MicroRNAs in Learning and Memory and Their Impact on Alzheimer’s Disease
Abstract
1. Introduction
1.1. MicroRNAs: Biogenesis/Functional Mechanism in Neurons
1.2. Learning and Memory
2. Role of miRNAs in Learning, Memory, and Cognitive Disorders
2.1. Direct Evidence of miRNA Regulation in Learning and Memory
2.1.1. miRNAs Involved in CREB-Dependent Transcription and CREB-Regulated miRNAs
2.1.2. Regulation of Fear Consolidation and Extinction by miRNAs—From Exposure to Inhibitory Learning
2.2. Direct Evidence of miRNA Involvement in Cognitive Impairment
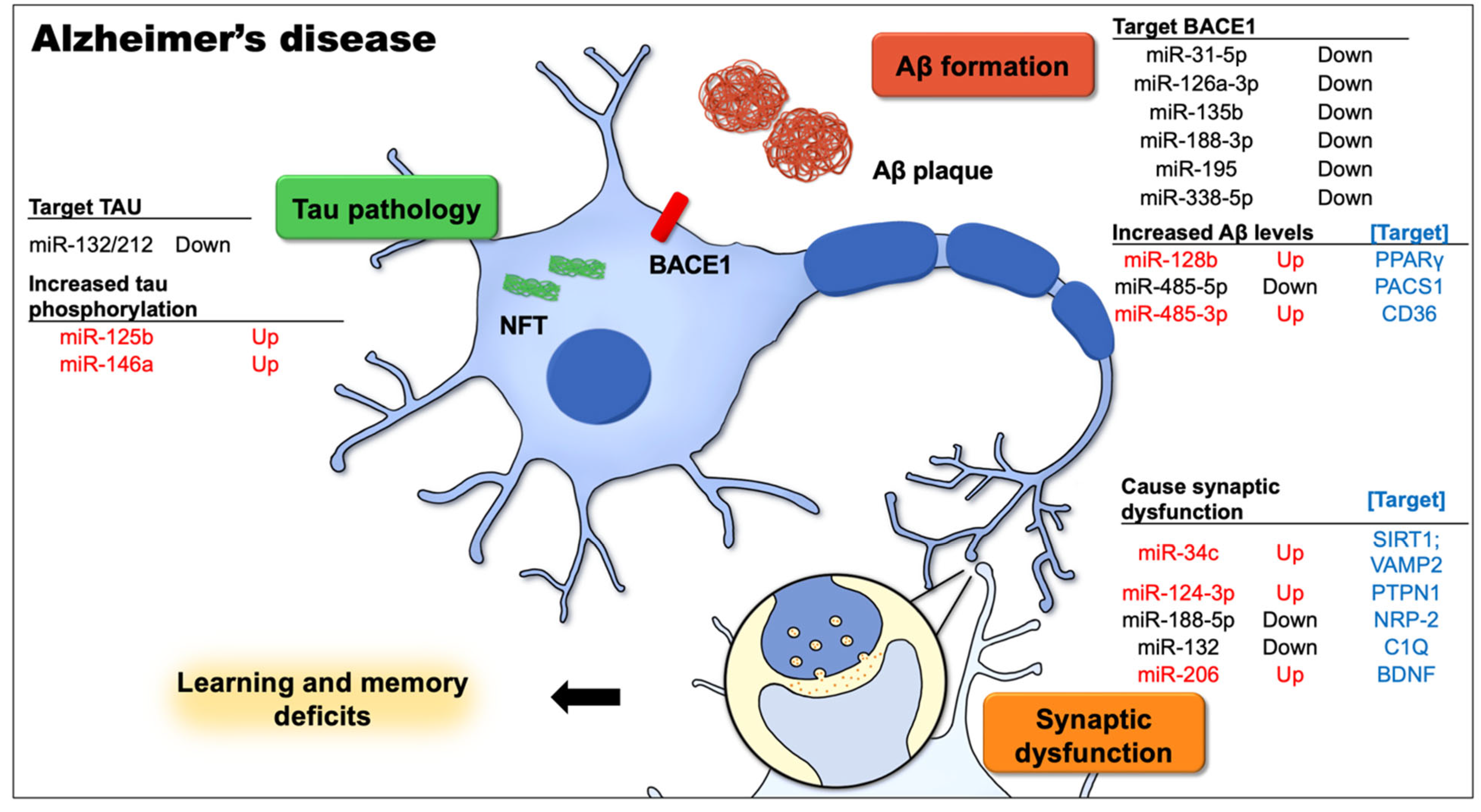
2.2.1. miRNA and AD
2.2.2. miRNAs Are Involved in Aβ Production and Metabolism
2.2.3. miRNAs Contribute to Abnormal Tau Protein Functions
2.2.4. miRNAs Mediate Synaptic Dysfunction
2.2.5. miRNAs Participate in Multiple Functions
2.2.6. miRNA and Other Neurodegenerative Disorders
2.2.7. miRNA and Post-Traumatic Stress Disorder
3. Preclinical Application
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, I.; Hansen, T.B. Biogenesis and Function of Ago-Associated RNAs. Trends Genet. 2017, 33, 208–219. [Google Scholar] [CrossRef]
- Sambandan, S.; Akbalik, G.; Kochen, L.; Rinne, J.; Kahlstatt, J.; Glock, C.; Tushev, G.; Alvarez-Castelao, B.; Heckel, A.; Schuman, E.M. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science 2017, 355, 634–637. [Google Scholar] [CrossRef]
- Wang, W.Y.; Kwon, E.J.; Tsai, L.H. MicroRNAs in learning, memory, and neurological diseases. Learn. Mem. 2012, 19, 359–368. [Google Scholar] [CrossRef]
- Wang, I.F.; Wang, Y.; Yang, Y.H.; Huang, G.J.; Tsai, K.J.; Shen, C.J. Activation of a hippocampal CREB-pCREB-miRNA-MEF2 axis modulates individual variation of spatial learning and memory capability. Cell Rep. 2021, 36, 109477. [Google Scholar] [CrossRef]
- Asok, A.; Leroy, F.; Rayman, J.B.; Kandel, E.R. Molecular Mechanisms of the Memory Trace. Trends Neurosci. 2019, 42, 14–22. [Google Scholar] [CrossRef]
- Vorhees, C.V.; Williams, M.T. Assessing spatial learning and memory in rodents. ILAR J. 2014, 55, 310–332. [Google Scholar] [CrossRef]
- Pitts, M.W. Barnes Maze Procedure for Spatial Learning and Memory in Mice. Bio Protoc 2018, 8. [Google Scholar] [CrossRef]
- Kim, W.B.; Cho, J.H. Encoding of contextual fear memory in hippocampal-amygdala circuit. Nat. Commun 2020, 11, 1382. [Google Scholar] [CrossRef]
- Lueptow, L.M. Novel Object Recognition Test for the Investigation of Learning and Memory in Mice. J. Vis. Exp. 2017. [Google Scholar] [CrossRef] [PubMed]
- Casellas, J. Inbred mouse strains and genetic stability: A review. Animal 2011, 5, 1–7. [Google Scholar] [CrossRef][Green Version]
- Vedell, P.T.; Svenson, K.L.; Churchill, G.A. Stochastic variation of transcript abundance in C57BL/6J mice. BMC Genom. 2011, 12, 167. [Google Scholar] [CrossRef]
- Tsai, K.J.; Chen, S.K.; Ma, Y.L.; Hsu, W.L.; Lee, E.H. sgk, a primary glucocorticoid-induced gene, facilitates memory consolidation of spatial learning in rats. Proc. Natl. Acad. Sci. USA 2002, 99, 3990–3995. [Google Scholar] [CrossRef]
- Bature, F.; Guinn, B.A.; Pang, D.; Pappas, Y. Signs and symptoms preceding the diagnosis of Alzheimer′s disease: A systematic scoping review of literature from 1937 to 2016. BMJ Open 2017, 7, e015746. [Google Scholar] [CrossRef]
- Sanabria-Castro, A.; Alvarado-Echeverria, I.; Monge-Bonilla, C. Molecular Pathogenesis of Alzheimer′s Disease: An Update. Ann. Neurosci. 2017, 24, 46–54. [Google Scholar] [CrossRef]
- Wei, W.; Wang, Z.Y.; Ma, L.N.; Zhang, T.T.; Cao, Y.; Li, H. MicroRNAs in Alzheimer′s Disease: Function and Potential Applications as Diagnostic Biomarkers. Front. Mol. Neurosci. 2020, 13, 160. [Google Scholar] [CrossRef] [PubMed]
- Bak, M.; Silahtaroglu, A.; Moller, M.; Christensen, M.; Rath, M.F.; Skryabin, B.; Tommerup, N.; Kauppinen, S. MicroRNA expression in the adult mouse central nervous system. RNA 2008, 14, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Issler, O.; Chen, A. Determining the role of microRNAs in psychiatric disorders. Nat. Rev. Neurosci 2015, 16, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Salta, E.; De Strooper, B. Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. 2017, 18, 627–640. [Google Scholar] [CrossRef]
- Agostini, M.; Tucci, P.; Steinert, J.R.; Shalom-Feuerstein, R.; Rouleau, M.; Aberdam, D.; Forsythe, I.D.; Young, K.W.; Ventura, A.; Concepcion, C.P.; et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc. Natl. Acad. Sci. USA 2011, 108, 21099–21104. [Google Scholar] [CrossRef]
- Dias, B.G.; Goodman, J.V.; Ahluwalia, R.; Easton, A.E.; Andero, R.; Ressler, K.J. Amygdala-dependent fear memory consolidation via miR-34a and Notch signaling. Neuron 2014, 83, 906–918. [Google Scholar] [CrossRef] [PubMed]
