The Search for Antibacterial Inhibitors Targeting Cell Division Protein FtsZ at Its Nucleotide and Allosteric Binding Sites
Abstract
1. Cell Division Protein FtsZ as an Antibacterial Target
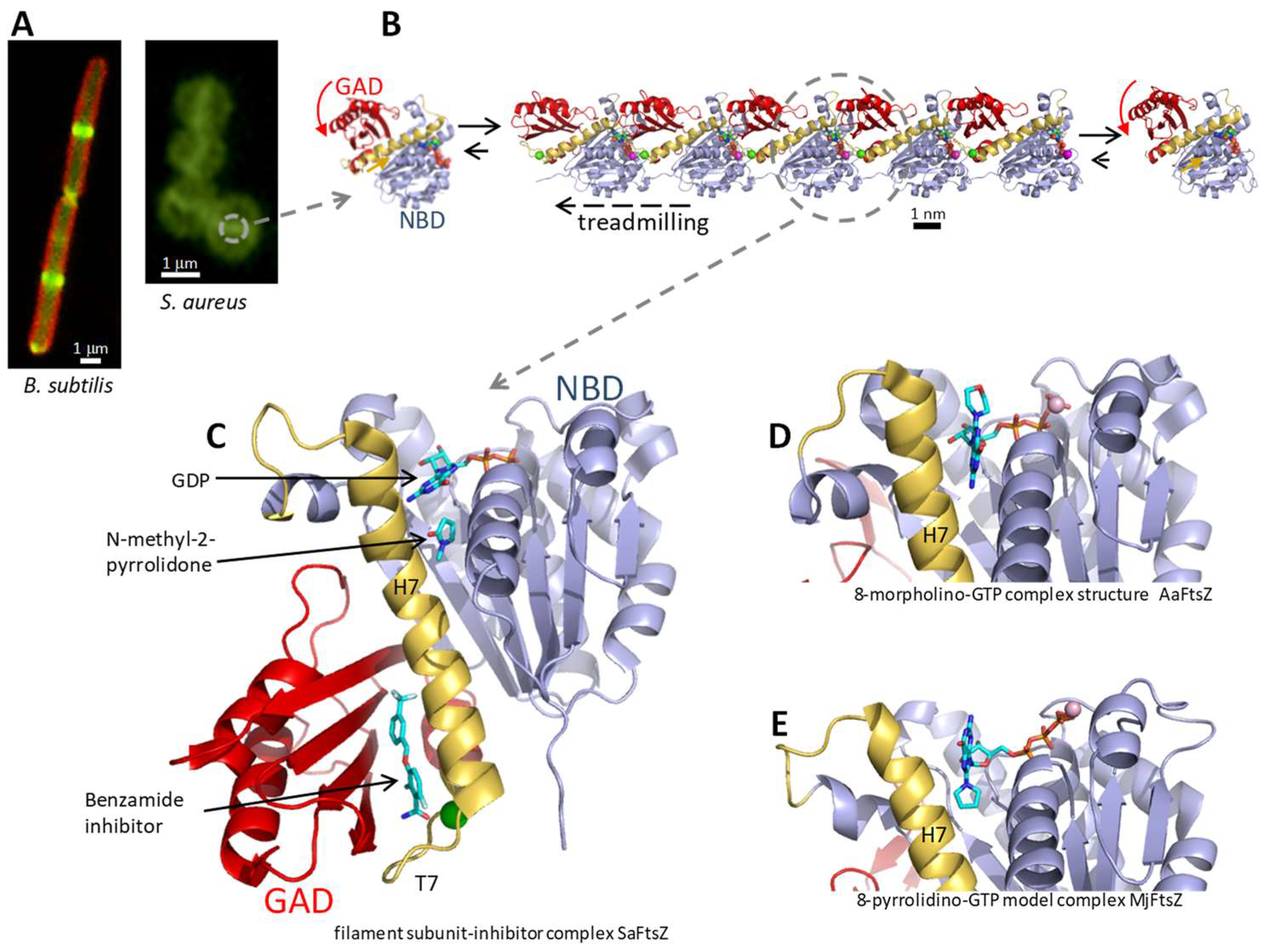
2. The FtsZ Nucleotide Binding Site
