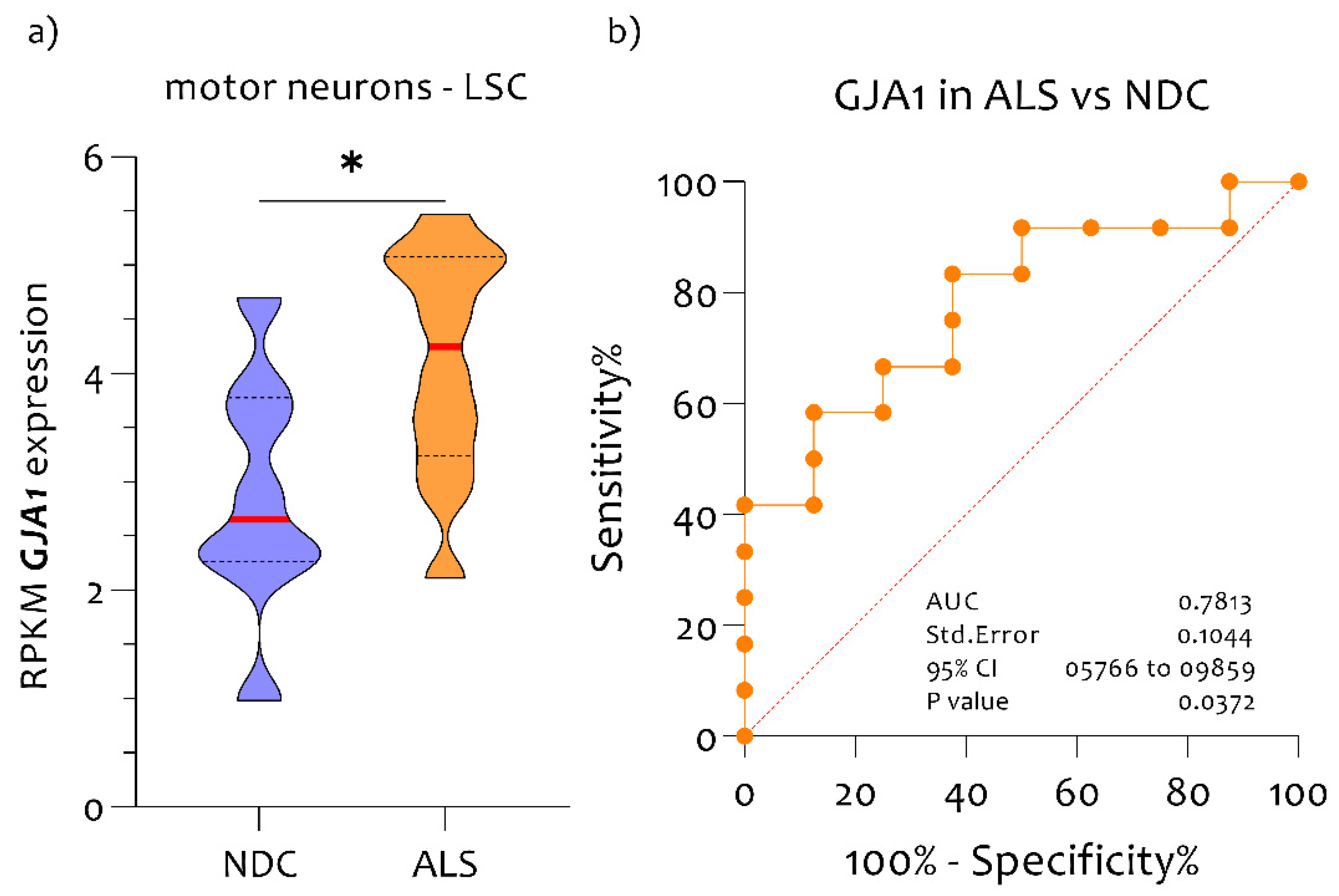
3.1. High Expression Levels of GJA1 in MNs of ALS Patients Compared to NDC
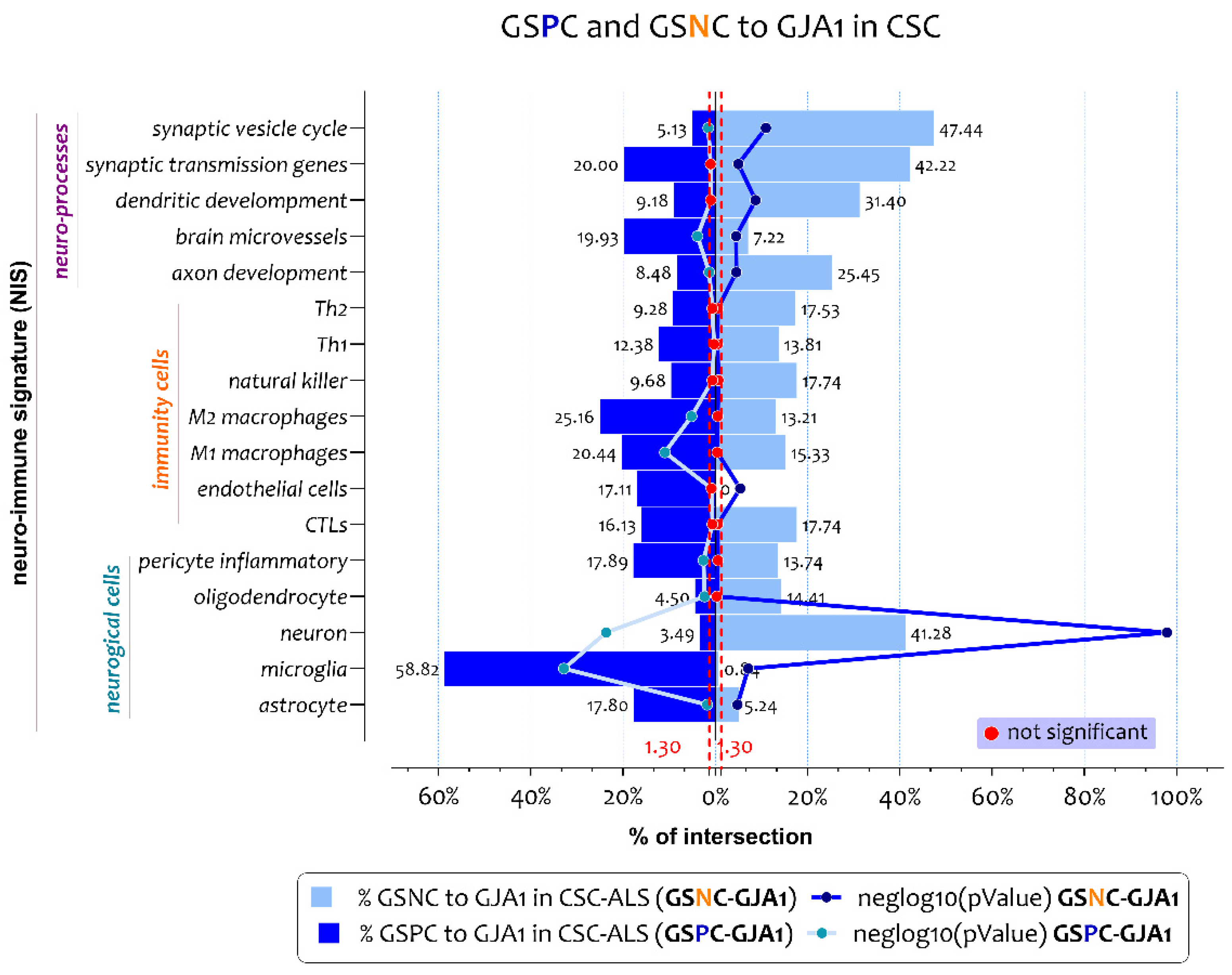
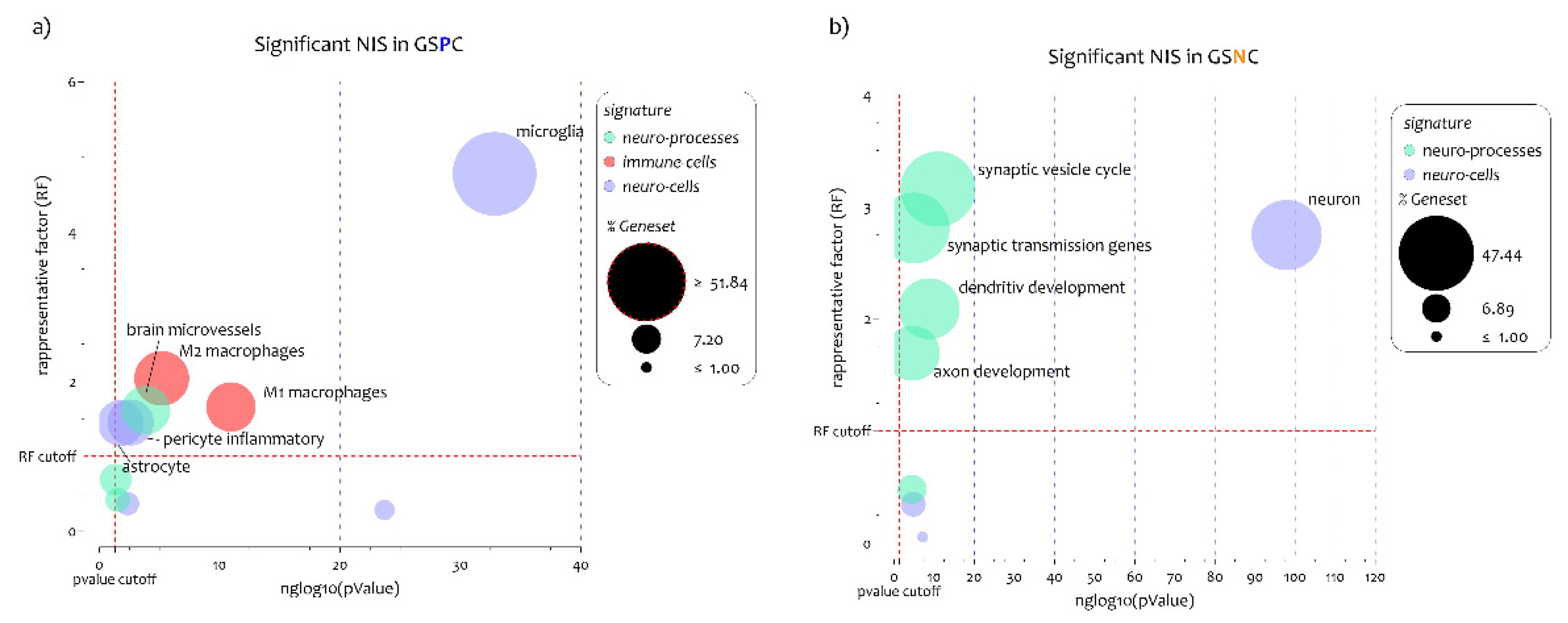
3.2. ALS Patients Exhibit Different Neuro-Immune Cellular Profile According to GJA1 Expression Levels
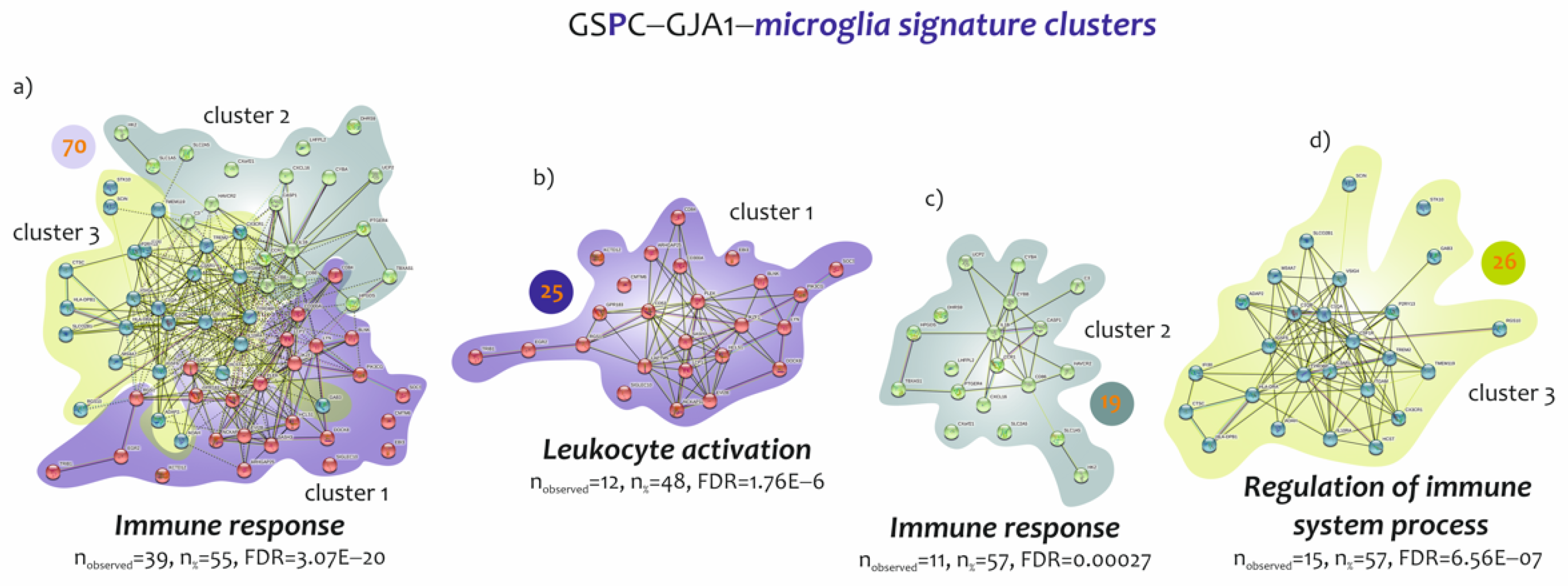
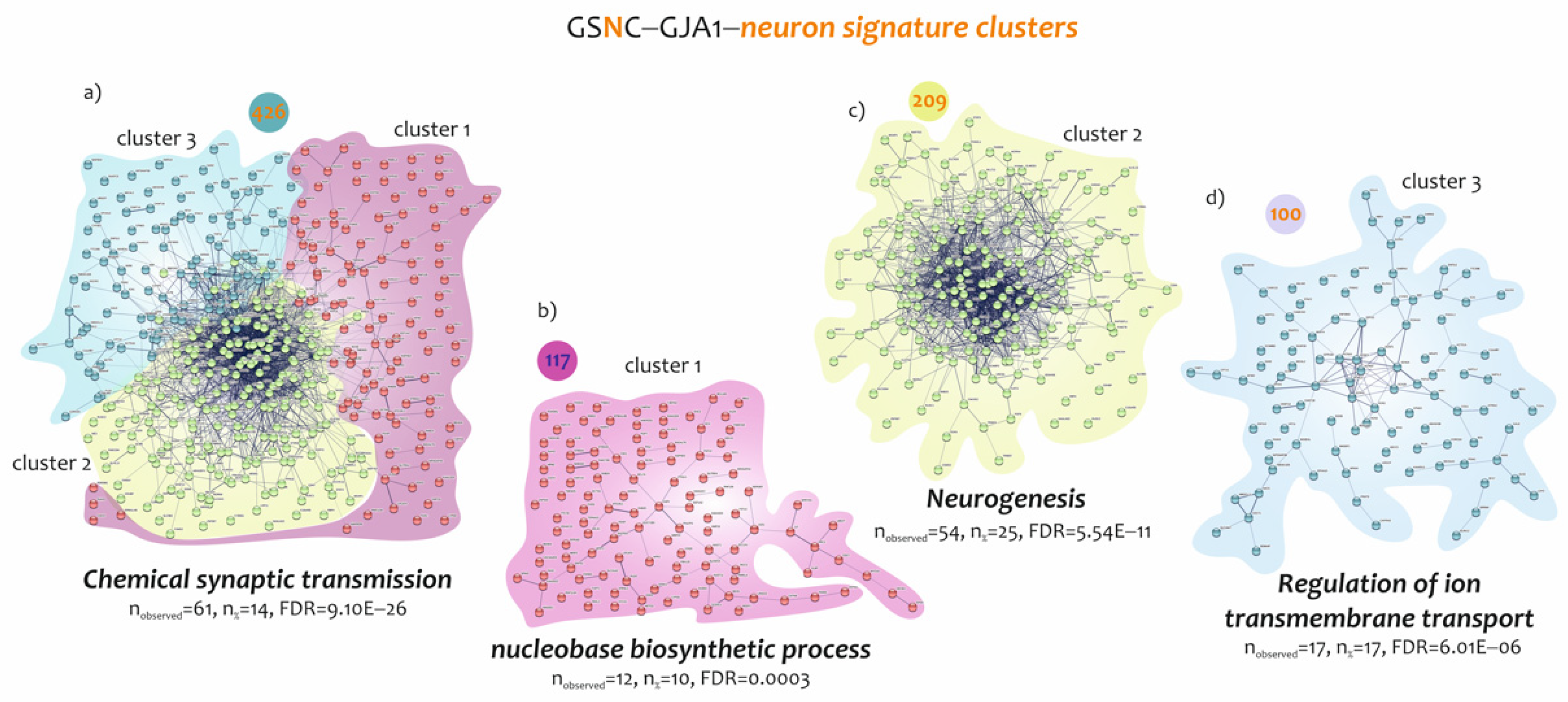
3.3. Biological Processes Identified by the Genes Belonging to Microglia and Neuron Signatures Determined by GJA1 Expression Levels in the Spinal Cord of ALS Patients
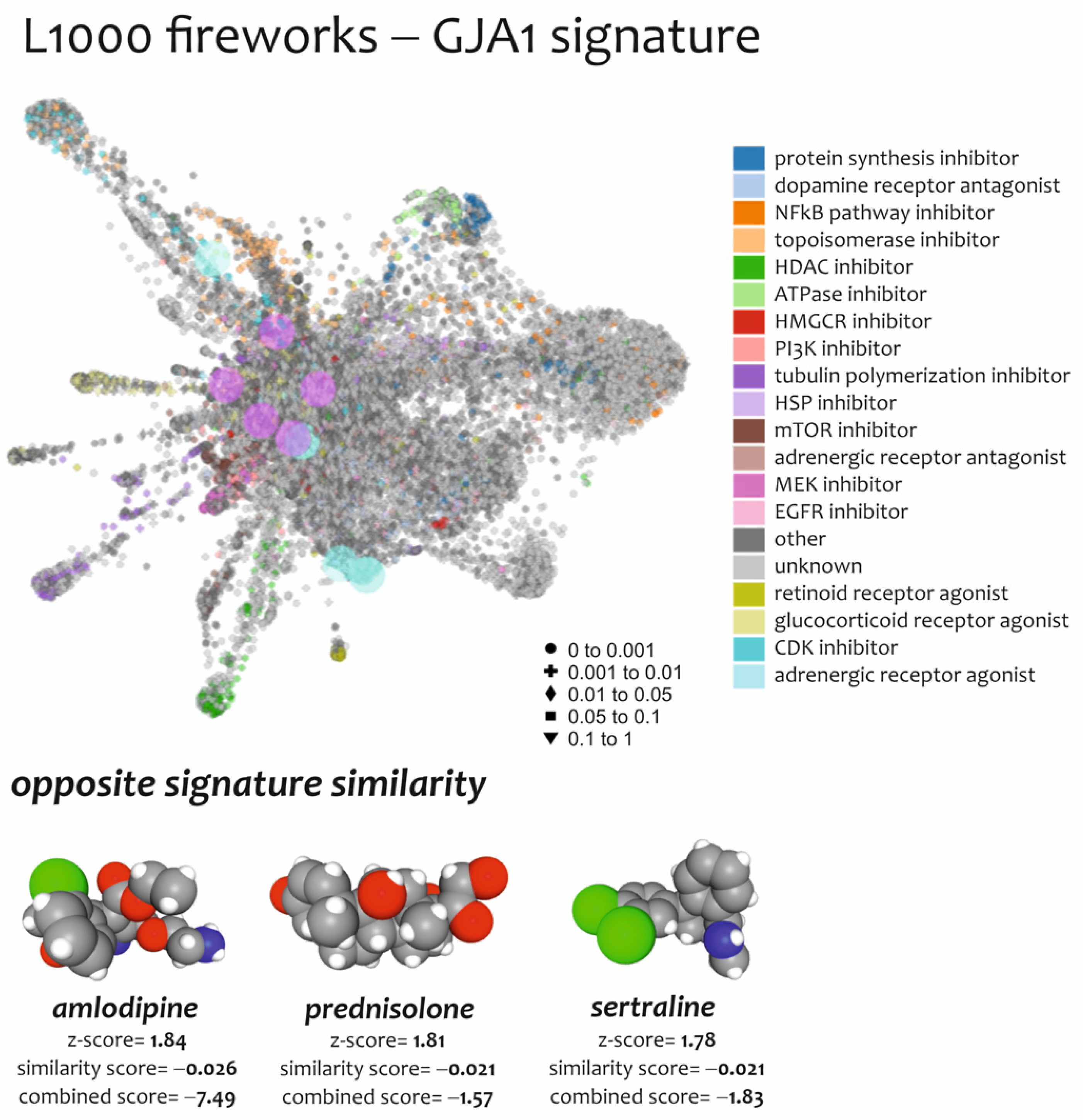
3.4. Effect of Drugs Mimicking and Opposing GJA1 Transcriptomic Signatures Obtained from Spinal Cord of ALS Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Approval
Abbreviations
| AD | Alzheimer’s disease |
| ALS | amyotrophic lateral sclerosis |
| AUC | area under the ROC curve |
| CNS | central nervous system |
| CSC | cervical spinal cord |
| CX | connexin |
| FDR | false discovery rate |
| GDA | genomic deconvolution analysis |
| GEO | gene expression omnibus |
| GJ | gap junction |
| GO | gene ontology |
| GSNC | genes significant negative correlated |
| GSNC-GJA1 | genes significantly negatively correlated to GJA1 |
| GSPC | genes significantly positively correlated |
| GSPC-GJA1 | genes significantly positively correlated to GJA1 |
| HC | hemichannel |
| LCM | laser capture microdissection |
| MD | microarray datasets |
| MeV | MultiExperiment Viewer |
| MNs | motoneurons |
| MOA | mode of action |
| MS | multiple sclerosis |
| NDC | not demented control subjects–healthy individuals |
| PD | Parkinson’s disease |
| SDEG | significantly different expressed genes |
References
- Glass, C.K.; Saijo, K.; Winner, B.; Marchetto, M.C.; Gage, F.H. Mechanisms underlying inflammation in neurodegeneration. Cell 2010, 140, 918–934. [Google Scholar] [CrossRef] [PubMed]
- Chio, A.; Logroscino, G.; Hardiman, O.; Swingler, R.; Mitchell, D.; Beghi, E.; Traynor, B.G.; Eurals, C. Prognostic factors in ALS: A critical review. Amyotroph. Lateral Scler. 2009, 10, 310–323. [Google Scholar] [CrossRef] [PubMed]
- Vicario, N.; Spitale, F.M.; Tibullo, D.; Giallongo, C.; Amorini, A.M.; Scandura, G.; Spoto, G.; Saab, M.W.; D’Aprile, S.; Alberghina, C.; et al. Clobetasol promotes neuromuscular plasticity in mice after motoneuronal loss via sonic hedgehog signaling, immunomodulation and metabolic rebalancing. Cell Death Dis. 2021, 12, 625. [Google Scholar] [CrossRef] [PubMed]
- Vicario, N.; Turnaturi, R.; Spitale, F.M.; Torrisi, F.; Zappala, A.; Gulino, R.; Pasquinucci, L.; Chiechio, S.; Parenti, C.; Parenti, R. Intercellular communication and ion channels in neuropathic pain chronicization. Inflamm. Res. 2020, 69, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Vicario, N.; Zappala, A.; Calabrese, G.; Gulino, R.; Parenti, C.; Gulisano, M.; Parenti, R. Connexins in the Central Nervous System: Physiological Traits and Neuroprotective Targets. Front. Physiol. 2017, 8, 1060. [Google Scholar] [CrossRef]
- Harcha, P.A.; Garces, P.; Arredondo, C.; Fernandez, G.; Saez, J.C.; van Zundert, B. Mast Cell and Astrocyte Hemichannels and Their Role in Alzheimer’s Disease, ALS, and Harmful Stress Conditions. Int. J. Mol. Sci. 2021, 22, 1924. [Google Scholar] [CrossRef]
- Huang, X.; Su, Y.; Wang, N.; Li, H.; Li, Z.; Yin, G.; Chen, H.; Niu, J.; Yi, C. Astroglial Connexins in Neurodegenerative Diseases. Front. Mol. Neurosci. 2021, 14, 657514. [Google Scholar] [CrossRef]
- Stavropoulos, F.; Georgiou, E.; Sargiannidou, I.; Kleopa, K.A. Dysregulation of Blood-Brain Barrier and Exacerbated Inflammatory Response in Cx47-Deficient Mice after Induction of EAE. Pharmaceuticals 2021, 14, 621. [Google Scholar] [CrossRef]
