Integration of miRNA:mRNA Co-Expression Revealed Crucial Mechanisms Modulated in Immunogenic Cancer Cell Death
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Reagents
2.3. RNA Purification, Sequencing, and Data Analysis
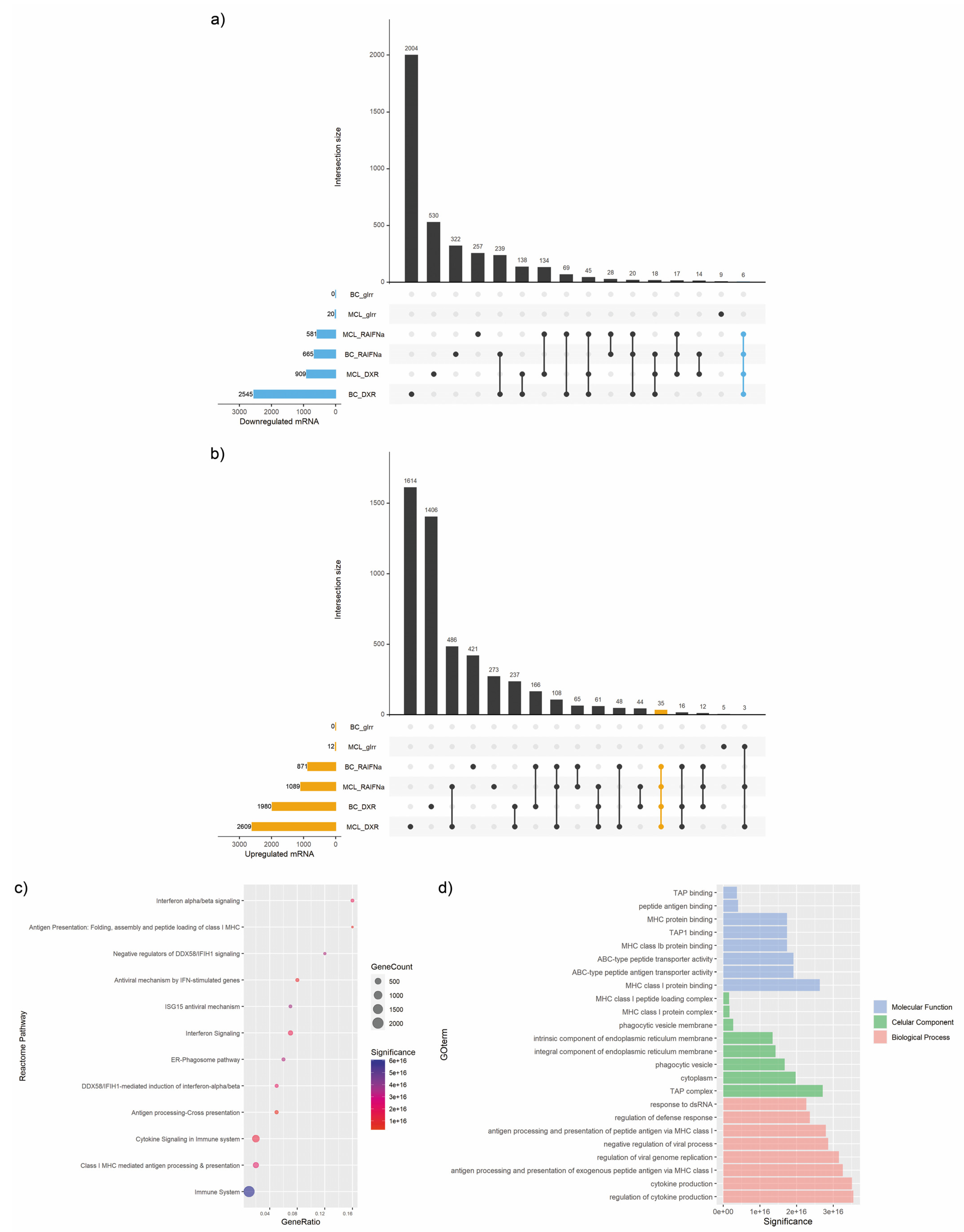
2.4. UpSet Plot
2.5. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR (RT-qPCR)
2.6. Performance and Visualization of Enrichment Analysis of Differentially Expressed Genes
2.7. Visualization and Analysis of miRNA—Targets Interactions
2.8. Gene Expression Analysis from GEO Database
2.9. Flow Cytometry
2.10. Oligonucleotides Transfection
2.11. Data Availability
2.12. Statistics
3. Results
3.1. miRNA Expression Signature in Immunogenic Cancer Cell Death
3.2. mRNA Expression Signature in Immunogenic Cancer Cell Death
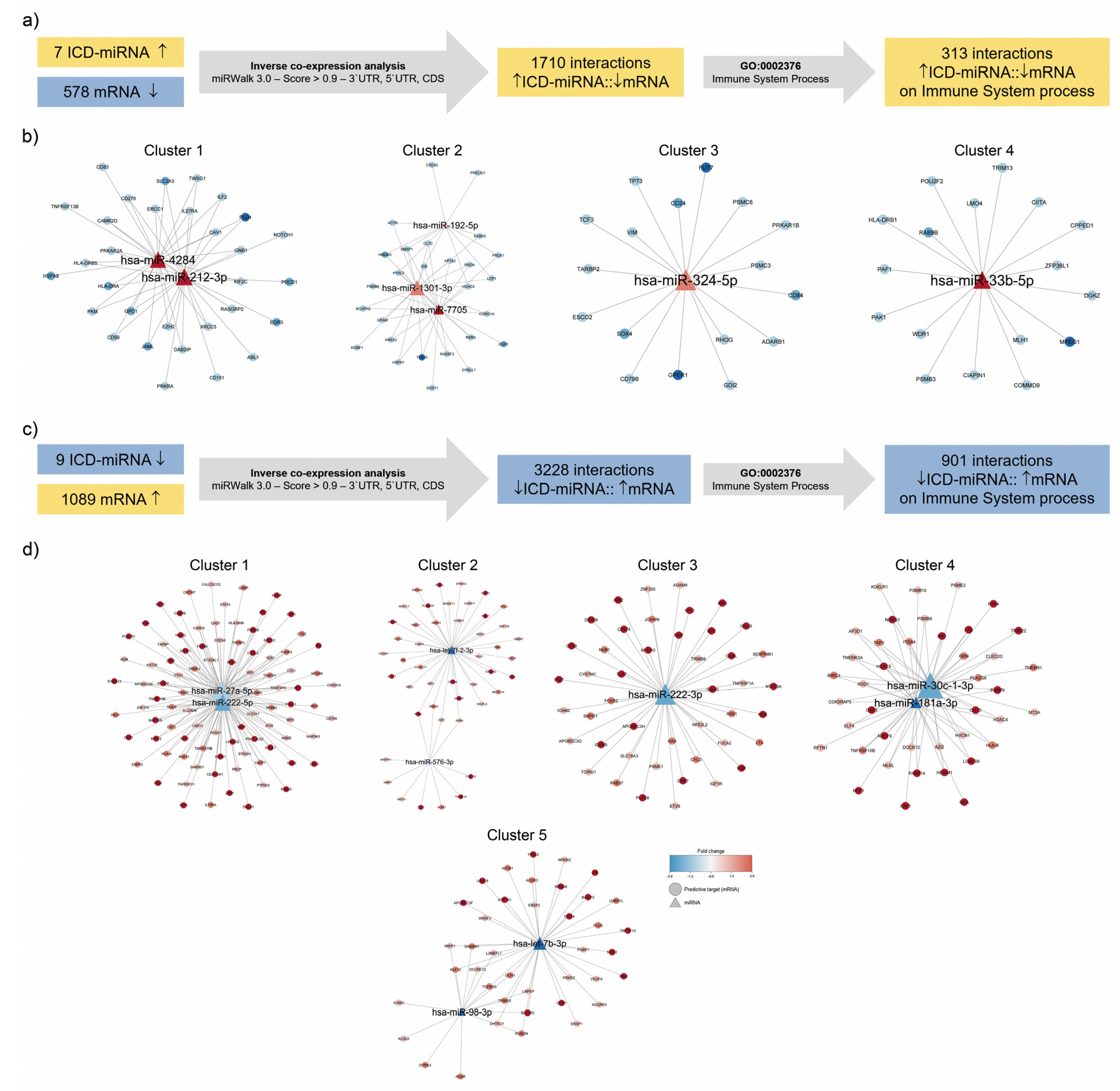
3.3. Identification of Immunogenic miRNA–mRNA Pairs by Inverse Co-Expression Networks of Immune System-Associated Genes
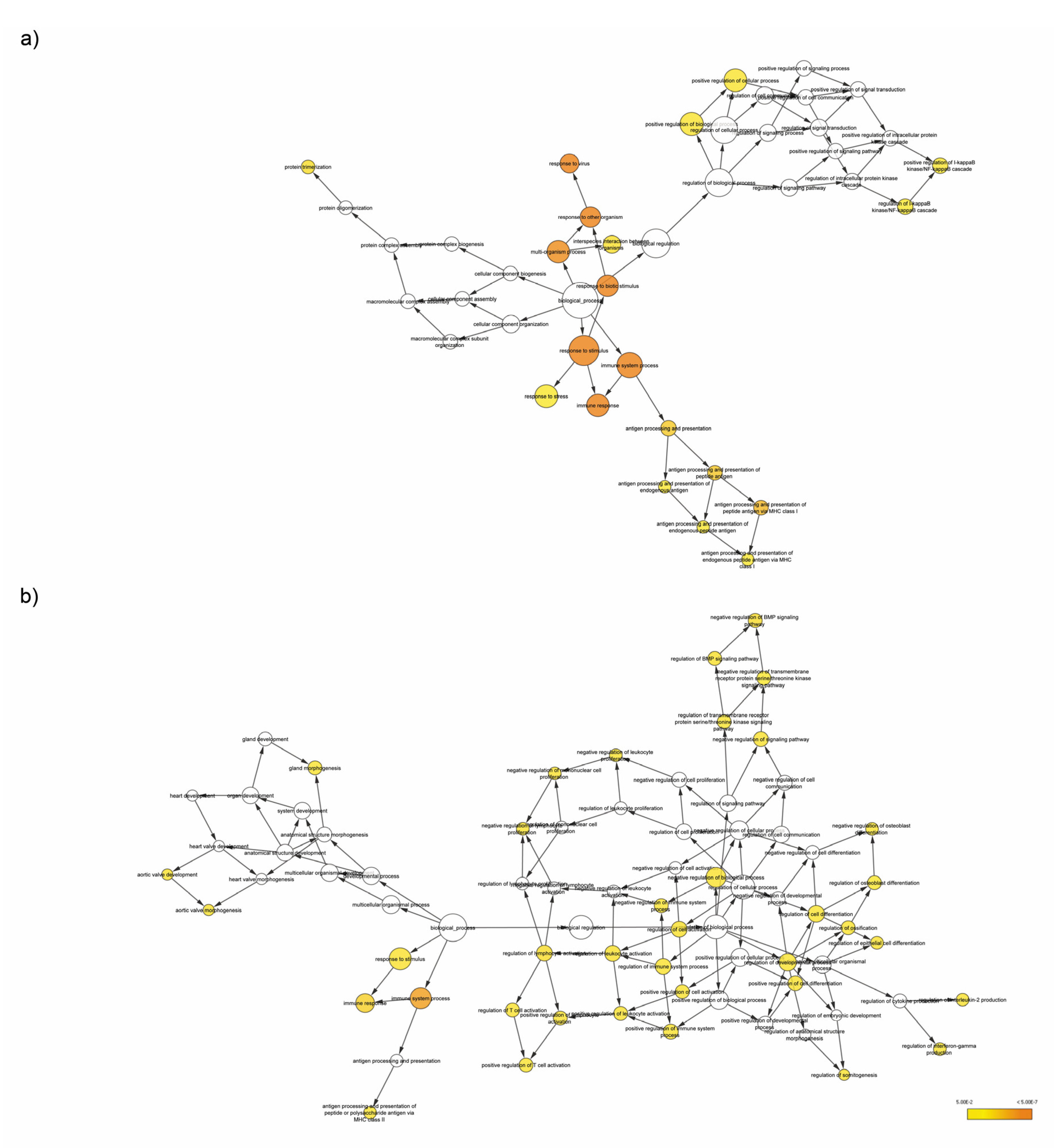
3.4. Enrichment Analysis of Target Genes of miRNA Members in Most Representative Clusters
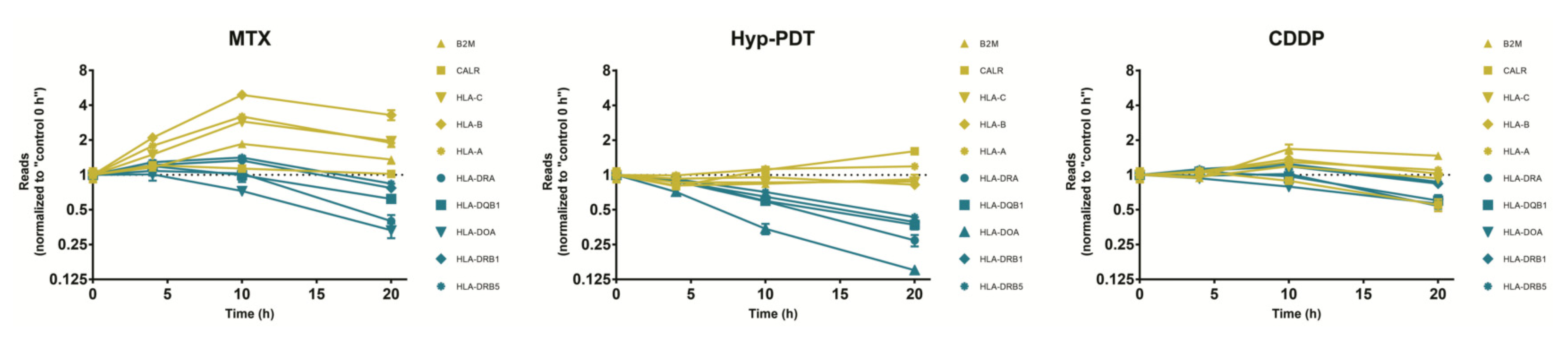
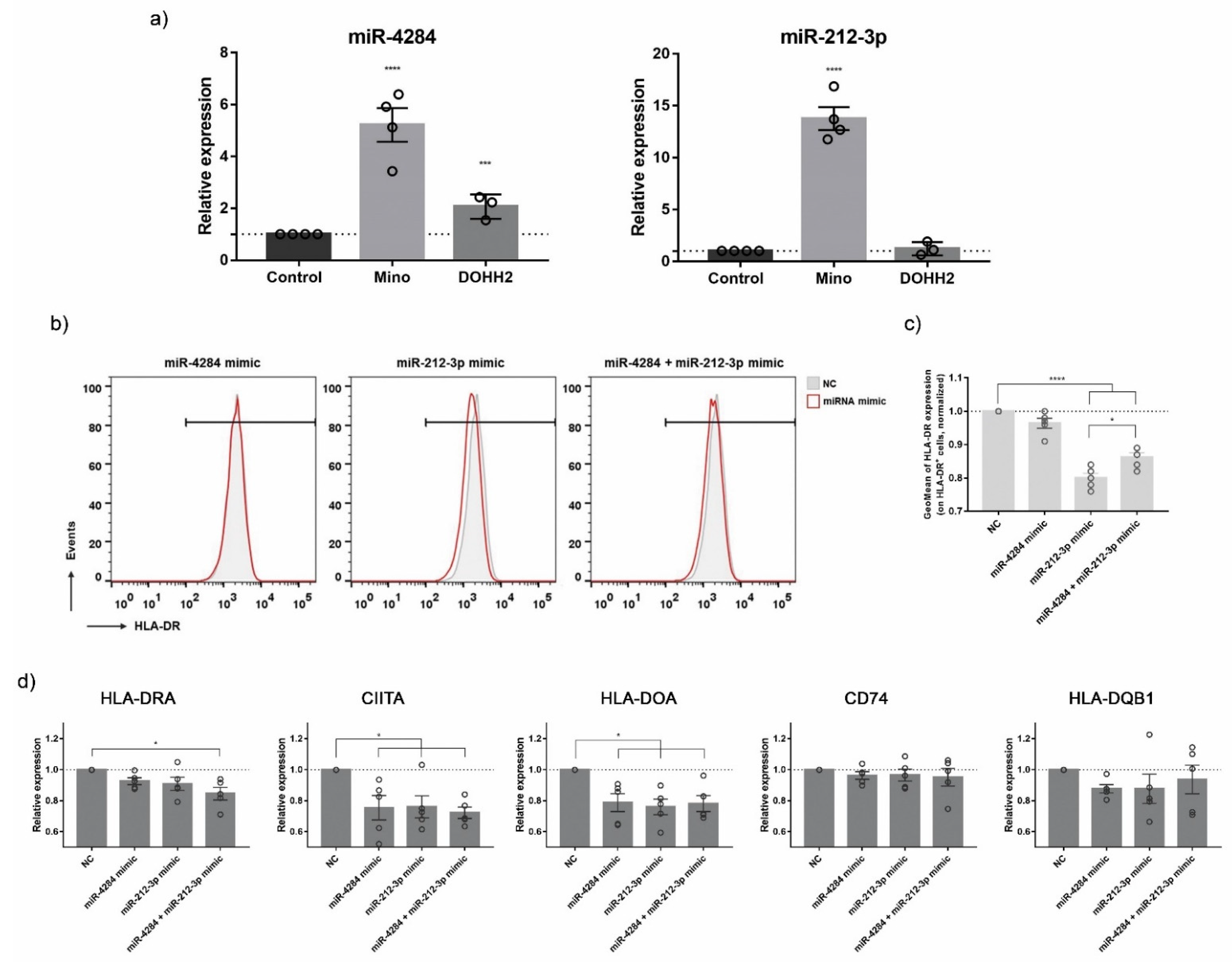
3.5. Validation of ICD-Mediated Decrease of Class II MHC Antigen Presentation Pathway via Upregulation of miR-212-3p
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dunn, G.; Old, L.; Schreiber, R.D. The Three Es of Cancer Immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer Immunoediting: From Immunosurveillance to Tumor Escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Paladini, L.; Fabris, L.; Bottai, G.; Raschioni, C.; Calin, G.A.; Santarpia, L. Targeting MicroRNAs as Key Modulators of Tumor Immune Response. J. Exp. Clin. Cancer Res. 2016, 35, 103. [Google Scholar] [CrossRef]
- Xing, Y.; Ruan, G.; Ni, H.; Qin, H.; Chen, S.; Gu, X.; Shang, J.; Zhou, Y.; Tao, X.; Zheng, L. Tumor Immune Microenvironment and Its Related MiRNAs in Tumor Progression. Front. Immunol. 2021, 12, 624725. [Google Scholar] [CrossRef]
- Hirschberger, S.; Hinske, L.C.; Kreth, S. MiRNAs: Dynamic Regulators of Immune Cell Functions in Inflammation and Cancer. Cancer Lett. 2018, 431, 11–21. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Lamberti, M.; Nigro, A.; Mentucci, F.; Rumie Vittar, N.; Casolaro, V.; Dal Col, J. Dendritic Cells and Immunogenic Cancer Cell Death: A Combination for Improving Antitumor Immunity. Pharmaceutics 2020, 12, 256. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Kepp, O.; Zitvogel, L. Immunogenic Cell Death in Cancer Therapy. Annu. Rev. Immunol. 2013, 31, 51–72. [Google Scholar] [CrossRef]
- Galluzzi, L.; Buqué, A.; Kepp, O.; Zitvogel, L.; Kroemer, G. Immunogenic Cell Death in Cancer and Infectious Disease. Nat. Rev. Immunol. 2017, 17, 97–111. [Google Scholar] [CrossRef]
- Bezu, L.; Gomes-de-Silva, L.; Dewitte, H.; Breckpot, K.; Fucikova, J.; Spisek, R.; Galluzzi, L.; Kepp, O.; Kroemer, G. Combinatorial Strategies for the Induction of Immunogenic Cell Death. Front. Immunol. 2015, 6, 187. [Google Scholar] [CrossRef] [PubMed]
- Legrand, A.J.; Konstantinou, M.; Goode, E.F.; Meier, P. The Diversification of Cell Death and Immunity: Memento Mori. Mol. Cell 2019, 76, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Krysko, D.; Garg, A.; Kaczmarek, A.; Krysko, O.; Agostinis, P.; Vandenabeele, P. Immunogenic Cell Death and DAMPs in Cancer Therapy. Nat. Rev. Cancer 2012, 12, 860–875. [Google Scholar] [CrossRef]
- Garg, A.D.; Galluzzi, L.; Apetoh, L.; Baert, T.; Birge, R.B.; Bravo-San Pedro, J.M.; Breckpot, K.; Brough, D.; Chaurio, R.; Cirone, M.; et al. Molecular and Translational Classifications of DAMPs in Immunogenic Cell Death. Front. Immunol. 2015, 6, 588. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Nowis, D.; Golab, J.; Vandenabeele, P.; Krysko, D.; Agostinis, P. Immunogenic Cell Death, DAMPs and Anticancer Therapeutics: An Emerging Amalgamation. Biochim. Biophys. Acta 2010, 1805, 53–71. [Google Scholar] [CrossRef]
