Prevention of Initial Bacterial Attachment by Osteopontin and Other Bioactive Milk Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Purification of Milk Proteins
2.2. Bacterial Culture
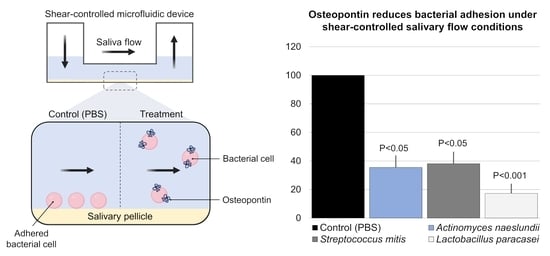
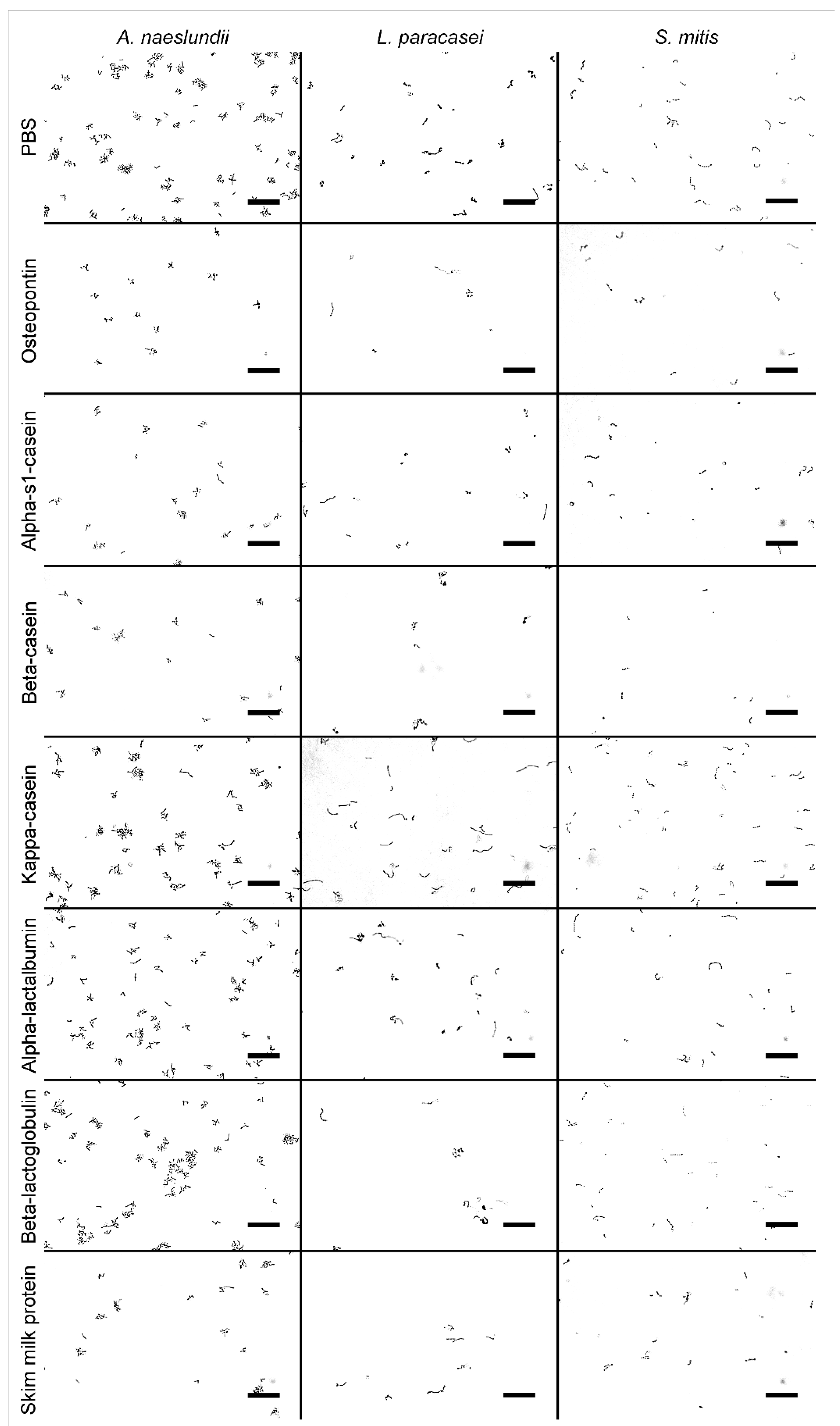
2.3. Bacterial Adhesion
2.4. Quantification of Bacterial Adhesion
2.5. Statistical Analysis
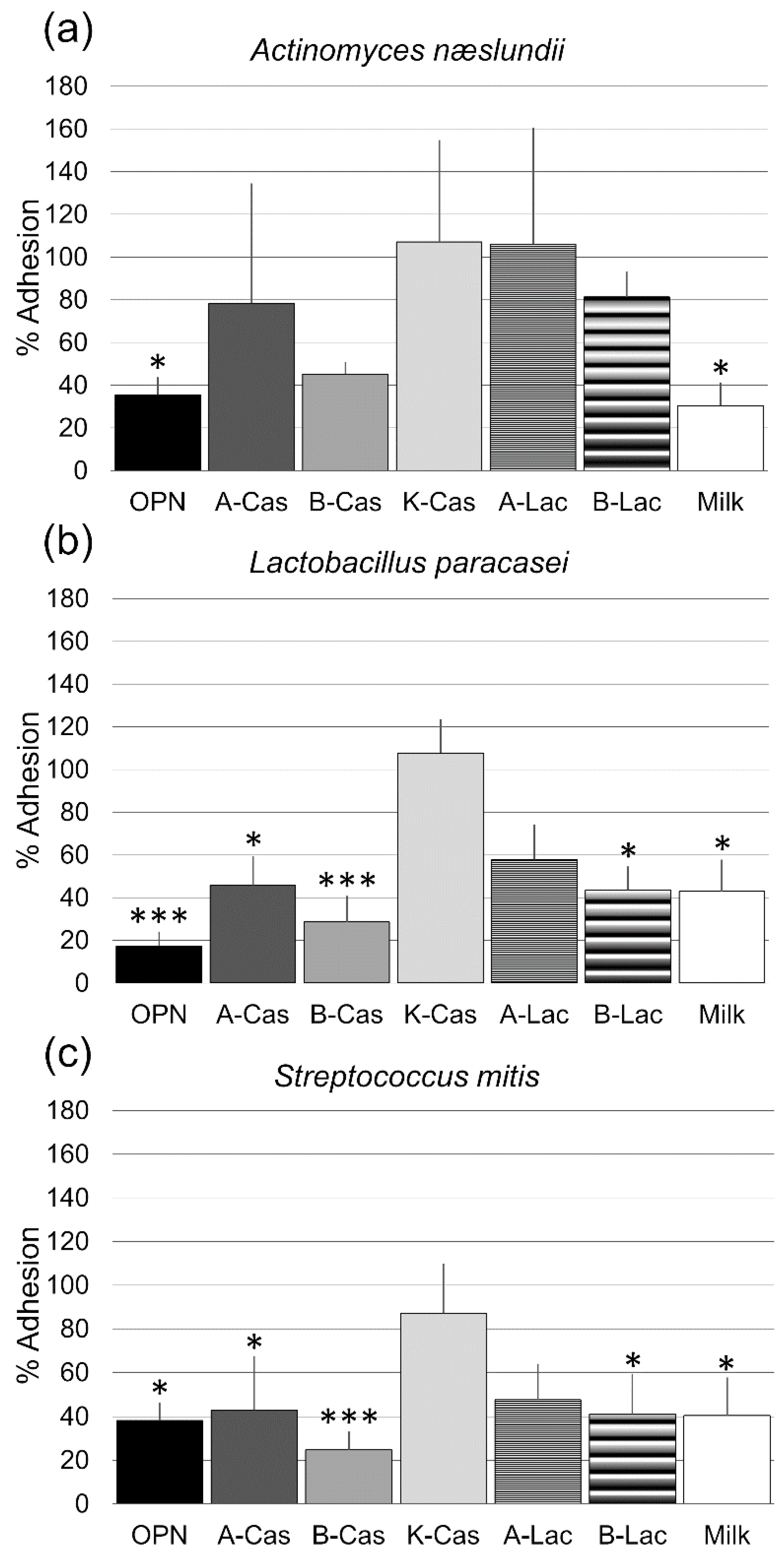
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruun, S.; Jacobsen, L.N.; Ze, X.; Husby, S.; Ueno, H.M.; Nojiri, K.; Kobayashi, S.; Kwon, J.; Liu, X.; Yan, S.; et al. Osteopontin levels in human milk vary across countries and within lactation period: Data from a multicenter study. J. Pediatr. Gastroenterol. Nutr. 2018, 67, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Icer, M.A.; Gezmen-Karadag, M. The multiple functions and mechanisms of osteopontin. Clin. Biochem. 2018, 59, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Schack, L.; Stapulionis, R.; Christensen, B.; Kofod-Olsen, E.; Skov Sørensen, U.B.; Vorup-Jensen, T.; Sørensen, E.S.; Höllsberg, P. Osteopontin enhances phagocytosis through a novel osteopontin receptor, the alphaXbeta2 integrin. J. Immunol. 2009, 182, 6943–6950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, M.F.; Zeng, G.; Neu, T.R.; Meyer, R.L.; Baelum, V.; Schlafer, S. Osteopontin adsorption to Gram-positive cells reduces adhesion forces and attachment to surfaces under flow. J. Oral Microbiol. 2017, 9, 1379826. [Google Scholar] [CrossRef]
- Schlafer, S.; Meyer, R.L.; Sutherland, D.S.; Städler, B. Effect of osteopontin on the initial adhesion of dental bacteria. J. Nat. Prod. 2012, 75, 2108–2112. [Google Scholar] [CrossRef]
- Schlafer, S.; Raarup, M.K.; Wejse, P.L.; Nyvad, B.; Städler, B.M.; Sutherland, D.S.; Birkedal, H.; Meyer, R.L. Osteopontin reduces biofilm formation in a multi-species model of dental biofilm. PLoS ONE 2012, 7, e41534. [Google Scholar] [CrossRef]