- Mollinari, C.; Racaniello, M.; Berry, A.; Pieri, M.; de Stefano, M.C.; Cardinale, A.; Zona, C.; Cirulli, F.; Garaci, E.; Merlo, D. miR-34a regulates cell proliferation, morphology and function of newborn neurons resulting in improved behavioural outcomes. Cell Death Dis. 2015, 6, e1622. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Engler-Chiurazzi, E.B.; Cavendish, J.Z.; Povroznik, J.M.; Russell, A.E.; Quintana, D.D.; Mathers, P.H.; Simpkins, J.W. Over-expression of miR-34a induces rapid cognitive impairment and Alzheimer′s disease-like pathology. Brain Res. 2019, 1721, 146327. [Google Scholar] [CrossRef] [PubMed]
- Zovoilis, A.; Agbemenyah, H.Y.; Agis-Balboa, R.C.; Stilling, R.M.; Edbauer, D.; Rao, P.; Farinelli, L.; Delalle, I.; Schmitt, A.; Falkai, P.; et al. microRNA-34c is a novel target to treat dementias. EMBO J. 2011, 30, 4299–4308. [Google Scholar] [CrossRef]
- Shi, Z.; Zhang, K.; Zhou, H.; Jiang, L.; Xie, B.; Wang, R.; Xia, W.; Yin, Y.; Gao, Z.; Cui, D.; et al. Increased miR-34c mediates synaptic deficits by targeting synaptotagmin 1 through ROS-JNK-p53 pathway in Alzheimer′s Disease. Aging Cell 2020, 19, e13125. [Google Scholar] [CrossRef]
- Rajasethupathy, P.; Fiumara, F.; Sheridan, R.; Betel, D.; Puthanveettil, S.V.; Russo, J.J.; Sander, C.; Tuschl, T.; Kandel, E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 2009, 63, 803–817. [Google Scholar] [CrossRef]
- Yang, Y.; Shu, X.; Liu, D.; Shang, Y.; Wu, Y.; Pei, L.; Xu, X.; Tian, Q.; Zhang, J.; Qian, K.; et al. EPAC null mutation impairs learning and social interactions via aberrant regulation of miR-124 and Zif268 translation. Neuron 2012, 73, 774–788. [Google Scholar] [CrossRef]
- Gao, J.; Wang, W.Y.; Mao, Y.W.; Graff, J.; Guan, J.S.; Pan, L.; Mak, G.; Kim, D.; Su, S.C.; Tsai, L.H. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 2010, 466, 1105–1109. [Google Scholar] [CrossRef]
- Hansen, K.F.; Sakamoto, K.; Aten, S.; Snider, K.H.; Loeser, J.; Hesse, A.M.; Page, C.E.; Pelz, C.; Arthur, J.S.; Impey, S.; et al. Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learn. Mem. 2016, 23, 61–71. [Google Scholar] [CrossRef]
- Wang, R.Y.; Phang, R.Z.; Hsu, P.H.; Wang, W.H.; Huang, H.T.; Liu, I.Y. In vivo knockdown of hippocampal miR-132 expression impairs memory acquisition of trace fear conditioning. Hippocampus 2013, 23, 625–633. [Google Scholar] [CrossRef]
- Hansen, K.F.; Sakamoto, K.; Wayman, G.A.; Impey, S.; Obrietan, K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE 2010, 5, e15497. [Google Scholar] [CrossRef] [PubMed]
- Scott, H.L.; Tamagnini, F.; Narduzzo, K.E.; Howarth, J.L.; Lee, Y.B.; Wong, L.F.; Brown, M.W.; Warburton, E.C.; Bashir, Z.I.; Uney, J.B. MicroRNA-132 regulates recognition memory and synaptic plasticity in the perirhinal cortex. Eur. J. Neurosci. 2012, 36, 2941–2948. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.F.; Karelina, K.; Sakamoto, K.; Wayman, G.A.; Impey, S.; Obrietan, K. miRNA-132: A dynamic regulator of cognitive capacity. Brain Struct. Funct. 2013, 218, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Shaltiel, G.; Hanan, M.; Wolf, Y.; Barbash, S.; Kovalev, E.; Shoham, S.; Soreq, H. Hippocampal microRNA-132 mediates stress-inducible cognitive deficits through its acetylcholinesterase target. Brain Struct. Funct. 2013, 218, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Griggs, E.M.; Young, E.J.; Rumbaugh, G.; Miller, C.A. MicroRNA-182 Regulates Amygdala-Dependent Memory Formation. J. Neurosci. 2013, 33, 1734–1740. [Google Scholar] [CrossRef] [PubMed]
- Woldemichael, B.T.; Jawaid, A.; Kremer, E.A.; Gaur, N.; Krol, J.; Marchais, A.; Mansuy, I.M. The microRNA cluster miR-183/96/182 contributes to long-term memory in a protein phosphatase 1-dependent manner. Nat. Commun. 2016, 7, 12594. [Google Scholar] [CrossRef]
- Xue, B.; Qu, Y.; Zhang, X.; Xu, X.F. miRNA-126a-3p Participates in Hippocampal Memory via Alzheimer′s Disease-Related Proteins. Cereb Cortex 2022, bhab515. [Google Scholar] [CrossRef]
- Sillivan, S.E.; Jamieson, S.; de Nijs, L.; Jones, M.; Snijders, C.; Klengel, T.; Joseph, N.F.; Krauskopf, J.; Kleinjans, J.; Vinkers, C.H.; et al. MicroRNA regulation of persistent stress-enhanced memory. Mol. Psychiatry 2019. [Google Scholar] [CrossRef]
- Lin, Q.; Wei, W.; Coelho, C.M.; Li, X.; Baker-Andresen, D.; Dudley, K.; Ratnu, V.S.; Boskovic, Z.; Kobor, M.S.; Sun, Y.E.; et al. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat. Neurosci. 2011, 14, 1115–1117. [Google Scholar] [CrossRef]
- Ronovsky, M.; Zambon, A.; Cicvaric, A.; Boehm, V.; Hoesel, B.; Moser, B.A.; Yang, J.; Schmid, J.A.; Haubensak, W.E.; Monje, F.J.; et al. A role for miR-132 in learned safety. Sci. Rep. 2019, 9, 528. [Google Scholar] [CrossRef]
- Kao, Y.C.; Wang, I.F.; Tsai, K.J. miRNA-34c Overexpression Causes Dendritic Loss and Memory Decline. Int. J. Mol. Sci. 2018, 19, 2323. [Google Scholar] [CrossRef] [PubMed]