- Rufo, N.; Korovesis, D.; van Eygen, S.; Derua, R.; Garg, A.D.; Finotello, F.; Vara-Perez, M.; Rožanc, J.; Dewaele, M.; de Witte, P.A.; et al. Stress-Induced Inflammation Evoked by Immunogenic Cell Death Is Blunted by the IRE1α Kinase Inhibitor KIRA6 through HSP60 Targeting. Cell Death Differ. 2022, 29, 230–245. [Google Scholar] [CrossRef]
- Lamberti, M.J.; Nigro, A.; Casolaro, V.; Rumie Vittar, N.B.; Dal Col, J. Damage-Associated Molecular Patterns Modulation by MicroRNA: Relevance on Immunogenic Cell Death and Cancer Treatment Outcome. Cancers 2021, 13, 2566. [Google Scholar] [CrossRef]
- Nigro, A.; Montico, B.; Casolaro, V.; Dal Col, J. A Novel Dendritic Cell-Based Vaccination Protocol to Stimulate Immunosurveillance of Aggressive Cancers. Methods Mol. Biol. 2019, 1884, 317–333. [Google Scholar] [CrossRef]
- Montico, B.; Nigro, A.; Casolaro, V.; Dal Col, J. Immunogenic Apoptosis as a Novel Tool for Anticancer Vaccine Development. Int. J. Mol. Sci. 2018, 19, 594. [Google Scholar] [CrossRef]
- Montico, B.; Lapenta, C.; Ravo, M.; Martorelli, D.; Muraro, E.; Zeng, B.; Comaro, E.; Spada, M.; Donati, S.; Santini, S.; et al. Exploiting a New Strategy to Induce Immunogenic Cell Death to Improve Dendritic Cell-Based Vaccines for Lymphoma Immunotherapy. Oncoimmunology 2017, 6, e1356964. [Google Scholar] [CrossRef]
- Hashim, A.; Rizzo, F.; Marchese, G.; Ravo, M.; Tarallo, R.; Nassa, G.; Giurato, G.; Santamaria, G.; Cordella, A.; Cantarella, C.; et al. RNA Sequencing Identifies Specific PIWI-Interacting Small Non-Coding RNA Expression Patterns in Breast Cancer. Oncotarget 2014, 5, 9901–9910. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome. Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Panero, R.; Rinaldi, A.; Memoli, D.; Nassa, G.; Ravo, M.; Rizzo, F.; Tarallo, R.; Milanesi, L.; Weisz, A.; Giurato, G. ISmaRT: A Toolkit for a Comprehensive Analysis of Small RNA-Seq Data. Bioinformatics 2017, 33, btw734. [Google Scholar] [CrossRef] [PubMed]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R Package for the Visualization of Intersecting Sets and Their Properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. G:Profiler—A Web Server for Functional Interpretation of Gene Lists (2016 Update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef]
- Villanueva, R.A.M.; Chen, Z.J. Ggplot2: Elegant Graphics for Data Analysis (2nd Ed.). Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Sticht, C.; de La Torre, C.; Parveen, A.; Gretz, N. MiRWalk: An Online Resource for Prediction of MicroRNA Binding Sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- León, L.E.; Calligaris, S.D. Visualization and Analysis of MiRNA–Targets Interactions Networks; Humana Press: New York, NY, USA, 2017; pp. 209–220. [Google Scholar]
- Wang, J.; Zhong, J.; Chen, G.; Li, M.; Wu, F.; Pan, Y. ClusterViz: A Cytoscape APP for Cluster Analysis of Biological Network. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015, 12, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape Plugin to Assess Overrepresentation of Gene Ontology Categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Newman, M. Modularity and Community Structure in Networks. PNAS 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [PubMed]
- Sheu-Gruttadauria, J.; Pawlica, P.; Klum, S.M.; Wang, S.; Yario, T.A.; Schirle Oakdale, N.T.; Steitz, J.A.; MacRae, I.J. Structural Basis for Target-Directed MicroRNA Degradation. Mol. Cell 2019, 75, 1243–1255.e7. [Google Scholar] [CrossRef]