2.1. Selective Inhibition of FtsZ Versus Tubulin by C8 Nucleotide Analogs
2.2. Fluorescent Nucleotide Analogs and Competition Assays for FtsZ Inhibitors
2.3. Synthetic Nucleotide-Replacing FtsZ Inhibitors
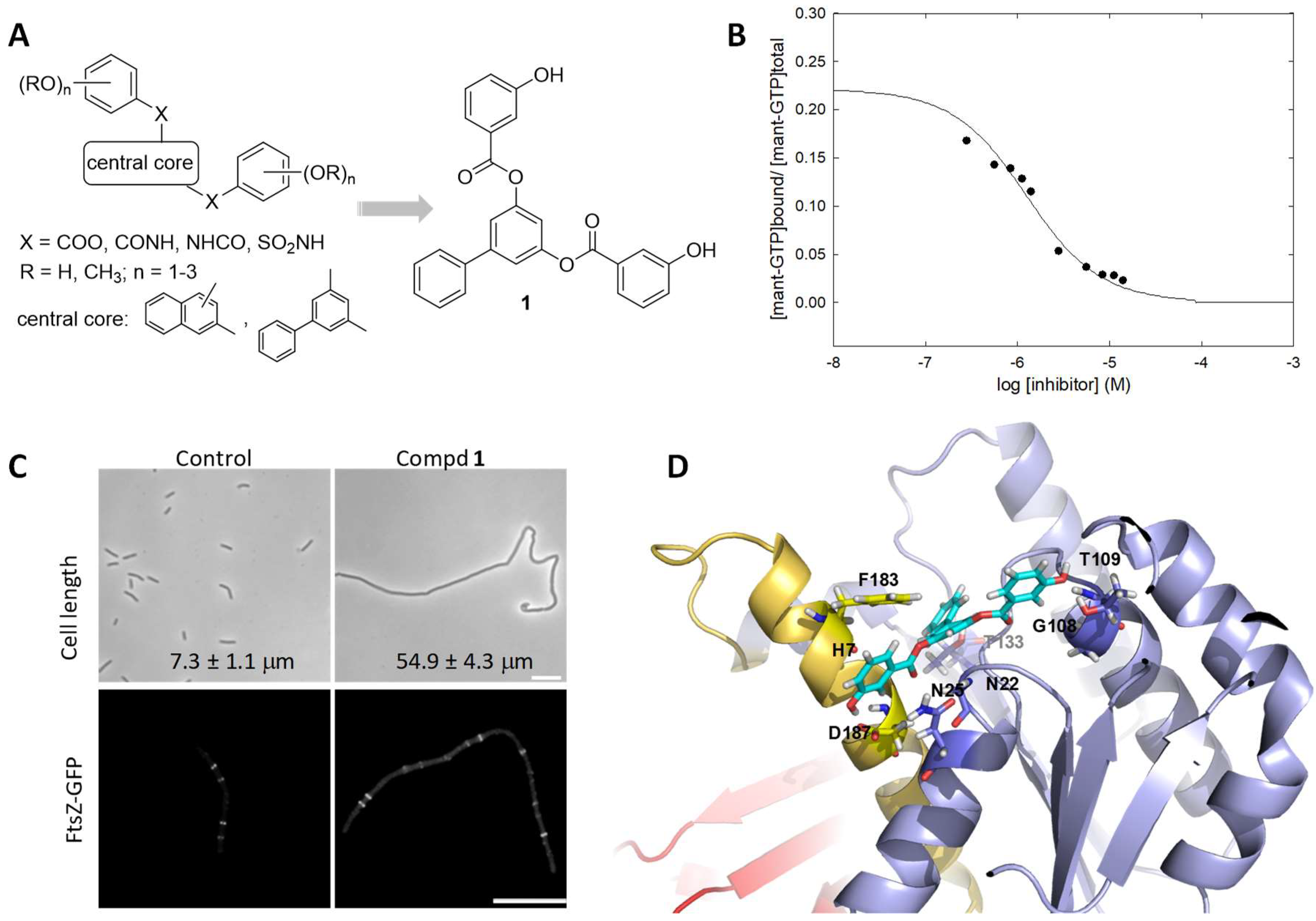
2.4. Antibacterial Activity of Nucleotide-Replacing FtsZ Inhibitors