- Vetere, G.; Barbato, C.; Pezzola, S.; Frisone, P.; Aceti, M.; Ciotti, M.; Cogoni, C.; Ammassari-Teule, M.; Ruberti, F. Selective Inhibition of miR-92 in Hippocampal Neurons Alters Contextual Fear Memory. Hippocampus 2014, 24, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer′s disease. EMBO J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef]
- Siegert, S.; Seo, J.; Kwon, E.J.; Rudenko, A.; Cho, S.; Wang, W.; Flood, Z.; Martorell, A.J.; Ericsson, M.; Mungenast, A.E.; et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat. Neurosci. 2015, 18, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, Z.M.; Tan, W.; Wang, X.; Li, Y.; Bai, B.; Li, Y.; Zhang, S.F.; Yan, H.L.; Chen, Z.L.; et al. Partial loss of psychiatric risk gene Mir137 in mice causes repetitive behavior and impairs sociability and learning via increased Pde10a. Nat. Neurosci. 2018, 21, 1689–1703. [Google Scholar] [CrossRef] [PubMed]
- Fregeac, J.; Moriceau, S.; Poli, A.; Nguyen, L.S.; Oury, F.; Colleaux, L. Loss of the neurodevelopmental disease-associated gene miR-146a impairs neural progenitor differentiation and causes learning and memory deficits. Mol. Autism 2020, 11, 22. [Google Scholar] [CrossRef]
- Ai, J.; Sun, L.H.; Che, H.; Zhang, R.; Zhang, T.Z.; Wu, W.C.; Su, X.L.; Chen, X.; Yang, G.; Li, K.; et al. MicroRNA-195 protects against dementia induced by chronic brain hypoperfusion via its anti-amyloidogenic effect in rats. J. Neurosci. 2013, 33, 3989–4001. [Google Scholar] [CrossRef]
- Capitano, F.; Camon, J.; Licursi, V.; Ferretti, V.; Maggi, L.; Scianni, M.; Del Vecchio, G.; Rinaldi, A.; Mannironi, C.; Limatola, C.; et al. MicroRNA-335-5p modulates spatial memory and hippocampal synaptic plasticity. Neurobiol. Learn. Mem. 2017, 139, 63–68. [Google Scholar] [CrossRef]
- Barros-Viegas, A.T.; Carmona, V.; Ferreiro, E.; Guedes, J.; Cardoso, A.M.; Cunha, P.; Pereira de Almeida, L.; Resende de Oliveira, C.; Pedro de Magalhaes, J.; Peca, J.; et al. miRNA-31 Improves Cognition and Abolishes Amyloid-beta Pathology by Targeting APP and BACE1 in an Animal Model of Alzheimer′s Disease. Mol. Ther Nucleic Acids 2020, 19, 1219–1236. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Zhang, X.; Du, G.; Fu, Q.; Huang, L. MicroRNA-107 prevents amyloid-beta-induced neurotoxicity and memory impairment in mice. Int. J. Mol. Med. 2018, 41, 1665–1672. [Google Scholar] [CrossRef]
- Wang, X.; Liu, D.; Huang, H.Z.; Wang, Z.H.; Hou, T.Y.; Yang, X.; Pang, P.; Wei, N.; Zhou, Y.F.; Dupras, M.J.; et al. A Novel MicroRNA-124/PTPN1 Signal Pathway Mediates Synaptic and Memory Deficits in Alzheimer′s Disease. Biol. Psychiatry 2018, 83, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Li, A.D.; Tong, L.; Xu, N.; Ye, Y.; Nie, P.Y.; Wang, Z.Y.; Ji, L.L. miR-124 regulates cerebromicrovascular function in APP/PS1 transgenic mice via C1ql3. Brain Res. Bull. 2019, 153, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Y.; Liu, P.; Bai, H.; Li, X.; Xiao, J.; Yuan, Q.; Geng, S.; Yin, H.; Zhang, H.; et al. MicroRNA-128 knockout inhibits the development of Alzheimer′s disease by targeting PPARgamma in mouse models. Eur. J. Pharmacol. 2019, 843, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Li, A.D.; Ji, L.L.; Ye, Y.; Wang, Z.Y.; Tong, L. miR-132 regulates the expression of synaptic proteins in APP/PS1 transgenic mice through C1q. Eur. J. Histochem. 2019, 63. [Google Scholar] [CrossRef]
- Deng, Y.; Zhang, J.; Sun, X.; Ma, G.; Luo, G.; Miao, Z.; Song, L. miR-132 improves the cognitive function of rats with Alzheimer′s disease by inhibiting the MAPK1 signal pathway. Exp. Ther. Med. 2020, 20, 159. [Google Scholar] [CrossRef]
- Smith, P.Y.; Hernandez-Rapp, J.; Jolivette, F.; Lecours, C.; Bisht, K.; Goupil, C.; Dorval, V.; Parsi, S.; Morin, F.; Planel, E.; et al. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 2015, 24, 6721–6735. [Google Scholar] [CrossRef]
- Zhang, Y.; Xing, H.; Guo, S.; Zheng, Z.; Wang, H.; Xu, D. MicroRNA-135b has a neuroprotective role via targeting of beta-site APP-cleaving enzyme 1. Exp. Ther. Med. 2016, 12, 809–814. [Google Scholar] [CrossRef]
- Tang, Y.; Bao, J.S.; Su, J.H.; Huang, W. MicroRNA-139 modulates Alzheimer′s-associated pathogenesis in SAMP8 mice by targeting cannabinoid receptor type 2. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef]
- Wang, G.; Huang, Y.; Wang, L.L.; Zhang, Y.F.; Xu, J.; Zhou, Y.; Lourenco, G.F.; Zhang, B.; Wang, Y.; Ren, R.J.; et al. MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer′s disease. Sci. Rep. 2016, 6, 26697. [Google Scholar] [CrossRef]
- Wu, Q.; Yuan, X.; Bai, J.; Han, R.; Li, Z.; Zhang, H.; Xiu, R. MicroRNA-181a protects against pericyte apoptosis via directly targeting FOXO1: Implication for ameliorated cognitive deficits in APP/PS1 mice. Aging 2019, 11, 6120–6133. [Google Scholar] [CrossRef]