- Zhao, Y.; Qi, Y.; Li, Q.; Quan, H.; Liu, D.; Zhou, H. Connexin43 inhibition attenuated dopaminergic neuronal loss in the lipopolysaccharide-induced mice model of Parkinson’s disease. Neurosci. Lett. 2022, 771, 136471. [Google Scholar] [CrossRef]
- Zhao, Y.; Yamasaki, R.; Yamaguchi, H.; Nagata, S.; Une, H.; Cui, Y.; Masaki, K.; Nakamuta, Y.; Iinuma, K.; Watanabe, M.; et al. Oligodendroglial connexin 47 regulates neuroinflammation upon autoimmune demyelination in a novel mouse model of multiple sclerosis. Proc. Natl. Acad. Sci. USA 2020, 117, 2160–2169. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Mizoguchi, H.; Doi, Y.; Jin, S.; Noda, M.; Liang, J.; Li, H.; Zhou, Y.; Mori, R.; Yasuoka, S.; et al. Blockade of gap junction hemichannel suppresses disease progression in mouse models of amyotrophic lateral sclerosis and Alzheimer’s disease. PLoS ONE 2011, 6, e21108. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wei, X.; Xiang, J.; Gao, J.; Wang, L.; You, J.; Cai, Y.; Cai, D. Protection of erythropoietin against ischemic neurovascular unit injuries through the effects of connexin43. Biochem. Biophys. Res. Commun. 2015, 458, 656–662. [Google Scholar] [CrossRef]
- Cibelli, A.; Stout, R.; Timmermann, A.; de Menezes, L.; Guo, P.; Maass, K.; Seifert, G.; Steinhauser, C.; Spray, D.C.; Scemes, E. Cx43 carboxyl terminal domain determines AQP4 and Cx30 endfoot organization and blood brain barrier permeability. Sci. Rep. 2021, 11, 24334. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yang, L.; Chen, J.; Chen, Y.; Zhang, L.; Wang, L.; Li, X.; Li, Y.; Yu, H. Inhibition of Connexin43 hemichannels with Gap19 protects cerebral ischemia/reperfusion injury via the JAK2/STAT3 pathway in mice. Brain Res. Bull. 2019, 146, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Cicirata, F.; Parenti, R.; Spinella, F.; Giglio, S.; Tuorto, F.; Zuffardi, O.; Gulisano, M. Genomic organization and chromosomal localization of the mouse Connexin36 (mCx36) gene. Gene 2000, 251, 123–130. [Google Scholar] [CrossRef]
- Almad, A.A.; Taga, A.; Joseph, J.; Gross, S.K.; Welsh, C.; Patankar, A.; Richard, J.P.; Rust, K.; Pokharel, A.; Plott, C.; et al. Cx43 hemichannels contribute to astrocyte-mediated toxicity in sporadic and familial ALS. Proc. Natl. Acad. Sci. USA 2022, 119, e2107391119. [Google Scholar] [CrossRef]
- Vicario, N.; Pasquinucci, L.; Spitale, F.M.; Chiechio, S.; Turnaturi, R.; Caraci, F.; Tibullo, D.; Avola, R.; Gulino, R.; Parenti, R.; et al. Simultaneous Activation of Mu and Delta Opioid Receptors Reduces Allodynia and Astrocytic Connexin 43 in an Animal Model of Neuropathic Pain. Mol. Neurobiol. 2019, 56, 7338–7354. [Google Scholar] [CrossRef]
- Karagiannis, A.; Sylantyev, S.; Hadjihambi, A.; Hosford, P.S.; Kasparov, S.; Gourine, A.V. Hemichannel-mediated release of lactate. J. Cereb. Blood Flow Metab. 2016, 36, 1202–1211. [Google Scholar] [CrossRef]
- Nagy, J.I.; Ionescu, A.V.; Lynn, B.D.; Rash, J.E. Coupling of astrocyte connexins Cx26, Cx30, Cx43 to oligodendrocyte Cx29, Cx32, Cx47: Implications from normal and connexin32 knockout mice. Glia 2003, 44, 205–218. [Google Scholar] [CrossRef]
- Sanchez, O.F.; Rodriguez, A.V.; Velasco-Espana, J.M.; Murillo, L.C.; Sutachan, J.J.; Albarracin, S.L. Role of Connexins 30, 36, and 43 in Brain Tumors, Neurodegenerative Diseases, and Neuroprotection. Cells 2020, 9, 846. [Google Scholar] [CrossRef]
- Orellana, J.A.; Saez, P.J.; Shoji, K.F.; Schalper, K.A.; Palacios-Prado, N.; Velarde, V.; Giaume, C.; Bennett, M.V.; Saez, J.C. Modulation of brain hemichannels and gap junction channels by pro-inflammatory agents and their possible role in neurodegeneration. Antioxid. Redox Signal. 2009, 11, 369–399. [Google Scholar] [CrossRef] [PubMed]