- Bernabe, E.; Marcenes, W.; Hernandez, C.R.; Bailey, J.; Abreu, L.G.; Alipour, V.; Amini, S.; Arabloo, J.; Arefi, Z.; Arora, A.; et al. Global, regional, and national levels and trends in burden of oral conditions from 1990 to 2017: A systematic analysis for the global burden of disease 2017 study. J. Dent. Res. 2020, 99, 362–373. [Google Scholar] [CrossRef]
- Jafer, M.; Patil, S.; Hosmani, J.; Bhandi, S.H.; Chalisserry, E.P.; Anil, S. Chemical plaque control strategies in the prevention of biofilm-associated oral diseases. J. Contemp. Dent. Pract. 2016, 17, 337–343. [Google Scholar] [CrossRef]
- Goh, C.E.; Trinh, P.; Colombo, P.C.; Genkinger, J.M.; Mathema, B.; Uhlemann, A.-C.; LeDuc, C.; Leibel, R.; Rosenbaum, M.; Paster, B.J.; et al. Association between nitrate-reducing oral bacteria and cardiometabolic outcomes: Results from ORIGINS. J. Am. Heart Assoc. 2019, 8, e013324. [Google Scholar] [CrossRef]
- Aimutis, W.R. Bioactive properties of milk proteins with particular focus on anticariogenesis. J. Nutr. 2004, 134, 989S–995S. [Google Scholar] [CrossRef] [Green Version]
- Danielsson Niemi, L.; Hernell, O.; Johansson, I. Human milk compounds inhibiting adhesion of mutans streptococci to host ligand-coated hydroxyapatite in vitro. Caries Res. 2009, 43, 171–178. [Google Scholar] [CrossRef]
- Cheaib, Z.; Rakmathulina, E.; Lussi, A.; Eick, S. Impact of Acquired Pellicle Modification on Adhesion of Early Colonizers. Caries Res. 2015, 49, 626–632. [Google Scholar] [CrossRef] [Green Version]
- Oho, T.; Mitoma, M.; Koga, T. Functional domain of bovine milk lactoferrin which inhibits the adherence of Streptococcus mutans cells to a salivary film. Infect. Immun. 2002, 70, 5279–5282. [Google Scholar] [CrossRef] [Green Version]
- Neeser, J.R.; Golliard, M.; Woltz, A.; Rouvet, M.; Dillmann, M.L.; Guggenheim, B. In vitro modulation of oral bacterial adhesion to saliva-coated hydroxyapatite beads by milk casein derivatives. Oral Microbiol. Immunol. 1994, 9, 193–201. [Google Scholar] [CrossRef]