- Lee, K.; Kim, H.; An, K.; Kwon, O.B.; Park, S.; Cha, J.H.; Kim, M.H.; Lee, Y.; Kim, J.H.; Cho, K.; et al. Replenishment of microRNA-188-5p restores the synaptic and cognitive deficits in 5XFAD Mouse Model of Alzheimer′s Disease. Sci. Rep. 2016, 6, 34433. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, M.; Teng, Z.; Tang, Y.P.; Chen, C. Synaptic and cognitive improvements by inhibition of 2-AG metabolism are through upregulation of microRNA-188-3p in a mouse model of Alzheimer′s disease. J. Neurosci. 2014, 34, 14919–14933. [Google Scholar] [CrossRef]
- Su, D.; Chai, Y.; Yang, J.; Wang, X.; Liu, Y.; Ma, J.; Tang, X.; Mishra, C.; Chandra, S.R.; Yue, W.; et al. Lentivirus-Carried microRNA-195 Rescues Memory Deficits of Alzheimer′s Disease Transgenic Mouse by Attenuating the Generation of Amyloid Plaques. Front. Pharmacol. 2021, 12, 633805. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Huang, M.; Guo, L.; Zhu, L.; Hou, J.; Zhang, L.; Pero, A.; Ng, S.; El Gaamouch, F.; Elder, G.; et al. MicroRNA-195 rescues ApoE4-induced cognitive deficits and lysosomal defects in Alzheimer′s disease pathogenesis. Mol. Psychiatry 2021, 26, 4687–4701. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Feng, S.; Ren, J.; Zhou, W. Upregulation of microRNA-196a improves cognitive impairment and alleviates neuronal damage in hippocampus tissues of Alzheimer′s disease through downregulating LRIG3 expression. J. Cell Biochem. 2019, 120, 17811–17821. [Google Scholar] [CrossRef] [PubMed]
- Higaki, S.; Muramatsu, M.; Matsuda, A.; Matsumoto, K.; Satoh, J.I.; Michikawa, M.; Niida, S. Defensive effect of microRNA-200b/c against amyloid-beta peptide-induced toxicity in Alzheimer′s disease models. PLoS ONE 2018, 13, e0196929. [Google Scholar] [CrossRef]
- Lee, S.T.; Chu, K.; Jung, K.H.; Kim, J.H.; Huh, J.Y.; Yoon, H.; Park, D.K.; Lim, J.Y.; Kim, J.M.; Jeon, D.; et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 2012, 72, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; Zhang, J.; He, F.P.; Bao, W.X.; Zheng, T.T.; Zhou, D.M.; Pan, H.Y.; Zhang, H.; Zhang, X.Q.; He, X.; et al. Down-regulated expression of microRNA-338-5p contributes to neuropathology in Alzheimer′s disease. FASEB J. 2019, 33, 4404–4417. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, D.; Zhou, H.; Wu, G.; He, Z.; Liao, W.; Li, Y.; Zhi, Y. MicroRNA-338-5p alleviates neuronal apoptosis via directly targeting BCL2L11 in APP/PS1 mice. Aging 2020, 12, 20728–20742. [Google Scholar] [CrossRef]
- He, C.; Su, C.; Zhang, W.; Wan, Q. miR-485-5p alleviates Alzheimer′s disease progression by targeting PACS1. Transl. Neurosci. 2021, 12, 335–345. [Google Scholar] [CrossRef]
- Koh, H.S.; Lee, S.; Lee, H.J.; Min, J.W.; Iwatsubo, T.; Teunissen, C.E.; Cho, H.J.; Ryu, J.H. Targeting MicroRNA-485-3p Blocks Alzheimer′s Disease Progression. Int. J. Mol. Sci. 2021, 22, 13136. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J.; Alexandrov, P.N. Regulation of complement factor H (CFH) by multiple miRNAs in Alzheimer′s disease (AD) brain. Mol. Neurobiol. 2012, 46, 11–19. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Gong, G.; Wang, Y.; Bian, M.; Yu, L.; Wei, C. MiR-124 acts as a target for Alzheimer′s disease by regulating BACE1. Oncotarget 2017, 8, 114065–114071. [Google Scholar] [CrossRef] [PubMed]
- Pichler, S.; Gu, W.; Hartl, D.; Gasparoni, G.; Leidinger, P.; Keller, A.; Meese, E.; Mayhaus, M.; Hampel, H.; Riemenschneider, M. The miRNome of Alzheimer′s disease: Consistent downregulation of the miR-132/212 cluster. Neurobiol. Aging 2017, 50, 167.e1–167.e10. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.; Bossers, K.; Janky, R.; Salta, E.; Frigerio, C.S.; Barbash, S.; Rothman, R.; Sierksma, A.S.; Thathiah, A.; Greenberg, D.; et al. Alteration of the microRNA network during the progression of Alzheimer′s disease. EMBO Mol. Med. 2013, 5, 1613–1634. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Veremeyko, T.; Wong, A.H.; El Fatimy, R.; Wei, Z.; Cai, W.; Krichevsky, A.M. Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer′s disease. Neurobiol. Aging 2017, 51, 156–166. [Google Scholar] [CrossRef]
- Sierksma, A.; Lu, A.; Salta, E.; Vanden Eynden, E.; Callaerts-Vegh, Z.; D′Hooge, R.; Blum, D.; Buee, L.; Fiers, M.; De Strooper, B. Deregulation of neuronal miRNAs induced by amyloid-beta or TAU pathology. Mol. Neurodegener. 2018, 13, 54. [Google Scholar] [CrossRef]
- Das, U.; Wang, L.; Ganguly, A.; Saikia, J.M.; Wagner, S.L.; Koo, E.H.; Roy, S. Visualizing APP and BACE-1 approximation in neurons yields insight into the amyloidogenic pathway. Nat. Neurosci 2016, 19, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer′s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef]
- Geng, L.; Zhang, T.; Liu, W.; Chen, Y. Inhibition of miR-128 Abates Abeta-Mediated Cytotoxicity by Targeting PPAR-gamma via NF-kappaB Inactivation in Primary Mouse Cortical Neurons and Neuro2a Cells. Yonsei Med. J. 2018, 59, 1096–1106. [Google Scholar] [CrossRef]