- Yawata, I.; Takeuchi, H.; Doi, Y.; Liang, J.; Mizuno, T.; Suzumura, A. Macrophage-induced neurotoxicity is mediated by glutamate and attenuated by glutaminase inhibitors and gap junction inhibitors. Life Sci. 2008, 82, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Jin, S.; Wang, J.; Zhang, G.; Kawanokuchi, J.; Kuno, R.; Sonobe, Y.; Mizuno, T.; Suzumura, A. Tumor necrosis factor-alpha induces neurotoxicity via glutamate release from hemichannels of activated microglia in an autocrine manner. J. Biol. Chem. 2006, 281, 21362–21368. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Jin, S.; Suzuki, H.; Doi, Y.; Liang, J.; Kawanokuchi, J.; Mizuno, T.; Sawada, M.; Suzumura, A. Blockade of microglial glutamate release protects against ischemic brain injury. Exp. Neurol. 2008, 214, 144–146. [Google Scholar] [CrossRef]
- De Vuyst, E.; Decrock, E.; Cabooter, L.; Dubyak, G.R.; Naus, C.C.; Evans, W.H.; Leybaert, L. Intracellular calcium changes trigger connexin 32 hemichannel opening. EMBO J. 2006, 25, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Kang, N.; Lovatt, D.; Torres, A.; Zhao, Z.; Lin, J.; Nedergaard, M. Connexin 43 hemichannels are permeable to ATP. J. Neurosci. 2008, 28, 4702–4711. [Google Scholar] [CrossRef]
- Dobrenis, K.; Chang, H.Y.; Pina-Benabou, M.H.; Woodroffe, A.; Lee, S.C.; Rozental, R.; Spray, D.C.; Scemes, E. Human and mouse microglia express connexin36, and functional gap junctions are formed between rodent microglia and neurons. J. Neurosci. Res. 2005, 82, 306–315. [Google Scholar] [CrossRef]
- Spitale, F.M.; Vicario, N.; Rosa, M.D.; Tibullo, D.; Vecchio, M.; Gulino, R.; Parenti, R. Increased expression of connexin 43 in a mouse model of spinal motoneuronal loss. Aging 2020, 12, 12598–12608. [Google Scholar] [CrossRef]
- Vicario, N.; Denaro, S.; Turnaturi, R.; Longhitano, L.; Spitale, F.M.; Spoto, S.; Marrazzo, A.; Zappala, A.; Tibullo, D.; Li Volti, G.; et al. Mu and Delta Opioid Receptor Targeting Reduces Connexin 43-Based Heterocellular Coupling during Neuropathic Pain. Int. J. Mol. Sci. 2022, 23, 5864. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar] [CrossRef]
- Durrenberger, P.F.; Fernando, F.S.; Magliozzi, R.; Kashefi, S.N.; Bonnert, T.P.; Ferrer, I.; Seilhean, D.; Nait-Oumesmar, B.; Schmitt, A.; Gebicke-Haerter, P.J.; et al. Selection of novel reference genes for use in the human central nervous system: A BrainNet Europe Study. Acta Neuropathol. 2012, 124, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Krach, F.; Batra, R.; Wheeler, E.C.; Vu, A.Q.; Wang, R.; Hutt, K.; Rabin, S.J.; Baughn, M.W.; Libby, R.T.; Diaz-Garcia, S.; et al. Transcriptome-pathology correlation identifies interplay between TDP-43 and the expression of its kinase CK1E in sporadic ALS. Acta Neuropathol. 2018, 136, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Castrogiovanni, P.; Li Volti, G.; Sanfilippo, C.; Tibullo, D.; Galvano, F.; Vecchio, M.; Avola, R.; Barbagallo, I.; Malaguarnera, L.; Castorina, S.; et al. Fasting and Fast Food Diet Play an Opposite Role in Mice Brain Aging. Mol. Neurobiol. 2018, 55, 6881–6893. [Google Scholar] [CrossRef] [PubMed]
- Sanfilippo, C.; Castrogiovanni, P.; Imbesi, R.; Tibullo, D.; Li Volti, G.; Barbagallo, I.; Vicario, N.; Musumeci, G.; Di Rosa, M. Middle-aged healthy women and Alzheimer’s disease patients present an overlapping of brain cell transcriptional profile. Neuroscience 2019, 406, 333–344. [Google Scholar] [CrossRef]