- Jackson, A.L.; Linsley, P.S. Recognizing and Avoiding SiRNA Off-Target Effects for Target Identification and Therapeutic Application. Nat. Rev. Drug Discov. 2010, 9, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; van den Hoorn, T.; Jongsma, M.L.M.; Bakker, M.J.; Hengeveld, R.; Janssen, L.; Cresswell, P.; Egan, D.A.; van Ham, M.; ten Brinke, A.; et al. A Genome-Wide Multidimensional RNAi Screen Reveals Pathways Controlling MHC Class II Antigen Presentation. Cell 2011, 145, 268–283. [Google Scholar] [CrossRef]
- Rufo, N.; Garg, A.D.; Agostinis, P. The Unfolded Protein Response in Immunogenic Cell Death and Cancer Immunotherapy. Trends Cancer 2017, 3, 643–658. [Google Scholar] [CrossRef]
- Workenhe, S.T.; Pol, J.; Kroemer, G. Tumor-Intrinsic Determinants of Immunogenic Cell Death Modalities. OncoImmunology 2021, 10, 1893466. [Google Scholar] [CrossRef]
- Yatim, N.; Cullen, S.; Albert, M. Dying Cells Actively Regulate Adaptive Immune Responses. Nat. Rev. Immunol. 2017, 17, 262–275. [Google Scholar] [CrossRef]
- Sistigu, A.; Yamazaki, T.; Vacchelli, E.; Chaba, K.; Enot, D.; Adam, J.; Vitale, I.; Goubar, A.; Baracco, E.; Remédios, C.; et al. Cancer Cell-Autonomous Contribution of Type I Interferon Signaling to the Efficacy of Chemotherapy. Nat. Med. 2014, 20, 1301–1309. [Google Scholar] [CrossRef]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liang, T.-T. CD59 Receptor Targeted Delivery of MiRNA-1284 and Cisplatin-Loaded Liposomes for Effective Therapeutic Efficacy against Cervical Cancer Cells. AMB Express 2020, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Phung, C.D.; Nguyen, H.T.; Choi, J.Y.; Pham, T.T.; Acharya, S.; Timilshina, M.; Chang, J.-H.; Kim, J.-H.; Jeong, J.-H.; Ku, S.K.; et al. Reprogramming the T Cell Response to Cancer by Simultaneous, Nanoparticle-Mediated PD-L1 Inhibition and Immunogenic Cell Death. J. Control. Release 2019, 315, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Bastian, I.N.; Long, M.D.; Dow, M.; Li, W.; Liu, T.; Ngu, R.K.; Antonucci, L.; Huang, J.Y.; Phung, Q.T.; et al. Activation of NF-ΚB and P300/CBP Potentiates Cancer Chemoimmunotherapy through Induction of MHC-I Antigen Presentation. Proc. Natl. Acad. Sciences. 2021, 118, e2025840118. [Google Scholar] [CrossRef]
- Takahashi, A.; Umemura, A.; Yano, K.; Okishio, S.; Kataoka, S.; Okuda, K.; Seko, Y.; Yamaguchi, K.; Moriguchi, M.; Okanoue, T.; et al. Tyrosine Kinase Inhibitors Stimulate HLA Class I Expression by Augmenting the IFNγ/STAT1 Signaling in Hepatocellular Carcinoma Cells. Front. Oncol. 2021, 11, 707473. [Google Scholar] [CrossRef]
- Trowsdale, J. Genomic Structure and Function in the MHC. Trends Genet. 1993, 9, 117–122. [Google Scholar] [CrossRef]
- Axelrod, M.L.; Cook, R.S.; Johnson, D.B.; Balko, J.M. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin. Cancer Res. 2019, 25, 2392–2402. [Google Scholar] [CrossRef]
- Roemer, M.G.M.; Redd, R.A.; Cader, F.Z.; Pak, C.J.; Abdelrahman, S.; Ouyang, J.; Sasse, S.; Younes, A.; Fanale, M.; Santoro, A.; et al. Major Histocompatibility Complex Class II and Programmed Death Ligand 1 Expression Predict Outcome After Programmed Death 1 Blockade in Classic Hodgkin Lymphoma. J. Clin. Oncol. 2018, 36, 942–950. [Google Scholar] [CrossRef]
- Forero, A.; Li, Y.; Chen, D.; Grizzle, W.E.; Updike, K.L.; Merz, N.D.; Downs-Kelly, E.; Burwell, T.C.; Vaklavas, C.; Buchsbaum, D.J.; et al. Expression of the MHC Class II Pathway in Triple-Negative Breast Cancer Tumor Cells Is Associated with a Good Prognosis and Infiltrating Lymphocytes. Cancer Immunol. Res. 2016, 4, 390–399. [Google Scholar] [CrossRef]
- Callahan, M.J.; Nagymanyoki, Z.; Bonome, T.; Johnson, M.E.; Litkouhi, B.; Sullivan, E.H.; Hirsch, M.S.; Matulonis, U.A.; Liu, J.; Birrer, M.J.; et al. Increased HLA-DMB Expression in the Tumor Epithelium Is Associated with Increased CTL Infiltration and Improved Prognosis in Advanced-Stage Serous Ovarian Cancer. Clin. Cancer Res. 2008, 14, 7667–7673. [Google Scholar] [CrossRef]