- Cha, D.J.; Mengel, D.; Mustapic, M.; Liu, W.; Selkoe, D.J.; Kapogiannis, D.; Galasko, D.; Rissman, R.A.; Bennett, D.A.; Walsh, D.M. miR-212 and miR-132 Are Downregulated in Neurally Derived Plasma Exosomes of Alzheimer′s Patients. Front. Neurosci. 2019, 13, 1208. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Wang, H.; Chen, K.; Cheng, P.; Gao, S.; Liu, J.; Li, X.; Sun, X. MicroRNA-34c Downregulation Ameliorates Amyloid-beta-Induced Synaptic Failure and Memory Deficits by Targeting VAMP2. J. Alzheimers Dis. 2015, 48, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.H.; Ban, T.; Liu, C.D.; Chen, Q.X.; Wang, X.; Yan, M.L.; Hu, X.L.; Su, X.L.; Bao, Y.N.; Sun, L.L.; et al. Activation of Cdk5/p25 and tau phosphorylation following chronic brain hypoperfusion in rats involves microRNA-195 down-regulation. J. Neurochem. 2015, 134, 1139–1151. [Google Scholar] [CrossRef]
- Liu, C.D.; Wang, Q.; Zong, D.K.; Pei, S.C.; Yan, Y.; Yan, M.L.; Sun, L.L.; Hao, Y.Y.; Mao, M.; Xing, W.J.; et al. Knockdown of microRNA-195 contributes to protein phosphatase-2A inactivation in rats with chronic brain hypoperfusion. Neurobiol. Aging 2016, 45, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, X.M.; Zhao, L.J.; Sun, L.L.; Yan, M.L.; Tian, Y.; Zhang, S.; Duan, M.J.; Zhao, H.M.; Li, W.R.; et al. MicroRNA-195 prevents dendritic degeneration and neuron death in rats following chronic brain hypoperfusion. Cell Death Dis. 2017, 8, e2850. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Li, J.; Huang, L.; Chen, X.; Li, D.; Wang, T.; Hu, C.; Xu, J.; Zhang, C.; Zen, K.; et al. Serum MicroRNA Profiles Serve as Novel Biomarkers for the Diagnosis of Alzheimer′s Disease. Dis. Markers 2015, 2015, 625659. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Chertkow, H.; Schipper, H.M.; Yuan, Z.; Shetty, V.; Jenkins, S.; Jones, T.; Wang, E. Increased microRNA-34c abundance in Alzheimer′s disease circulating blood plasma. Front. Mol. Neurosci. 2014, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Jain, G.; Stuendl, A.; Rao, P.; Berulava, T.; Pena Centeno, T.; Kaurani, L.; Burkhardt, S.; Delalle, I.; Kornhuber, J.; Hull, M.; et al. A combined miRNA-piRNA signature to detect Alzheimer′s disease. Transl. Psychiatry 2019, 9, 250. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C.; Zhang, Y. An investigation of microRNA-103 and microRNA-107 as potential blood-based biomarkers for disease risk and progression of Alzheimer′s disease. J. Clin. Lab. Anal. 2020, 34, e23006. [Google Scholar] [CrossRef]
- Sheinerman, K.S.; Tsivinsky, V.G.; Crawford, F.; Mullan, M.J.; Abdullah, L.; Umansky, S.R. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging 2012, 4, 590–605. [Google Scholar] [CrossRef]
- Xie, B.; Liu, Z.; Jiang, L.; Liu, W.; Song, M.; Zhang, Q.; Zhang, R.; Cui, D.; Wang, X.; Xu, S. Increased Serum miR-206 Level Predicts Conversion from Amnestic Mild Cognitive Impairment to Alzheimer′s Disease: A 5-Year Follow-up Study. J. Alzheimers Dis. 2017, 55, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Kenny, A.; McArdle, H.; Calero, M.; Rabano, A.; Madden, S.F.; Adamson, K.; Forster, R.; Spain, E.; Prehn, J.H.M.; Henshall, D.C.; et al. Elevated Plasma microRNA-206 Levels Predict Cognitive Decline and Progression to Dementia from Mild Cognitive Impairment. Biomolecules 2019, 9, 734. [Google Scholar] [CrossRef] [PubMed]
- Gascon, E.; Lynch, K.; Ruan, H.; Almeida, S.; Verheyden, J.M.; Seeley, W.W.; Dickson, D.W.; Petrucelli, L.; Sun, D.; Jiao, J.; et al. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat. Med. 2014, 20, 1444–1451. [Google Scholar] [CrossRef] [PubMed]
- Minones-Moyano, E.; Porta, S.; Escaramis, G.; Rabionet, R.; Iraola, S.; Kagerbauer, B.; Espinosa-Parrilla, Y.; Ferrer, I.; Estivill, X.; Marti, E. MicroRNA profiling of Parkinson′s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011, 20, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.; Peplow, P.V. Altered microRNA expression in animal models of Huntington′s disease and potential therapeutic strategies. Neural Regen. Res. 2021, 16, 2159–2169. [Google Scholar] [CrossRef]
- Liu, T.; Im, W.; Mook-Jung, I.; Kim, M. MicroRNA-124 slows down the progression of Huntington′s disease by promoting neurogenesis in the striatum. Neural Regen. Res. 2015, 10, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, M.; Takahashi, M.; Fujita, H.; Chiyo, T.; Popiel, H.A.; Watanabe, S.; Furuya, H.; Murata, M.; Wada, K.; Okada, T.; et al. Supplemental Treatment for Huntington′s Disease with miR-132 that Is Deficient in Huntington′s Disease Brain. Mol. Ther. Nucleic Acids 2018, 11, 79–90. [Google Scholar] [CrossRef]