- Xiao, J.; Cao, H.; Chen, J. False discovery rate control incorporating phylogenetic tree increases detection power in microbiome-wide multiple testing. Bioinformatics 2017, 33, 2873–2881. [Google Scholar] [CrossRef]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Zuberi, K.; Franz, M.; Rodriguez, H.; Montojo, J.; Lopes, C.T.; Bader, G.D.; Morris, Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013, 41, W115–W122. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Chang, J.T.; Nevins, J.R. GATHER: A systems approach to interpreting genomic signatures. Bioinformatics 2006, 22, 2926–2933. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Roussos, P.; McKenzie, A.; Zhou, X.; Kajiwara, Y.; Brennand, K.J.; De Luca, G.C.; Crary, J.F.; Casaccia, P.; Buxbaum, J.D.; et al. Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer’s disease. Genome Med. 2016, 8, 104. [Google Scholar] [CrossRef] [PubMed]
- Guijarro-Munoz, I.; Compte, M.; Alvarez-Cienfuegos, A.; Alvarez-Vallina, L.; Sanz, L. Lipopolysaccharide activates Toll-like receptor 4 (TLR4)-mediated NF-kappaB signaling pathway and proinflammatory response in human pericytes. J. Biol. Chem. 2014, 289, 2457–2468. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.R.; Baldwin, D.; Ma, Y.; Ouyang, W.; Gurney, A.; Martin, F.; Fong, S.; van Lookeren Campagne, M.; Godowski, P.; Williams, P.M.; et al. Immune response in silico (IRIS): Immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005, 6, 319–331. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Wang, Z.; Lachmann, A.; Keenan, A.B.; Ma’ayan, A. L1000FWD: Fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics 2018, 34, 2150–2152. [Google Scholar] [CrossRef]
- Hillman, C.H.; Erickson, K.I.; Kramer, A.F. Be smart, exercise your heart: Exercise effects on brain and cognition. Nat. Rev. Neurosci. 2008, 9, 58–65. [Google Scholar] [CrossRef]
- Buchman, A.S.; Boyle, P.A.; Yu, L.; Shah, R.C.; Wilson, R.S.; Bennett, D.A. Total daily physical activity and the risk of AD and cognitive decline in older adults. Neurology 2012, 78, 1323–1329. [Google Scholar] [CrossRef]
- Box, G.E.; Tiao, G.C. Bayesian Inference in Statistical Analysis; Wiley: Hoboken, NJ, USA, 1992. [Google Scholar] [CrossRef]
- Zetterberg, H.; Bozzetta, E.; Favole, A.; Corona, C.; Cavarretta, M.C.; Ingravalle, F.; Blennow, K.; Pocchiari, M.; Meloni, D. Neurofilaments in blood is a new promising preclinical biomarker for the screening of natural scrapie in sheep. PLoS ONE 2019, 14, e0226697. [Google Scholar] [CrossRef]
- Catrogiovanni, P.; Musumeci, G.; Giunta, S.; Imbesi, R.; Di Rosa, M. The expression levels of CHI3L1 and IL15Ralpha correlate with TGM2 in duodenum biopsies of patients with celiac disease. Inflamm. Res. 2020, 69, 925–935. [Google Scholar] [CrossRef]
- Sanfilippo, C.; Pinzone, M.R.; Cambria, D.; Longo, A.; Palumbo, M.; Di Marco, R.; Condorelli, F.; Nunnari, G.; Malaguarnera, L.; Di Rosa, M. OAS Gene Family Expression Is Associated with HIV-Related Neurocognitive Disorders. Mol. Neurobiol. 2018, 55, 1905–1914. [Google Scholar] [CrossRef]
- Sanfilippo, C.; Nunnari, G.; Calcagno, A.; Malaguarnera, L.; Blennow, K.; Zetterberg, H.; Di Rosa, M. The chitinases expression is related to Simian Immunodeficiency Virus Encephalitis (SIVE) and in HIV encephalitis (HIVE). Virus Res. 2017, 227, 220–230. [Google Scholar] [CrossRef][Green Version]
- Sanfilippo, C.; Malaguarnera, L.; Di Rosa, M. Chitinase expression in Alzheimer’s disease and non-demented brains regions. J. Neurol. Sci. 2016, 369, 242–249. [Google Scholar] [CrossRef]
- Di Rosa, M.; Giallongo, C.; Romano, A.; Tibullo, D.; Li Volti, G.; Musumeci, G.; Barbagallo, I.; Imbesi, R.; Castrogiovanni, P.; Palumbo, G.A. Immunoproteasome Genes Are Modulated in CD34(+) JAK2(V617F) Mutated Cells from Primary Myelofibrosis Patients. Int. J. Mol. Sci. 2020, 21, 2926. [Google Scholar] [CrossRef]
- Di Rosa, M.; Sanfilippo, C.; Libra, M.; Musumeci, G.; Malaguarnera, L. Different pediatric brain tumors are associated with different gene expression profiling. Acta Histochem. 2015, 117, 477–485. [Google Scholar] [CrossRef]