- Oldford, S.A.; Robb, J.D.; Codner, D.; Gadag, V.; Watson, P.H.; Drover, S. Tumor Cell Expression of HLA-DM Associates with a Th1 Profile and Predicts Improved Survival in Breast Carcinoma Patients. Int. Immunol. 2006, 18, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Michel, S.; Linnebacher, M.; Alcaniz, J.; Voss, M.; Wagner, R.; Dippold, W.; Becker, C.; von Knebel Doeberitz, M.; Ferrone, S.; Kloor, M. Lack of HLA Class II Antigen Expression in Microsatellite Unstable Colorectal Carcinomas Is Caused by Mutations in HLA Class II Regulatory Genes. Int. J. Cancer 2010, 127, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Younger, A.R.; Amria, S.; Jeffrey, W.A.; Mahdy, A.E.M.; Goldstein, O.G.; Norris, J.S.; Haque, A. HLA Class II Antigen Presentation by Prostate Cancer Cells. Prostate Cancer Prostatic Dis. 2008, 11, 334–341. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yi, R.; Hong, S.; Zhang, Y.; Lin, A.; Ying, H.; Zou, W.; Wang, Q.; Wei, T.; Cheng, Q.; Zhu, W.; et al. MHC-II Signature Correlates With Anti-Tumor Immunity and Predicts Anti-PD-L1 Response of Bladder Cancer. Front. Cell Dev. Biol. 2022, 10, 757137. [Google Scholar] [CrossRef]
- Johnson, D.B.; Bordeaux, J.M.; Kim, J.-Y.; Vaupel, C.A.; Rimm, D.L.; Ho, T.H.; Joseph, R.W.; Daud, A.I.; Conry, R.M.; Gaughan, E.M.; et al. Quantitative Spatial Profiling of PD-1/PD-L1 Interaction and HLA-DR/IDO-1 Predicts Improved Outcomes of Anti-PD-1 Therapies in Metastatic Melanoma. Clin. Cancer Res. 2018, 24, 5250–5260. [Google Scholar] [CrossRef]
- Shen, F.-F.; Pan, Y.; Li, J.-Z.; Zhao, F.; Yang, H.-J.; Li, J.-K.; Gao, Z.-W.; Su, J.-F.; Duan, L.-J.; Lun, S.-M.; et al. High Expression of HLA-DQA1 Predicts Poor Outcome in Patients with Esophageal Squamous Cell Carcinoma in Northern China. Medicine 2019, 98, e14454. [Google Scholar] [CrossRef]
- Moretti, S.; Pinzi, C.; Berti, E.; Spallanzani, A.; Chiarugi, A.; Boddi, V.; Reali, U.M.; Giannotti, B. In Situ Expression of Transforming Growth Factor ?? Is Associated with Melanoma Progression and Correlates with Ki67, HLA-DR and beta 3 Integrin Expression. Melanoma Res. 1997, 7, 313–321. [Google Scholar] [CrossRef]
- Maruhashi, T.; Okazaki, I.; Sugiura, D.; Takahashi, S.; Maeda, T.K.; Shimizu, K.; Okazaki, T. LAG-3 Inhibits the Activation of CD4+ T Cells That Recognize Stable PMHCII through Its Conformation-Dependent Recognition of PMHCII. Nat. Immunol. 2018, 19, 1415–1426. [Google Scholar] [CrossRef]
- Andrews, L.P.; Cillo, A.R.; Karapetyan, L.; Kirkwood, J.M.; Workman, C.J.; Vignali, D.A.A. Molecular Pathways and Mechanisms of LAG-3 in Cancer Therapy. Clin. Cancer Res. 2022. [Google Scholar] [CrossRef]
- Long, L.; Zhang, X.; Chen, F.; Pan, Q.; Phiphatwatchara, P.; Zeng, Y.; Chen, H. The Promising Immune Checkpoint LAG-3: From Tumor Microenvironment to Cancer Immunotherapy. Genes Cancer 2018, 9, 176–189. [Google Scholar] [CrossRef]
- Liang, B.; Workman, C.; Lee, J.; Chew, C.; Dale, B.M.; Colonna, L.; Flores, M.; Li, N.; Schweighoffer, E.; Greenberg, S.; et al. Regulatory T Cells Inhibit Dendritic Cells by Lymphocyte Activation Gene-3 Engagement of MHC Class II. J. Immunol. 2008, 180, 5916–5926. [Google Scholar] [CrossRef] [PubMed]
- Hemon, P.; Jean-Louis, F.; Ramgolam, K.; Brignone, C.; Viguier, M.; Bachelez, H.; Triebel, F.; Charron, D.; Aoudjit, F.; Al-Daccak, R.; et al. MHC Class II Engagement by Its Ligand LAG-3 (CD223) Contributes to Melanoma Resistance to Apoptosis. J. Immunol. 2011, 186, 5173–5183. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Nixon, M.J.; Wang, Y.; Wang, D.Y.; Castellanos, E.; Estrada, M.V.; Ericsson-Gonzalez, P.I.; Cote, C.H.; Salgado, R.; Sanchez, V.; et al. Tumor-Specific MHC-II Expression Drives a Unique Pattern of Resistance to Immunotherapy via LAG-3/FCRL6 Engagement. JCI Insight 2018, 3, 120360. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-K.; Lin, K.; Xu, T.; He, B.; Pan, Y.; Sun, H.; Peng, H.; Hu, X. MicroRNA Expression Profiles Predict Progression and Clinical Outcome in Lung Adenocarcinoma. OncoTargets Ther. 2016, 9, 5679–5692. [Google Scholar] [CrossRef] [PubMed]