- Snijders, C.; de Nijs, L.; Baker, D.G.; Hauger, R.L.; van den Hove, D.; Kenis, G.; Nievergelt, C.M.; Boks, M.P.; Vermetten, E.; Gage, F.H.; et al. MicroRNAs in Post-traumatic Stress Disorder. Curr. Top. Behav. Neurosci. 2018, 38, 23–46. [Google Scholar] [CrossRef]
- Giridharan, V.V.; Thandavarayan, R.A.; Fries, G.R.; Walss-Bass, C.; Barichello, T.; Justice, N.J.; Reddy, M.K.; Quevedo, J. Newer insights into the role of miRNA a tiny genetic tool in psychiatric disorders: Focus on post-traumatic stress disorder. Transl. Psychiatry 2016, 6, e954. [Google Scholar] [CrossRef]
- Maurel, O.M.; Torrisi, S.A.; Barbagallo, C.; Purrello, M.; Salomone, S.; Drago, F.; Ragusa, M.; Leggio, G.M. Dysregulation of miR-15a-5p, miR-497a-5p and miR-511-5p Is Associated with Modulation of BDNF and FKBP5 in Brain Areas of PTSD-Related Susceptible and Resilient Mice. Int. J. Mol. Sci. 2021, 22, 5157. [Google Scholar] [CrossRef]
- Chen, R.J.; Kelly, G.; Sengupta, A.; Heydendael, W.; Nicholas, B.; Beltrami, S.; Luz, S.; Peixoto, L.; Abel, T.; Bhatnagar, S. MicroRNAs as biomarkers of resilience or vulnerability to stress. Neuroscience 2015, 305, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Torrisi, S.A.; Lavanco, G.; Maurel, O.M.; Gulisano, W.; Laudani, S.; Geraci, F.; Grasso, M.; Barbagallo, C.; Caraci, F.; Bucolo, C.; et al. A novel arousal-based individual screening reveals susceptibility and resilience to PTSD-like phenotypes in mice. Neurobiol. Stress 2021, 14, 100286. [Google Scholar] [CrossRef] [PubMed]
- Solich, J.; Kolasa, M.; Faron-Gorecka, A.; Hajto, J.; Piechota, M.; Dziedzicka-Wasylewska, M. MicroRNA Let-7e in the Mouse Prefrontal Cortex Differentiates Restraint-Stress-Resilient Genotypes from Susceptible Genotype. Int. J. Mol. Sci. 2021, 22, 9439. [Google Scholar] [CrossRef] [PubMed]
- Zendjabil, M. Circulating microRNAs as novel biomarkers of Alzheimer′s disease. Clin. Chim. Acta 2018, 484, 99–104. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, T.; Xiao, J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018, 34, 267–274. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Bai, S.F.; Yan, J.Q. Blood circulating miRNAs as biomarkers of Alzheimer′s disease: A systematic review and meta-analysis. Biomark. Med. 2019, 13, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J. New approaches to symptomatic treatments for Alzheimer′s disease. Mol. Neurodegener. 2021, 16, 2. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Aisen, P.; Lemere, C.; Atri, A.; Sabbagh, M.; Salloway, S. Aducanumab produced a clinically meaningful benefit in association with amyloid lowering. Alzheimers Res. Ther. 2021, 13, 98. [Google Scholar] [CrossRef]
- Hampel, H.; O′Bryant, S.E.; Molinuevo, J.L.; Zetterberg, H.; Masters, C.L.; Lista, S.; Kiddle, S.J.; Batrla, R.; Blennow, K. Blood-based biomarkers for Alzheimer disease: Mapping the road to the clinic. Nat. Rev. Neurol. 2018, 14, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Jun, S.; Rellick, S.; Quintana, D.D.; Cavendish, J.Z.; Simpkins, J.W. Expression of microRNA-34a in Alzheimer′s disease brain targets genes linked to synaptic plasticity, energy metabolism, and resting state network activity. Brain Res. 2016, 1646, 139–151. [Google Scholar] [CrossRef]
- Chen, J.; Qi, Y.; Liu, C.F.; Lu, J.M.; Shi, J.; Shi, Y. MicroRNA expression data analysis to identify key miRNAs associated with Alzheimer’s disease. J. Gene Med. 2018, 20, e3014. [Google Scholar] [CrossRef] [PubMed]
- Tian, N.; Cao, Z.; Zhang, Y. MiR-206 decreases brain-derived neurotrophic factor levels in a transgenic mouse model of Alzheimer′s disease. Neurosci. Bull. 2014, 30, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
| miRNA | Change Direction | Method | Fold Change | Location | Behavioral Tasks | Effects in Learning/Memory | Reference |
|---|---|---|---|---|---|---|---|
| miR-34a | Down | miRNA sponge | - | Basolateral amygdala | Auditory fear conditioning | Impaired | [21] |
| Up | Viral overexpression | - | Lateral ventricle | MWM | Enhanced | [22] | |
| Up | Transgenic overepxression | Thousands | Whole brain | T-maze | Impaired | [23] | |
| miR-34c | Up | Viral overexpression | Hippocampus | MWM | Impaired | [41] | |
| miR-92 | Down | miRNA sponge | - | Hippocampus | CFC | Impaired | [42] |
| miR-125b | Up | Injection of mimic | 2 | Hippocampus | CFC | Impaired | [43] |
| miR-126a-3p | Up | Viral overexpression | 1.5~2 | Hippocampus | CFC; MWM; NOR | Enhanced | [37] |
| miR-126a-3p | Down | Virus-mediated interference | 0.4 | Impaired | |||
| miR-128b | Down | Virus-mediated interference | - | Infralimbic prefrontal cortex | Contextual fear extintion | Impaired | [39] |
| Up | Viral overexpression | - | Enhanced | ||||
| miR-132-3p | Up | Transgenic overepxression | 1.5 | Hippocampus | Barnes maze | Enhanced | [33] |
| Up | 3 | NOR; Barnes maze | Impaired | ||||
| Up | Viral overexpression | 2 | Perirhinal cortex | NOR | Impaired | [32] | |