- Liu, C.C.; Guo, Y.; Vrindten, K.L.; Lau, W.W.; Sparks, R.; Tsang, J.S. OMiCC: An expanded and enhanced platform for meta-analysis of public gene expression data. STAR Protoc. 2022, 3, 101474. [Google Scholar] [CrossRef]
- Ruffo, P.; Cavallaro, S.; Conforti, F.L. The Advent of Omics Sciences in Clinical Trials of Motor Neuron Diseases. J. Pers. Med. 2022, 12, 758. [Google Scholar] [CrossRef]
- De Bock, M.; Leybaert, L.; Giaume, C. Connexin Channels at the Glio-Vascular Interface: Gatekeepers of the Brain. Neurochem. Res. 2017, 42, 2519–2536. [Google Scholar] [CrossRef]
- Kielian, T. Glial connexins and gap junctions in CNS inflammation and disease. J. Neurochem. 2008, 106, 1000–1016. [Google Scholar] [CrossRef]
- Kirichenko, E.Y.; Skatchkov, S.N.; Ermakov, A.M. Structure and Functions of Gap Junctions and Their Constituent Connexins in the Mammalian CNS. Biochem. (Mosc.) Suppl. Ser. A Membr. Cell Biol. 2021, 15, 107–119. [Google Scholar] [CrossRef]
- Dharmadasa, T.; Matamala, J.M.; Huynh, W.; Zoing, M.C.; Kiernan, M.C. Motor neurone disease. Handb. Clin. Neurol. 2018, 159, 345–357. [Google Scholar] [CrossRef]
- Diaz-Amarilla, P.; Olivera-Bravo, S.; Trias, E.; Cragnolini, A.; Martinez-Palma, L.; Cassina, P.; Beckman, J.; Barbeito, L. Phenotypically aberrant astrocytes that promote motoneuron damage in a model of inherited amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 18126–18131. [Google Scholar] [CrossRef]
- Freitas-Andrade, M.; Naus, C.C. Astrocytes in neuroprotection and neurodegeneration: The role of connexin43 and pannexin1. Neuroscience 2016, 323, 207–221. [Google Scholar] [CrossRef]
- Eugenin, E.A.; Eckardt, D.; Theis, M.; Willecke, K.; Bennett, M.V.; Saez, J.C. Microglia at brain stab wounds express connexin 43 and in vitro form functional gap junctions after treatment with interferon-gamma and tumor necrosis factor-alpha. Proc. Natl. Acad. Sci. USA 2001, 98, 4190–4195. [Google Scholar] [CrossRef]
- Shaikh, S.B.; Uy, B.; Perera, A.; Nicholson, L.F. AGEs-RAGE mediated up-regulation of connexin43 in activated human microglial CHME-5 cells. Neurochem. Int. 2012, 60, 640–651. [Google Scholar] [CrossRef]
- Gajardo-Gomez, R.; Labra, V.C.; Orellana, J.A. Connexins and Pannexins: New Insights into Microglial Functions and Dysfunctions. Front. Mol. Neurosci. 2016, 9, 86. [Google Scholar] [CrossRef]
- Genc, B.; Jara, J.H.; Lagrimas, A.K.; Pytel, P.; Roos, R.P.; Mesulam, M.M.; Geula, C.; Bigio, E.H.; Ozdinler, P.H. Apical dendrite degeneration, a novel cellular pathology for Betz cells in ALS. Sci. Rep. 2017, 7, 41765. [Google Scholar] [CrossRef]
- Rinaldi, F.; Hartfield, E.M.; Crompton, L.A.; Badger, J.L.; Glover, C.P.; Kelly, C.M.; Rosser, A.E.; Uney, J.B.; Caldwell, M.A. Cross-regulation of Connexin43 and beta-catenin influences differentiation of human neural progenitor cells. Cell Death Dis. 2014, 5, e1017. [Google Scholar] [CrossRef] [PubMed]
- Brocardo, L.; Acosta, L.E.; Piantanida, A.P.; Rela, L. Beneficial and Detrimental Remodeling of Glial Connexin and Pannexin Functions in Rodent Models of Nervous System Diseases. Front. Cell. Neurosci. 2019, 13, 491. [Google Scholar] [CrossRef] [PubMed]
- Hosli, L.; Binini, N.; Ferrari, K.D.; Thieren, L.; Looser, Z.J.; Zuend, M.; Zanker, H.S.; Berry, S.; Holub, M.; Mobius, W.; et al. Decoupling astrocytes in adult mice impairs synaptic plasticity and spatial learning. Cell Rep. 2022, 38, 110484. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Wang, X.; Hao, Y.; Qiu, L.; Lou, Y.; Zhang, Y.; Ma, D.; Feng, J. The Multifaceted Role of Astrocyte Connexin 43 in Ischemic Stroke Through Forming Hemichannels and Gap Junctions. Front. Neurol. 2020, 11, 703. [Google Scholar] [CrossRef]