- Qi, B.; Liu, S.G.; Qin, X.G.; Yao, W.J.; Lu, J.G.; Guo, L.; Wang, T.Y.; Li, H.C.; Zhao, B.S. Overregulation of MicroRNA-212 in the Poor Prognosis of Esophageal Cancer Patients. Genet. Mol. Res. 2014, 13, 7800–7807. [Google Scholar] [CrossRef]
- Sun, S.M.; Rockova, V.; Bullinger, L.; Dijkstra, M.K.; Döhner, H.; Löwenberg, B.; Jongen-Lavrencic, M. The Prognostic Relevance of MiR-212 Expression with Survival in Cytogenetically and Molecularly Heterogeneous AML. Leukemia 2013, 27, 100–106. [Google Scholar] [CrossRef]
- Wu, W.-Y. Potentially Predictive MicroRNAs of Gastric Cancer with Metastasis to Lymph Node. World J. Gastroenterol. 2011, 17, 3645. [Google Scholar] [CrossRef]
- Tamaddon, G.; Geramizadeh, B.; Karimi, M.H.; Mowla, S.J.; Abroun, S. MiR-4284 and MiR-4484 as Putative Biomarkers for Diffuse Large B-Cell Lymphoma. Iran. J. Med. Sci. 2016, 41, 334–339. [Google Scholar]
- Yang, F.; Nam, S.; Brown, C.E.; Zhao, R.; Starr, R.; Horne, D.A.; Malkas, L.H.; Jove, R.; Hickey, R.J. A Novel Berbamine Derivative Inhibits Cell Viability and Induces Apoptosis in Cancer Stem-Like Cells of Human Glioblastoma, via Up-Regulation of MiRNA-4284 and JNK/AP-1 Signaling. PLoS ONE 2014, 9, e94443. [Google Scholar] [CrossRef]
- Li, Y.; Shen, Z.; Jiang, H.; Lai, Z.; Wang, Z.; Jiang, K.; Ye, Y.; Wang, S. MicroRNA-4284 Promotes Gastric Cancer Tumorigenicity by Targeting Ten-Eleven Translocation 1. Mol. Med. Rep. 2018, 17, 6569–6575. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, W.; Luan, Q.; Liu, Y. MiR-4284 Promotes Cell Proliferation, Migration, and Invasion in Non-Small Cell Lung Cancer Cells and Is Associated with Postoperative Prognosis. Cancer Manag. Res. 2021, 13, 5865–5872. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yu, B.; Jiang, L. MiR-212-3p Mediates Apoptosis and Invasion of Esophageal Squamous Cell Carcinoma through Inhibition of the Wnt/β-Catenin Signaling Pathway by Targeting SOX4. J. Thorac. Dis. 2020, 12, 4357–4367. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zou, Z.-Z.; Wen, M.; Xie, Y.-Z.; Peng, K.-J.; Luo, T.; Liu, S.-Y.; Gu, Q.; Li, J.-J.; Luo, Z.-Y. ZLM-7 Inhibits the Occurrence and Angiogenesis of Breast Cancer through MiR-212-3p/Sp1/VEGFA Signal Axis. Mol. Med. 2020, 26, 109. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Li, Y.; He, Z.; Chen, D.; Hong, Q.; You, J. Upregulation of Linc-ROR Promotes the Proliferation, Migration, and Invasion of Gastric Cancer Cells Through MiR-212-3p/FGF7 Axis. Cancer Manag. Res. 2021, 13, 899–912. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Wang, X.; Liang, Y.; Luo, D.; Han, D.; Li, C.; Chen, T.; Zhang, H.; Liu, Y.; et al. LINC01977 Promotes Breast Cancer Progression and Chemoresistance to Doxorubicin by Targeting MiR-212-3p/GOLM1 Axis. Front. Oncol. 2021, 11, 657094. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-K.; Liu, Y.-K.; Li, J.-W.; Liu, Y.-T.; Li, Y.-F.; Li, B.-H. Circ-Sirt1 Inhibits Growth and Invasion of Gastric Cancer by Sponging MiR-132-3p/MiR-212-3p and Upregulating Sirt1 Expression. Neoplasma 2021, 68, 780–787. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, L.; Zhang, Y.; Li, L.; Fei, Y.; Zhang, X.; Lin, G. Association between MiR-212-3p and SOX11, and the Effects of MiR-212-3p on Cell Proliferation and Migration in Mantle Cell Lymphoma. Oncol. Lett. 2021, 22, 709. [Google Scholar] [CrossRef]
- Perumal, N.; Kanchan, R.K.; Doss, D.; Bastola, N.; Atri, P.; Chirravuri-Venkata, R.; Thapa, I.; Vengoji, R.; Maurya, S.K.; Klinkebiel, D.; et al. MiR-212-3p Functions as a Tumor Suppressor Gene in Group 3 Medulloblastoma via Targeting Nuclear Factor I/B (NFIB). Acta Neuropathol. Commun. 2021, 9, 195. [Google Scholar] [CrossRef]