| Down | Virus-mediated interference | Hippocampus | Trace fear conditioning | Impaired | [30] | ||
| Up | Injection of mimic | 2 | basolateral amygdala | Learned safety | Enhanced | [40] | |
| Down | Knock out (KO) | - | Impaired | ||||
| miR-132/212 | Down | Knock out (KO) | excitatory forebrain | NOR; CFC; Barnes maze | Impaired | [29] | |
| miR-134 | Up | Viral overexpression | 6 | Hippocampus | CFC | Impaired | [28] |
| miR-137 | Up | Viral overexpression | - | Hippocampus | CFC; MWM | Impaired | [44] |
| Down | conditional KO | 0.5 | Hippocampus | MWM; Barnes maze | Impaired | [45] | |
| miR-146a | Down | Virus-mediated interference | - | Hippocampus | CFC; NOR; Object location memory test | Impaired | [46] |
| miR-182 | Up | Injection of mimic | 4 | Lateral amygdala | Auditory fear conditioning | Impaired | [35] |
| miR-183/96/182 | Up | Viral overexpression | 4~12 | Hippocampus | NOR | Enhanced | [36] |
| Down | miRNA sponge | 0.5 | Impaired | ||||
| miR-195 | Down | Injection of antagomir | 0.5 | Hippocampus | MWM | Impaired | [47] |
| miR-335-5p | Up | Injection of mimic | - | Hippocampus | MWM | Impaired | [48] |
| miR-466f-3p | Up | Viral overexpression | 1.5 | Hippocampus | MWM; NOR; Barnes maze | Enhanced | [5] |
| Down | miRNA sponge | - | Impaired |
| miRNA | Change Direction | Method | Fold Change | Location | Behavioral Tasks | Effects in Learning/Memory | Strains of Disease Model | Reference |
|---|---|---|---|---|---|---|---|---|
| miR-31-5p | Up | Viral overexpression | 3 | Hippocampus | T-maze; NOR; Barnes maze | Enhanced | 3xTg-AD | [49] |
| miR-34c | Down | Injection of inhibitor | - | CFC | Rescued and enhanced | APP/PS1-21 | [24] | |
| Down | Injection of inhibitor | 0.5 | Third ventricle | MWM | Rescued | SAMP8 | [25] | |
| miR-107 | Down | Injection of mimic | 2 | Hippocampus | MWM | Rescued | Aβ ICV injection model | [50] |
| miR-124 | Down | Injection of LNA-probe | 0.4 | Hippocampus | MWM | Rescued | EPAC−/− | [27] |
| Up | Viral overexpression | - | Impaired | EPAC+/+ | ||||
| Up | Viral overexpression | - | Hippocampus | MWM | Impaired | Tg2576 | [51] | |
| Down | Injection of antagomir | - | Rescued | |||||
| miR-124-3p | Up | Viral overexpression | 5~10 | Lateral ventricle | MWM | Rescued | APP/PS1 | [52] |
| miR-126a-3p | Up | Viral overexpression | 1.5~2 | Hippocampus | CFC; MWM; NOR | Enhanced | APP/PS1 | [37] |
| miR-128b | Down | Knock out (KO) | * Cerebral cortex | MWM | Rescued | 3xTg-AD | [53] | |
| miR-132-3p | Up | Viral overexpression | 2.5 | Lateral ventricle | MWM | Rescued | APP/PS1 | [54] |
| Up | Viral overexpression | 1.5 | Hippocampus | MWM | Rescued | Aβ ICV injection model | [55] | |
| miR-132/212 | Up | Injection of mimic | - | Ventricles | Barnes maze | Rescued | 3xTg-AD | [56] |
| miR-134 | Down | Injection of LNA-probe | 0.6 | Rescued | SIRT1 mutant | [28] | ||
| miR-135b | Up | Injection of mimic | 5 | Third ventricle | Y-maze | Rescued | SAMP8 | [57] |
| miR-135b-5p | Up | Viral overexpression | basolateral amygdala | Acute restraint stress and auditory fear conditioning | Enhanced | Stress resilient mice | [38] | |
| Down | Injection of inhibitor | Rescued | Stress susceptible mice | |||||
| miR-139 | Up | Injection of mimic | 2 | Hippocampus | CFC; MWM; NOR | Impaired | SAMP8 | [58] |
| Down | Injection of inhibitor | 0.4 | Rescued | |||||
| Down | Injection of inhibitor | - | Hippocampus | Y-maze; MWM | Rescued | 5xFAD | [59] | |
| miR-181a | Up | Viral overexpression | 4 | Hippocampus | MWM | Rescued | APP/PS1 | [60] |
| miR-188-5p | Up | Viral overexpression | - | Hippocampus | T-maze; CFC | Rescued | 5xFAD | [61] |
| miR-188-3p | Up | Viral overexpression | - | Hippocampus | MWM | Rescued | 5xFAD | [62] |
| miR-195 | Up | Injection of mimic | 2 | Rescued | A chronic brain hypoperfusion model | [47] | ||
| Up | Viral overexpression | - | Hippocampus | MWM | Rescued | APP/PS1 | [63] | |
| Up | Viral overexpression | - | Hippocampus | NOR | Rescued | ApoE4KI +/- 5XFAD | [64] | |
| miR-196a | Up | Viral overexpression | 1.6 | Hippocampus | MWM | Rescued | Aβ ICV injection model | [65] |
| miR-200b/c | Up | Injection of mimic | - | Lateral ventricle | Barnes maze | Rescued | Aβ ICV injection model | [66] |
| miR-206 | Down | Injection of antagomir | - | Cerebral ventricles | CFC; Y-maze | Rescued | Tg2576 | [67] |
| miR-338-5p | Up | Viral overexpression | 2 | Hippocampus | MWM | Rescued | 5xFAD | [68] |
| Up | Viral overexpression | 4~5 | Hippocampus | MWM | Rescued | APP/PS1 | [69] | |
| miR-485-5p | Up | Viral overexpression | 4 | Hippocampus | MWM; CFC | Rescued | APP/PS1 | [70] |
| miR-485-3p | Down | Injection of inhibitor | - | Lateral ventricle | Y-maze | Rescued | 5xFAD | [71] |