- Schüpbach, P.; Neeser, J.R.; Golliard, M.; Rouvet, M.; Guggenheim, B. Incorporation of caseinoglycomacropeptide and caseinophosphopeptide into the salivary pellicle inhibits adherence of mutans streptococci. J. Dent. Res. 1996, 75, 1779–1788. [Google Scholar] [CrossRef] [Green Version]
- Vacca Smith, A.M.; Bowen, W.H. The effects of milk and kappa-casein on salivary pellicle formed on hydroxyapatite discs in situ. Caries Res. 2000, 34, 88–93. [Google Scholar] [CrossRef]
- Benoit, M.R.; Conant, C.G.; Ionescu-Zanetti, C.; Schwartz, M.; Matin, A. New device for high-throughput viability screening of flow biofilms. Appl. Environ. Microbiol. 2010, 76, 4136–4142. [Google Scholar] [CrossRef] [Green Version]
- Dawes, C.; Watanabe, S.; Biglow-Lecomte, P.; Dibdin, G.H. Estimation of the velocity of the salivary film at some different locations in the mouth. J. Dent. Res. 1989, 68, 1479–1482. [Google Scholar] [CrossRef]
- Rasmussen, L.K.; Petersen, T.E. Purification of disulphide-linked alpha s2- and kappa-casein from bovine milk. J. Dairy Res. 1991, 58, 187–193. [Google Scholar] [CrossRef]
- Rasmussen, L.K.; Højrup, P.; Petersen, T.E. The multimeric structure and disulfide-bonding pattern of bovine kappa-casein. Eur. J. Biochem. 1992, 207, 215–222. [Google Scholar] [CrossRef]
- Turck, D.; Castenmiller, J.; de Henauw, S.; Hirsch-Ernst, K.I.; Kearney, J.; Maciuk, A.; Mangelsdorf, I.; McArdle, H.J.; Naska, A.; Pelaez, C.; et al. Safety of bovine milk osteopontin as a Novel food pursuant to Regulation (EU) 2015/2283. EFSA J. 2022, 20, e07137. [Google Scholar] [CrossRef]
- de Jong, M.H.; van der Hoeven, J.S.; van OS, J.H.; Olijve, J.H. Growth of oral Streptococcus species and Actinomyces viscosus in human saliva. Appl. Environ. Microbiol. 1984, 47, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Daims, H.; Lücker, S.; Wagner, M. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 2006, 8, 200–213. [Google Scholar] [CrossRef]