- Wang, H.; Wang, D.; Wei, Q.; Li, C.; Li, C.; Yang, J. Long Non-Coding RNAs PGM5-AS1 Upregulates Decorin (DCN) to Inhibit Cervical Cancer Progression by Sponging MiR-4284. Bioengineered 2022, 13, 9872–9884. [Google Scholar] [CrossRef]
- Ding, G.; Zhou, L.; Shen, T.; Cao, L. IFN-γ Induces the Upregulation of RFXAP via Inhibition of MiR-212-3p in Pancreatic Cancer Cells: A Novel Mechanism for IFN-γ Response. Oncol. Lett. 2018, 15, 3760–3765. [Google Scholar] [CrossRef]
- Ding, G.; Zhou, L.; Qian, Y.; Fu, M.; Chen, J.; Chen, J.; Xiang, J.; Wu, Z.; Jiang, G.; Cao, L. Pancreatic Cancer-Derived Exosomes Transfer MiRNAs to Dendritic Cells and Inhibit RFXAP Expression via MiR-212-3p. Oncotarget 2015, 6, 29877–29888. [Google Scholar] [CrossRef] [PubMed]
- van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and Activity of MicroRNA-Loaded Minicells in Patients with Recurrent Malignant Pleural Mesothelioma: A First-in-Man, Phase 1, Open-Label, Dose-Escalation Study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.-K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I Study of MRX34, a Liposomal MiR-34a Mimic, Administered Twice Weekly in Patients with Advanced Solid Tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]


| ID | miRBase Accesion | Fold Change | p-Value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mino | MDA-MB-231 | Mino | MDA-MB-231 | ||||||||||
| gIrr vs. Untreated | DXR vs. Untreated | IFNα/RA vs. Untreated | gIrr vs. Untreated | DXR vs. Untreated | IFNα/RA vs. Untreated | gIrr vs. Untreated | DXR vs. Untreated | IFNα/RA vs. Untreated | gIrr vs. Untreated | DXR vs. Untreated | IFNα/RA vs. Untreated | ||
| hsa-let-7b-3p | MIMAT0004482 | NS | −2.37 | −3.26 | NS | −1.97 | −1.68 | NS | 1.20 × 10−70 | 1.07 × 10−137 | NS | 7.71 × 10−58 | 1.14 × 10−34 |
| hsa-let-7f-2-3p | MIMAT0004487 | NS | −1.66 | −3.71 | NS | −1.42 | −1.45 | NS | 3.93 × 10−43 | 1.10 × 10−233 | NS | 1.56 × 10−24 | 5.83 × 10−26 |
| hsa-miR-576-3p | MIMAT0004796 | NS | −1.44 | −1.89 | NS | −1.38 | −1.38 | NS | 4.30 × 10−2 | 4.28 × 10−5 | NS | 2.41 × 10−2 | 2.77 × 10−2 |
| hsa-miR-98-3p | MIMAT0022842 | −1.25 | −1.47 | −22.34 | NS | −1.34 | −2.29 | 1.08 × 10−4 | 1.67 × 10−14 | 1.00 × 10−169 | NS | 6.61 × 10−17 | 1.04 × 10−102 |
| hsa-miR-30c-1-3p | MIMAT0004674 | −1.15 | −2.20 | −2.40 | NS | −1.60 | −1.79 | 1.36 × 10−2 | 5.77 × 10−60 | 8.42 × 10−85 | NS | 4.11 × 10−23 | 5.49 × 10−32 |
| hsa-miR-181a-3p | MIMAT0000270 | −1.14 | −2.46 | −3.71 | NS | −1.78 | −1.52 | 6.43 × 10−18 | 0.00 | 0.00 | NS | 0.00 | 1.01 × 10−165 |
| hsa-miR-27a-5p | MIMAT0004501 | −1.12 | −1.88 | −1.89 | NS | −1.55 | −1.43 | 4.81 × 10−2 | 3.57 × 10−46 | 2.91 × 10−56 | NS | 6.96 × 10−46 | 1.75 × 10−30 |
| hsa-miR-222-3p | MIMAT0000279 | 1.09 | −1.32 | −2.14 | −1.06 | −1.54 | −2.26 | 1.37 × 10−3 | 2.89 × 10−35 | 7.25 × 10−256 | 4.89 × 10−2 | 8.52 × 10−96 | 0.00 |
| hsa-miR-222-5p | MIMAT0004569 | 2.03 | −4.15 | −2.94 | 1.12 | −2.08 | −2.34 | 2.11 × 10−65 | 8.41 × 10−84 | 5.46 × 10−84 | 1.39 × 10−8 | 0.00 | 0.00 |
| hsa-miR-212-3p | MIMAT0000269 | NS | 3.59 | 48.61 | NS | 1.70 | 1.56 | NS | 4.89 × 10−3 | 3.21 × 10−28 | NS | 1.28 × 10−6 | 1.02 × 10−4 |
| hsa-miR-4284 | MIMAT0016915 | NS | 6.08 | 11.55 | NS | 1.33 | 1.70 | NS | 8.03 × 10−15 | 2.08 × 10−29 | NS | 7.29 × 10−3 | 2.05 × 10−8 |
| hsa-miR-33b-5p | MIMAT0003301 | NS | 1.71 | 7.66 | NS | 1.34 | 7.36 | NS | 1.90 × 10−36 | 0.00 | NS | 6.39 × 10−13 | 0.00 |
| hsa-miR-7705 | MIMAT0030020 | NS | 2.44 | 6.48 | NS | 1.70 | 3.30 | NS | 1.57 × 10−7 | 1.12 × 10−39 | NS | 1.98 × 10−38 | 1.70 × 10−203 |
| hsa-miR-1301-3p | MIMAT0005797 | NS | 1.59 | 2.78 | NS | 1.85 | 1.64 | NS | 9.14 × 10−21 | 2.66 × 10−121 | NS | 5.24 × 10−13 | 3.32 × 10−8 |
| hsa-miR-324-5p | MIMAT0000761 | NS | 1.50 | 2.40 | NS | 1.30 | 1.39 | NS | 7.82 × 10−11 | 4.45 × 10−57 | NS | 3.33 × 10−7 | 2.21 × 10−10 |