| miRNAs | Reported Changes | Strains | Behavioral Task in Rodents | Target Genes | Function | Patient Samples (Brain) | References | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Brain | Peripheral Body Fluid | Animal Models | Huamn Brain | Human Peripheral Body Fluid | ||||||
| miR-31-5p | Down | 3xTg-AD | T-maze; NOR; Barnes maze | APP/BACE1 | Reduces plaque load and intraneuronal Aβ | serum | [49] | [86] | ||
| miR-34c | Up | APP/PS1-21 | CFC | SIRT1; VAMP2 | Induces synaptic failure and memory deficits | Hippocampus | plasma; CSF | [24] | [24,82] | [25,87,88] |
| Up | SAMP8 | MWM | SYT1/ROS-JNK-p53 pathway | Mediates synaptic and memory deficits by targeting SYT1 | * serum | [25] | [25] | |||
| miR-107 | Down | Aβ ICV injection model | MWM | BDNF-TrkB and AKT pathways | Prevents Aβ-induced cell death, Aβ and Tau accumlation | Neocortex | plasma | [50] | [72] | [89] |
| miR-124-3p | Down | APP/PS1 | MWM | C1QL3 | Cerebromicrovascular impairments, breakdown of BBB and promote angiogenesis | Neocortex; hippocampus | [52] | [72,73,75] | ||
| Up | Tg2576 | MWM | PTPN1 | Impairs synaptic transmission by reducing AMPA trafficking | Temporal cortex; hippocampus | [51] | [51] | |||
| miR-126a-3p | Down | APP/PS1 | CFC; MWM; NOR | EFHD2/BACE1 | Reduces Aβ plaque area and neuroinflammation | Temporal cortex | [37] | [72,74] | ||
| miR-128b | Up | 3×Tg-AD | MWM | PPARγ | Its knockout suppresses amyloid plaque formation, Aβ generation and neuroinflammation | Cerebral cortex | plasma | [53] | [53] | [80] |
| miR-132/212 | Down | 3XTg-ADKO | Barnes maze | TAU | Regulation of Aβ metabolism and tau pathology | Temporal cortex; prefrontal cortex; hippocampus | plasma | [56] | [56,74,75,76] | [81] |
| miR-132 | Down | Aβ ICV injection model | MWM | MAPK1 | Inhibits iNOS expression and oxidative stress | [55] | ||||
| - | APP/PS1 | MWM | C1Q | Induces synaptic proteins (PSD95, Synapsin-1, p-Synapsin) expression | [54] | |||||
| miR-135b | Down | SAMP8 | Y-maze | BACE1 | Promotes proliferation and neuroprotective effect | Prefrontal cortex | serum | [57] | [75] | [57] |
| miR-139 | Up | SAMP8 | CFC; MWM; NOR | CB2 | Decreases responses to proinflammatory stimuli | Neocortex | [58] | [72] | ||
| miR-146a | Up | 5xFAD | Y-maze; MWM | ROCK1/PTEN signal pathway | Induces tau hyperphosphorylation | Temporal lobe neocortex; hippocampus | serum | [59] | [72,77] | [86] |
| miR-181a | Down | APP/PS1 | MWM | FOXO1 | Ameliorates amyloid plaque deposition, decelerates pericyte apoptosis and BBB breakdown | Neocortex | * plasma | [60] | [72] | [90] |
| miR-188-5p | Down | 5xFAD | CFC; T-maze | NRP-2 | Reduces Aβ-mediated reduction in dendritic spine density and basal synaptic transmission | Cortex; hippocampus | [61] | [61] | ||
| miR-188-3p | Down | 5xFAD | MWM | BACE1 | Reduces Aβ formation and suppresses neuroinflammation | temporal lobe | [62] | [62] | ||
| miR-195 | Down | ApoE4KI +/− 5XFAD | NOR | ApoE-synj1-PIP2 pathway | Rescues lysosomal defects in neurons and cognitive deficits in mice | Parietal cortex | * CSF | [64] | [64] | [64] |
| Down | APP/PS1 | MWM | Prevents Aβ Deposition by Inhibiting APP Expression | [63] | ||||||
| miR-196a | Down | Aβ ICV injection model | MWM | LRIG3 | Stimulates neuronal survival and inhibits its apoptosis through represses oxidative stress and inflammation | [65] | ||||
| miR-206 | Up | Tg2576 | CFC; Y-maze | BDNF | Reduces dendritic spine density | Temporal cortex | * serum/plasma | [67] | [67] | [91,92] |
| miR-338-5p | Down | 5xFAD | MWM | BACE1 | Decreases Aβ formation and reduces neuroinflammation | Hippocampus | [68] | [68] | ||
| APP/PS1 | MWM | BCL2L11 | Attenuates amyloid plaque deposition, attenuates neuron apoptosis | [69] | ||||||
| miR-485-5p | Down | APP/PS1 | MWM; CFC | PACS1 | Represses Aβ40-induced pericyte apoptosis | Prefrontal cortex | [70] | [75] | ||
| miR-485-3p | Up | 5xFAD | Y-maze | CD36 | Induces Aβ plaque accumulation, tau pathology development, neuroinflammation, and cognitive decline | Frontal cortex; precentral gyrus | CSF; plasma | [71] | [71] | [71] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, I.-F.; Ho, P.-C.; Tsai, K.-J. MicroRNAs in Learning and Memory and Their Impact on Alzheimer’s Disease. Biomedicines 2022, 10, 1856. https://doi.org/10.3390/biomedicines10081856
Wang I-F, Ho P-C, Tsai K-J. MicroRNAs in Learning and Memory and Their Impact on Alzheimer’s Disease. Biomedicines. 2022; 10(8):1856. https://doi.org/10.3390/biomedicines10081856
Chicago/Turabian StyleWang, I-Fang, Pei-Chuan Ho, and Kuen-Jer Tsai. 2022. "MicroRNAs in Learning and Memory and Their Impact on Alzheimer’s Disease" Biomedicines 10, no. 8: 1856. https://doi.org/10.3390/biomedicines10081856
APA StyleWang, I.-F., Ho, P.-C., & Tsai, K.-J. (2022). MicroRNAs in Learning and Memory and Their Impact on Alzheimer’s Disease. Biomedicines, 10(8), 1856. https://doi.org/10.3390/biomedicines10081856