- Gulino, R.; Vicario, N.; Giunta, M.A.S.; Spoto, G.; Calabrese, G.; Vecchio, M.; Gulisano, M.; Leanza, G.; Parenti, R. Neuromuscular Plasticity in a Mouse Neurotoxic Model of Spinal Motoneuronal Loss. Int. J. Mol. Sci. 2019, 20, 1500. [Google Scholar] [CrossRef]
- Yacila, G.; Sari, Y. Potential therapeutic drugs and methods for the treatment of amyotrophic lateral sclerosis. Curr. Med. Chem. 2014, 21, 3583–3593. [Google Scholar] [CrossRef][Green Version]
- Kato, H.; Naito, M.; Saito, T.; Hideyama, T.; Suzuki, Y.; Kimura, T.; Kwak, S.; Aizawa, H. Prolyl Isomerase Pin1 Expression in the Spinal Motor Neurons of Patients With Sporadic Amyotrophic Lateral Sclerosis. J. Clin. Neurol. 2022, 18, 463–469. [Google Scholar] [CrossRef]
- Pinto, S.; Swash, M.; De Carvalho, M. Compensatory metabolic and central respiratory drive mechanisms in ALS. Amyotroph. Lateral Scler. Front. Degener. 2021, 22, 585–587. [Google Scholar] [CrossRef]
- Giusto, E.; Codrich, M.; de Leo, G.; Francardo, V.; Coradazzi, M.; Parenti, R.; Gulisano, M.; Vicario, N.; Gulino, R.; Leanza, G. Compensatory changes in degenerating spinal motoneurons sustain functional sparing in the SOD1-G93A mouse model of amyotrophic lateral sclerosis. J. Comp. Neurol. 2020, 528, 231–243. [Google Scholar] [CrossRef]
- Hur, S.K.; Hunter, M.; Dominique, M.A.; Farag, M.; Cotton-Samuel, D.; Khan, T.; Trojanowski, J.Q.; Spiller, K.J.; Lee, V.M. Slow motor neurons resist pathological TDP-43 and mediate motor recovery in the rNLS8 model of amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 2022, 10, 75. [Google Scholar] [CrossRef] [PubMed]
- Orellana, J.A.; Sanchez, H.A.; Schalper, K.A.; Figueroa, V.; Saez, J.C. Regulation of intercellular calcium signaling through calcium interactions with connexin-based channels. Adv. Exp. Med. Biol. 2012, 740, 777–794. [Google Scholar] [CrossRef] [PubMed]
- Morello, G.; Spampinato, A.G.; Cavallaro, S. Neuroinflammation and ALS: Transcriptomic Insights into Molecular Disease Mechanisms and Therapeutic Targets. Mediat. Inflamm. 2017, 2017, 7070469. [Google Scholar] [CrossRef]
- Froger, N.; Orellana, J.A.; Calvo, C.F.; Amigou, E.; Kozoriz, M.G.; Naus, C.C.; Saez, J.C.; Giaume, C. Inhibition of cytokine-induced connexin43 hemichannel activity in astrocytes is neuroprotective. Mol. Cell. Neurosci. 2010, 45, 37–46. [Google Scholar] [CrossRef] [PubMed]
- De Bock, M.; Wang, N.; Decrock, E.; Bol, M.; Gadicherla, A.K.; Culot, M.; Cecchelli, R.; Bultynck, G.; Leybaert, L. Endothelial calcium dynamics, connexin channels and blood-brain barrier function. Prog. Neurobiol. 2013, 108, 1–20. [Google Scholar] [CrossRef]
- Abudara, V.; Roux, L.; Dallerac, G.; Matias, I.; Dulong, J.; Mothet, J.P.; Rouach, N.; Giaume, C. Activated microglia impairs neuroglial interaction by opening Cx43 hemichannels in hippocampal astrocytes. Glia 2015, 63, 795–811. [Google Scholar] [CrossRef]
- Batova, S.; DeWever, J.; Godfraind, T.; Balligand, J.L.; Dessy, C.; Feron, O. The calcium channel blocker amlodipine promotes the unclamping of eNOS from caveolin in endothelial cells. Cardiovasc. Res. 2006, 71, 478–485. [Google Scholar] [CrossRef]
- Lukic-Panin, V.; Kamiya, T.; Zhang, H.; Hayashi, T.; Tsuchiya, A.; Sehara, Y.; Deguchi, K.; Yamashita, T.; Abe, K. Prevention of neuronal damage by calcium channel blockers with antioxidative effects after transient focal ischemia in rats. Brain Res. 2007, 1176, 143–150. [Google Scholar] [CrossRef]
- Miziak, B.; Blaszczyk, B.; Czuczwar, S.J. Some Candidate Drugs for Pharmacotherapy of Alzheimer’s Disease. Pharmaceuticals 2021, 14, 458. [Google Scholar] [CrossRef]