| hsa-miR-192-5p | MIMAT0000222 | NS | 1.67 | 1.51 | NS | 2.44 | 1.43 | NS | 9.77 × 10−229 | 2.97 × 10−149 | NS | 0.00 | 7.08 × 10−120 |
| ID | Fold Change | p-Value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mino | MDA-MB-231 | Mino | MDA-MB-231 | |||||||||
| gIrr vs. Untreated | DXR vs. Untreated | IFNα/RA vs. Untreated | gIrr vs. Untreated | DXR vs. Untreated | IFNα/RA vs. Untreated | gIrr vs. Untreated | DXR vs. Untreated | IFNα/RA vs. Untreated | gIrr vs. Untreated | DXR vs. Untreated | IFNα/RA vs. Untreated | |
| DPYSL2 | 1.10 | −1.69 | −1.89 | −1.01 | −3.84 | −2.84 | 1.00 | 1.81 × 10−2 | 3.63 × 10−3 | 1.00 | 2.91 × 10−14 | 2.00 × 10−8 |
| ENC1 | 1.14 | −1.52 | −2.35 | −1.04 | −1.46 | −1.53 | 1.00 | 1.61 × 10−2 | 7.31 × 10−8 | 1.00 | 4.42 × 10−3 | 2.09 × 10−3 |
| MEGF9 | −1.36 | −1.36 | −1.33 | −1.04 | −1.80 | −1.50 | 1.78 × 10−1 | 1.74 × 10−2 | 2.70 × 10−2 | 1.00 | 1.48 × 10−10 | 7.77 × 10−5 |
| NIN | −1.12 | −1.42 | −1.38 | 1.05 | −1.55 | −1.74 | 1.00 | 4.30 × 10−3 | 8.08 × 10−3 | 1.00 | 1.66 × 10−5 | 7.53 × 10−8 |
| PPM1F | 1.15 | −1.44 | −1.42 | −1.02 | −1.86 | −1.48 | 1.00 | 1.56 × 10−2 | 2.28 × 10−2 | 1.00 | 7.03 × 10−8 | 3.48 × 10−3 |
| TTC3 | −1.05 | −1.49 | −1.39 | 1.02 | −1.50 | −1.65 | 1.00 | 1.22 × 10−2 | 4.92 × 10−2 | 1.00 | 4.65 × 10−3 | 5.93 × 10−4 |
| ADM | −1.05 | 1.35 | 1.40 | −1.02 | 2.68 | 2.02 | 1.00 | 2.70 × 10−2 | 1.47 × 10−2 | 1.00 | 4.98 × 10−29 | 9.12 × 10−14 |
| ANKRD33B | −1.02 | 1.34 | 1.49 | 1.15 | 2.45 | 1.67 | 1.00 | 2.54 × 10−2 | 1.29 × 10−3 | 1.00 | 3.86 × 10−16 | 4.89 × 10−5 |
| CEACAM1 | −2.06 | 4.19 | 6.52 | 2.92 | 9.15 | 26.13 | NS | 4.04 × 10−3 | 2.62 × 10−5 | 1.00 | 6.84 × 10−3 | 3.32 × 10−5 |
| DDX58 | 1.02 | 9.96 | 10.85 | 1.05 | 2.66 | 6.46 | 1.00 | 2.96 × 10−6 | 1.49 × 10−7 | 1.00 | 3.94 × 10−2 | 2.13 × 10−5 |
| HERC5 | −1.45 | 149.92 | 213.69 | 1.06 | 3.60 | 5.23 | 1.00 | 1.61 × 10−5 | 3.92 × 10−7 | 1.00 | 8.48 × 10−3 | 7.83 × 10−4 |
| HLA-B | 1.03 | 2.12 | 2.47 | 1.04 | 1.60 | 2.95 | 1.00 | 2.98 × 10−4 | 9.62 × 10−7 | 1.00 | 1.50 × 10−2 | 5.89 × 10−10 |
| HLA-E | 1.04 | 2.61 | 2.52 | 1.04 | 2.32 | 2.21 | 1.00 | 2.62 × 10−3 | 2.66 × 10−3 | 1.00 | 2.84 × 10−3 | 1.10 × 10−2 |
| IFI27L1 | 1.11 | 1.42 | 1.58 | −1.07 | 1.42 | 1.40 | 1.00 | 3.11 × 10−2 | 2.98 × 10−3 | 1.00 | 5.86 × 10−3 | 2.19 × 10−2 |
| IFI44 | −1.61 | 39.21 | 3133.84 | 1.09 | 5.48 | 18.69 | 1.00 | 7.36 × 10−3 | 6.20 × 10−13 | 1.00 | 2.05 × 10−2 | 2.54 × 10−5 |
| IFI6 | 1.25 | 66.39 | 135.73 | 1.12 | 7.39 | 112.41 | 1.00 | 2.86 × 10−9 | 2.19 × 10−14 | 1.00 | 1.47 × 10−3 | 2.61 × 10−16 |
| IFIH1 | −1.11 | 6.94 | 11.16 | 1.06 | 3.06 | 15.55 | 1.00 | 2.04 × 10−5 | 2.76 × 10−9 | 1.00 | 9.59 × 10−3 | 1.31 × 10−12 |
| IFIT1 | −1.60 | 374.39 | 2665.15 | 1.03 | 5.84 | 27.03 | 1.00 | 2.96 × 10−6 | 2.60 × 10−12 | 1.00 | 1.74 × 10−2 | 1.90 × 10−6 |
| IFIT2 | 1.07 | 99.54 | 140.41 | 1.02 | 7.65 | 5.51 | 1.00 | 6.41 × 10−7 | 4.33 × 10−9 | 1.00 | 3.48 × 10−3 | 3.48 × 10−2 |
| IFIT3 | −1.02 | 21.47 | 81.05 | −1.04 | 4.85 | 10.30 | 1.00 | 2.45 × 10−7 | 8.54 × 10−17 | 1.00 | 2.30 × 10−3 | 4.08 × 10−6 |
| ISG15 | 1.15 | 11.46 | 29.74 | 1.09 | 2.62 | 12.29 | 1.00 | 2.53 × 10−7 | 5.05 × 10−16 | 1.00 | 4.01 × 10−2 | 1.02 × 10−9 |
| ISG20 | −1.24 | 7.94 | 14.92 | 1.36 | 6.74 | 16.63 | 1.00 | 3.36 × 10−5 | 9.48 × 10−10 | 1.00 | 6.96 × 10−5 | 1.79 × 10−9 |
| MDK | 1.02 | 5.67 | 5.04 | 1.03 | 2.83 | 2.92 | 1.00 | 1.03 × 10−3 | 1.33 × 10−3 | 1.00 | 2.44 × 10−2 | 3.82 × 10−2 |
| MX2 | 1.00 | 3.66 | 6.58 | −1.27 | 2.30 | 40.51 | 1.00 | 1.99 × 10−5 | 1.26 × 10−12 | 1.00 | 7.06 × 10−3 | 1.79 × 10−45 |
| OASL | −1.50 | 426.45 | 710.61 | −1.04 | 11.42 | 10.78 | 1.00 | 9.62 × 10−7 | 5.52 × 10−9 | 1.00 | 1.86 × 10−4 | 6.20 × 10−4 |
| PELI1 | 1.04 | 1.38 | 1.43 | −1.05 | 1.63 | 1.47 | 1.00 | 1.23 × 10−2 | 4.10 × 10−3 | 1.00 | 4.44 × 10−4 | 2.15 × 10−2 |
| PIM2 | −1.05 | 1.34 | 1.56 | −1.01 | 1.67 | 2.39 | 1.00 | 2.63 × 10−2 | 2.73 × 10−4 | 1.00 | 5.06 × 10−5 | 6.56 × 10−13 |