- R Core Team. The R Project for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Reynolds, E.C.; Wong, A. Effect of adsorbed protein on hydroxyapatite zeta potential and Streptococcus mutans adherence. Infect. Immun. 1983, 39, 1285–1290. [Google Scholar] [CrossRef] [Green Version]
- Vacca-Smith, A.M.; van Wuyckhuyse, B.C.; Tabak, L.A.; Bowen, W.H. The effect of milk and casein proteins on the adherence of Streptococcus mutans to saliva-coated hydroxyapatite. Arch. Oral Biol. 1994, 39, 1063–1069. [Google Scholar] [CrossRef]
- Latousakis, D.; MacKenzie, D.A.; Telatin, A.; Juge, N. Serine-rich repeat proteins from gut microbes. Gut Microbes 2020, 11, 102–117. [Google Scholar] [CrossRef]
- Busscher, H.J.; van der Mei, H.C. Physico-chemical interactions in initial microbial adhesion and relevance for biofilm formation. Adv. Dent. Res. 1997, 11, 24–32. [Google Scholar] [CrossRef]
- McNab, R.; Jenkinson, H.F. Gene disruption identifies a 290 kDa cell-surface polypeptide conferring hydrophobicity and coaggregation properties in Streptococcus gordonii. Mol. Microbiol. 1992, 6, 2939–2949. [Google Scholar] [CrossRef]
- Morris, E.J.; Ganeshkumar, N.; McBride, B.C. Cell surface components of Streptococcus sanguis: Relationship to aggregation, adherence, and hydrophobicity. J. Bacteriol. 1985, 164, 255–262. [Google Scholar] [CrossRef] [Green Version]
- Jenkinson, H.F. Genetic analysis of adherence by oral streptococci. J. Ind. Microbiol. 1995, 15, 186–192. [Google Scholar] [CrossRef]
- Głąb, T.K.; Boratyński, J. Potential of casein as a carrier for biologically active agents. Top. Curr. Chem. 2017, 375, 71. [Google Scholar] [CrossRef] [Green Version]
- Hallberg, K.; Holm, C.; Ohman, U.; Strömberg, N. Actinomyces naeslundii displays variant fimP and fimA fimbrial subunit genes corresponding to different types of acidic proline-rich protein and beta-linked galactosamine binding specificity. Infect. Immun. 1998, 66, 4403–4410. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Khah, M.K.; Slavnic, S.; Johansson, I.; Strömberg, N. Different type 1 fimbrial genes and tropisms of commensal and potentially pathogenic Actinomyces spp. with different salivary acidic proline-rich protein and statherin ligand specificities. Infect. Immun. 2001, 69, 7224–7233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbons, R.J.; Hay, D.I.; Cisar, J.O.; Clark, W.B. Adsorbed salivary proline-rich protein 1 and statherin: Receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces. Infect. Immun. 1988, 56, 2990–2993. [Google Scholar] [CrossRef] [Green Version]
- Guggenheim, B.; Schmid, R.; Aeschlimann, J.M.; Berrocal, R.; Neeser, J.R. Powdered milk micellar casein prevents oral colonization by Streptococcus sobrinus and dental caries in rats: A basis for the caries-protective effect of dairy products. Caries Res. 1999, 33, 446–454. [Google Scholar]
- Pereira, P.C. Milk nutritional composition and its role in human health. Nutrition 2014, 30, 619–627. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kristensen, M.F.; Sørensen, E.S.; Del Rey, Y.C.; Schlafer, S. Prevention of Initial Bacterial Attachment by Osteopontin and Other Bioactive Milk Proteins. Biomedicines 2022, 10, 1922. https://doi.org/10.3390/biomedicines10081922
Kristensen MF, Sørensen ES, Del Rey YC, Schlafer S. Prevention of Initial Bacterial Attachment by Osteopontin and Other Bioactive Milk Proteins. Biomedicines. 2022; 10(8):1922. https://doi.org/10.3390/biomedicines10081922
Chicago/Turabian StyleKristensen, Mathilde Frost, Esben Skipper Sørensen, Yumi Chokyu Del Rey, and Sebastian Schlafer. 2022. "Prevention of Initial Bacterial Attachment by Osteopontin and Other Bioactive Milk Proteins" Biomedicines 10, no. 8: 1922. https://doi.org/10.3390/biomedicines10081922
APA StyleKristensen, M. F., Sørensen, E. S., Del Rey, Y. C., & Schlafer, S. (2022). Prevention of Initial Bacterial Attachment by Osteopontin and Other Bioactive Milk Proteins. Biomedicines, 10(8), 1922. https://doi.org/10.3390/biomedicines10081922