- Sharma, N.; Jamwal, S.; Kumar, P. Beneficial effect of antidepressants against rotenone induced Parkinsonism like symptoms in rats. Pathophysiology 2016, 23, 123–134. [Google Scholar] [CrossRef]
- Fidilio, A.; Grasso, M.; Turnaturi, R.; Caruso, G.; Spitale, F.M.; Vicario, N.; Parenti, R.; Spoto, S.; Musso, N.; Marrazzo, A.; et al. The Multimodal MOPr/DOPr Agonist LP2 Reduces Allodynia in Chronic Constriction Injured Rats by Rescue of TGF-beta1 Signalling. Front. Pharm. 2021, 12, 749365. [Google Scholar] [CrossRef] [PubMed]
- Diem, R.; Sattler, M.B.; Merkler, D.; Demmer, I.; Maier, K.; Stadelmann, C.; Ehrenreich, H.; Bahr, M. Combined therapy with methylprednisolone and erythropoietin in a model of multiple sclerosis. Brain 2005, 128, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Viscomi, M.T.; Florenzano, F.; Latini, L.; Amantea, D.; Bernardi, G.; Molinari, M. Methylprednisolone treatment delays remote cell death after focal brain lesion. Neuroscience 2008, 154, 1267–1282. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xie, K.; Wang, C.; Bi, J. Oxidative stress induced by lipid peroxidation is related with inflammation of demyelination and neurodegeneration in multiple sclerosis. Eur. Neurol. 2014, 72, 249–254. [Google Scholar] [CrossRef]
- Zhang, H.K.; Ye, Y.; Zhao, Z.N.; Li, K.J.; Du, Y.; Hu, Q.M.; He, J.F. Neuroprotective effects of gypenosides in experimental autoimmune optic neuritis. Int. J. Ophthalmol. 2017, 10, 541–549. [Google Scholar] [CrossRef]
- Cheadle, C.; Vawter, M.P.; Freed, W.J.; Becker, K.G. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 2003, 5, 73–81. [Google Scholar] [CrossRef]
- Care, M.A.; Barrans, S.; Worrillow, L.; Jack, A.; Westhead, D.R.; Tooze, R.M. A microarray platform-independent classification tool for cell of origin class allows comparative analysis of gene expression in diffuse large B-cell lymphoma. PLoS ONE 2013, 8, e55895. [Google Scholar] [CrossRef]
- Wang, J.; Coombes, K.R.; Highsmith, W.E.; Keating, M.J.; Abruzzo, L.V. Differences in gene expression between B-cell chronic lymphocytic leukemia and normal B cells: A meta-analysis of three microarray studies. Bioinformatics 2004, 20, 3166–3178. [Google Scholar] [CrossRef]
- Reddy, T.B.; Riley, R.; Wymore, F.; Montgomery, P.; DeCaprio, D.; Engels, R.; Gellesch, M.; Hubble, J.; Jen, D.; Jin, H.; et al. TB database: An integrated platform for tuberculosis research. Nucleic Acids Res. 2009, 37, D499–D508. [Google Scholar] [CrossRef]
- Le Cao, K.A.; Rohart, F.; McHugh, L.; Korn, O.; Wells, C.A. YuGene: A simple approach to scale gene expression data derived from different platforms for integrated analyses. Genomics 2014, 103, 239–251. [Google Scholar] [CrossRef]
- Chen, Q.R.; Song, Y.K.; Wei, J.S.; Bilke, S.; Asgharzadeh, S.; Seeger, R.C.; Khan, J. An integrated cross-platform prognosis study on neuroblastoma patients. Genomics 2008, 92, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Yasrebi, H.; Sperisen, P.; Praz, V.; Bucher, P. Can survival prediction be improved by merging gene expression data sets? PLoS ONE 2009, 4, e7431. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, R.; El-Ashram, S.; Bie, R.; Dawood, H.; Kos, A. Clustering by fast search and merge of local density peaks for gene expression microarray data. Sci. Rep. 2017, 7, 45602. [Google Scholar] [CrossRef] [PubMed]
- Cheadle, C.; Cho-Chung, Y.S.; Becker, K.G.; Vawter, M.P. Application of z-score transformation to Affymetrix data. Appl. Bioinform. 2003, 2, 209–217. [Google Scholar]
- Feng, C.; Wu, J.; Yang, F.; Qiu, M.; Hu, S.; Guo, S.; Wu, J.; Ying, X.; Wang, J. Expression of Bcl-2 is a favorable prognostic biomarker in lung squamous cell carcinoma. Oncol. Lett. 2018, 15, 6925–6930. [Google Scholar] [CrossRef]