| PLEKHA4 | −1.34 | 14.67 | 51.65 | −1.23 | 3.87 | 3.33 | NS | 2.57 × 10−2 | 1.42 × 10−4 | 1.00 | 1.86 × 10−10 | 9.16 × 10−8 |
| PNPLA2 | −1.10 | 1.55 | 1.62 | −1.01 | 1.56 | 1.49 | 1.00 | 1.73 × 10−2 | 4.45 × 10−3 | 1.00 | 1.09 × 10−10 | 9.63 × 10−8 |
| PSMB9 | 1.07 | 1.91 | 3.05 | 1.10 | 1.55 | 2.36 | 1.00 | 1.39 × 10−3 | 2.48 × 10−11 | 1.00 | 1.12 × 10−2 | 6.13 × 10−8 |
| RNF114 | 1.18 | 1.53 | 1.31 | 1.01 | 1.45 | 1.49 | 1.00 | 5.61 × 10−4 | 3.88 × 10−2 | 1.00 | 2.04 × 10−4 | 1.56 × 10−4 |
| RSAD2 | −3.26 | 41.51 | 214.37 | −2.47 | 18.77 | 491.58 | NaN | 5.07 × 10−3 | 4.02 × 10−6 | 1.00 | 1.84 × 10−2 | 2.40 × 10−8 |
| RTP4 | −1.23 | 4.07 | 4.89 | −1.12 | 8.17 | 43.22 | NaN | 4.34 × 10−3 | 5.59 × 10−4 | 1.00 | 1.66 × 10−3 | 1.02 × 10−9 |
| SAMD9 | −1.14 | 8.24 | 6.89 | −1.09 | 3.14 | 6.83 | 1.00 | 3.60 × 10−5 | 6.22 × 10−5 | 1.00 | 2.01 × 10−2 | 4.13 × 10−5 |
| SCAMP1-AS1 | 1.03 | 2.26 | 2.30 | 1.01 | 2.04 | 3.42 | NS | 2.12 × 10−2 | 2.40 × 10−2 | 1.00 | 4.89 × 10−2 | 2.82 × 10−4 |
| SRGN | −1.08 | 1.77 | 1.71 | −1.03 | 1.54 | 1.66 | 1.00 | 6.71 × 10−3 | 1.07 × 10−2 | 1.00 | 2.87 × 10−2 | 1.60 × 10−2 |
| ST3GAL1 | −1.08 | 1.40 | 1.51 | 1.07 | 1.40 | 1.53 | 1.00 | 4.71 × 10−3 | 1.67 × 10−4 | 1.00 | 8.86 × 10−4 | 4.33 × 10−5 |
| TAP1 | 1.04 | 3.09 | 4.10 | 1.05 | 2.12 | 3.53 | 1.00 | 2.05 × 10−5 | 2.42 × 10−9 | 1.00 | 1.99 × 10−3 | 5.53 × 10−8 |
| TAP2 | −1.24 | 1.34 | 2.30 | −1.01 | 1.93 | 1.46 | 9.03 × 10−1 | 2.55 × 10−2 | 3.75 × 10−15 | 1.00 | 3.22 × 10−14 | 1.60 × 10−4 |
| ZNF600 | 1.07 | 1.84 | 1.55 | −1.00 | 1.76 | 1.56 | 1.00 | 4.32 × 10−3 | 5.28 × 10−2 | 1.00 | 1.92 × 10−3 | 4.58 × 10−2 |
| ZNFX1 | −1.04 | 2.05 | 2.84 | −1.01 | 1.56 | 2.91 | 1.00 | 2.34 × 10−3 | 3.73 × 10−7 | 1.00 | 4.48 × 10−2 | 4.86 × 10−8 |
| Cluster | miRNAs | Nodes | Edges | Modularity |
|---|---|---|---|---|
| 1 | hsa-miR-4284 hsa-miR-212-3p | 33 | 53 | 0.482 |
| 2 | hsa-miR-192-5p hsa-miR-1301-3p hsa-miR-7705 | 29 | 23 | 0.53 |
| 3 | hsa-miR-324-5p | 19 | 18 | 0.277 |
| 4 | hsa-miR-33b-5p | 18 | 17 | 0.246 |
| Cluster | miRNAs | Nodes | Edges | Modularity |
|---|---|---|---|---|
| 1 | hsa-miR-27a-5p hsa-miR-222-5p | 93 | 149 | 0.466 |
| 2 | hsa-miR-576-3p hsa-let-7f-2-3p | 48 | 50 | 0.314 |
| 3 | hsa-miR-222-3p | 46 | 45 | 0.250 |
| 4 | hsa-miR-30c-1-3p hsa-miR-181a-3p | 43 | 71 | 0.283 |
| 5 | hsa-miR-98-3p hsa-let-7b-3p | 42 | 51 | 0.319 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamberti, M.J.; Montico, B.; Ravo, M.; Nigro, A.; Giurato, G.; Iorio, R.; Tarallo, R.; Weisz, A.; Stellato, C.; Steffan, A.; et al. Integration of miRNA:mRNA Co-Expression Revealed Crucial Mechanisms Modulated in Immunogenic Cancer Cell Death. Biomedicines 2022, 10, 1896. https://doi.org/10.3390/biomedicines10081896
Lamberti MJ, Montico B, Ravo M, Nigro A, Giurato G, Iorio R, Tarallo R, Weisz A, Stellato C, Steffan A, et al. Integration of miRNA:mRNA Co-Expression Revealed Crucial Mechanisms Modulated in Immunogenic Cancer Cell Death. Biomedicines. 2022; 10(8):1896. https://doi.org/10.3390/biomedicines10081896
Chicago/Turabian StyleLamberti, María Julia, Barbara Montico, Maria Ravo, Annunziata Nigro, Giorgio Giurato, Roberta Iorio, Roberta Tarallo, Alessandro Weisz, Cristiana Stellato, Agostino Steffan, and et al. 2022. "Integration of miRNA:mRNA Co-Expression Revealed Crucial Mechanisms Modulated in Immunogenic Cancer Cell Death" Biomedicines 10, no. 8: 1896. https://doi.org/10.3390/biomedicines10081896
APA StyleLamberti, M. J., Montico, B., Ravo, M., Nigro, A., Giurato, G., Iorio, R., Tarallo, R., Weisz, A., Stellato, C., Steffan, A., Dolcetti, R., Casolaro, V., Faè, D. A., & Dal Col, J. (2022). Integration of miRNA:mRNA Co-Expression Revealed Crucial Mechanisms Modulated in Immunogenic Cancer Cell Death. Biomedicines, 10(8), 1896. https://doi.org/10.3390/biomedicines10081896