- Kang, C.; Huo, Y.; Xin, L.; Tian, B.; Yu, B. Feature selection and tumor classification for microarray data using relaxed Lasso and generalized multi-class support vector machine. J. Theor. Biol. 2019, 463, 77–91. [Google Scholar] [CrossRef]
| N° | Dataset | Platform | Org | Samples | Sample Origin | Platform | NDC | ALS | Ref |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GSE26927 | Microarray | Human | 19 | Spinal cord | GPL6255 | 9 | 10 | [31] |
| 2 | GSE76220 | RNA-seq | Human | 22 | Spinal cord MNs | GPL9115 | 9 | 13 | [32] |
| Samples | Genes | R |
|---|---|---|
| Unique Genes significantly positively correlated (GSPC) to GJA1 | 2542 | 0.50 < R < 0.91 |
| Unique Genes significantly negatively correlated (GSNC) to GJA1 | 3110 | 0.50 < R < 0.96 |
| Signatures | N° | Cells and Processes | Source | Unique Genes |
|---|---|---|---|---|
| Neural cells | 1 | Astrocyte | GSE67835 | 177 |
| 2 | Microglia | GSE67835 | 93 | |
| 3 | Neuron | GSE67835 | 974 | |
| 4 | Oligodendrocyte | GSE67835 | 95 | |
| 5 | Pericyte immune activated | GSE46236 | 206 | |
| Immune cells | 6 | M1 macrophages | GSE5099 | 674 |
| 7 | M2 macrophages | GSE5099 | 132 | |
| 8 | Natural killer (KN) | GSE22886 | 114 | |
| 9 | TH1 | GSE22886 | 191 | |
| 10 | TH2 | GSE22886 | 85 | |
| 11 | Endothelial cells | GSE67835 | 55 | |
| 12 | CTLs | GSE22886 | 62 | |
| Biological processes | 13 | Brain microvessels | GSE22886 | 291 |
| 14 | Host virus interaction | KEGG | 279 | |
| 15 | Inflammatory response | KEGG | 704 | |
| 16 | Synaptic transmission genes | GO:0098814 | 45 | |
| 17 | Synaptic vesicle cycle | KEGG | 78 |
| N° | Drugs | Score Similarity | Z-Score | Combined Score (CS) | Mechanism of Action (MOA) | Indication(s) |
|---|---|---|---|---|---|---|
| 1 | Amlodipine | −0.026 | 1.84 | −7.49 | Calcium channel blocker | Hypertension |
| 2 | Sertraline | −0.021 | 1.78 | −1.83 | Selective serotonin reuptake inhibitor | Depression; Obsessive–compulsive disorder; panic disorder; post-traumatic stress disorder |
| 3 | Prednisolone | −0.021 | 1.81 | −1.57 | Synthetic glucocorticoid with anti-inflammatory and immunomodulatory effect | Adrenergic agent; anti-inflammatory drug; antineoplastic agent; immunosuppressive agent |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vicario, N.; Castrogiovanni, P.; Imbesi, R.; Giallongo, S.; Mannino, G.; Furno, D.L.; Giuffrida, R.; Zappalà, A.; Li Volti, G.; Tibullo, D.; et al. GJA1/CX43 High Expression Levels in the Cervical Spinal Cord of ALS Patients Correlate to Microglia-Mediated Neuroinflammatory Profile. Biomedicines 2022, 10, 2246. https://doi.org/10.3390/biomedicines10092246
Vicario N, Castrogiovanni P, Imbesi R, Giallongo S, Mannino G, Furno DL, Giuffrida R, Zappalà A, Li Volti G, Tibullo D, et al. GJA1/CX43 High Expression Levels in the Cervical Spinal Cord of ALS Patients Correlate to Microglia-Mediated Neuroinflammatory Profile. Biomedicines. 2022; 10(9):2246. https://doi.org/10.3390/biomedicines10092246
Chicago/Turabian StyleVicario, Nunzio, Paola Castrogiovanni, Rosa Imbesi, Sebastiano Giallongo, Giuliana Mannino, Debora Lo Furno, Rosario Giuffrida, Agata Zappalà, Giovanni Li Volti, Daniele Tibullo, and et al. 2022. "GJA1/CX43 High Expression Levels in the Cervical Spinal Cord of ALS Patients Correlate to Microglia-Mediated Neuroinflammatory Profile" Biomedicines 10, no. 9: 2246. https://doi.org/10.3390/biomedicines10092246
APA StyleVicario, N., Castrogiovanni, P., Imbesi, R., Giallongo, S., Mannino, G., Furno, D. L., Giuffrida, R., Zappalà, A., Li Volti, G., Tibullo, D., Di Rosa, M., & Parenti, R. (2022). GJA1/CX43 High Expression Levels in the Cervical Spinal Cord of ALS Patients Correlate to Microglia-Mediated Neuroinflammatory Profile. Biomedicines, 10(9), 2246. https://doi.org/10.3390/biomedicines10092246