3. The Filamenting Phenotype and the Cytological Profile of FtsZ Inhibitors
3.1. The Importance of Assessing Membrane Integrity
3.2. The Cytological Profile of FtsZ Inhibitors
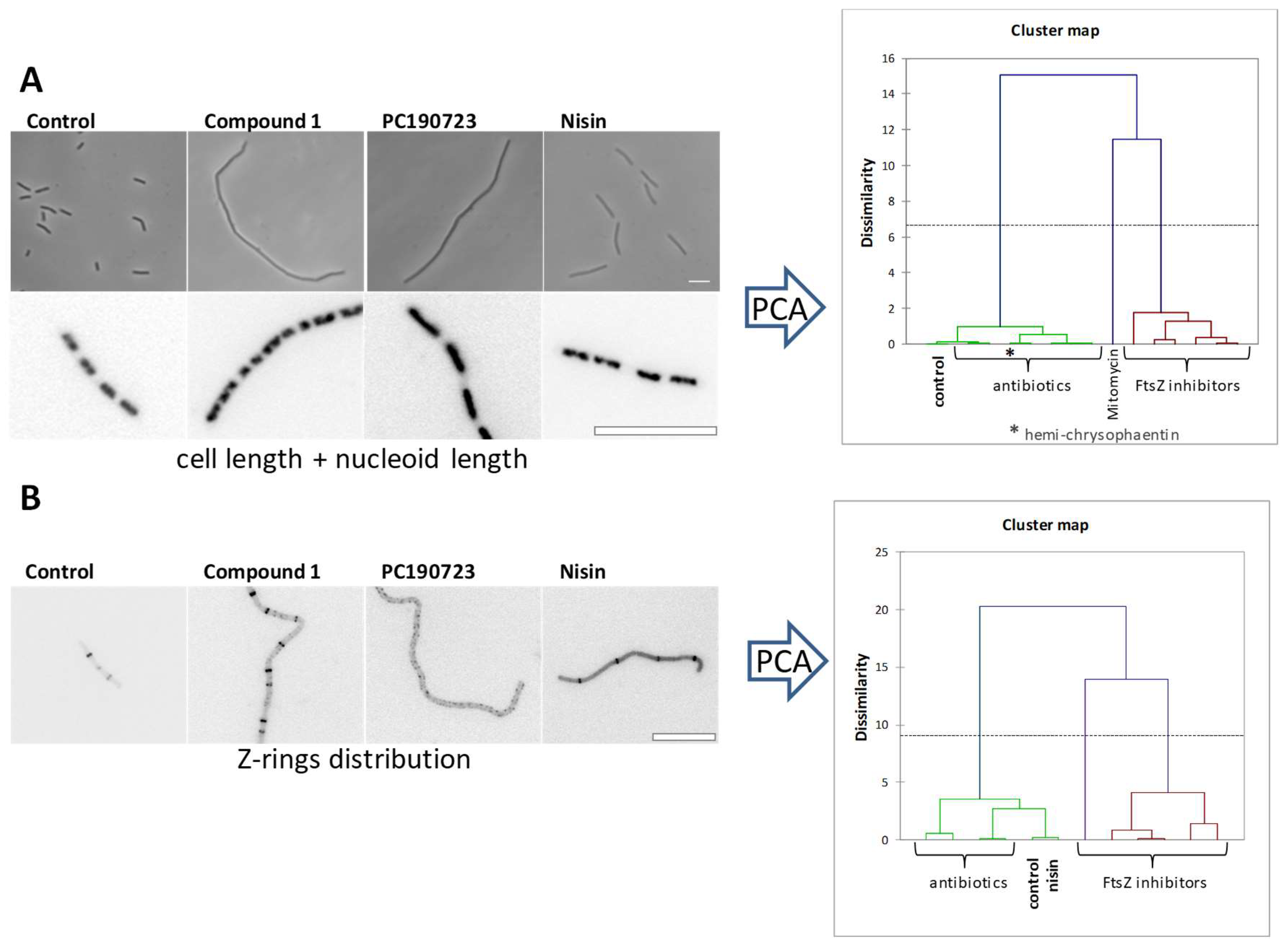
4. The FtsZ Interdomain Cleft
4.1. The Reference Antibacterial FtsZ Inhibitor PC190723
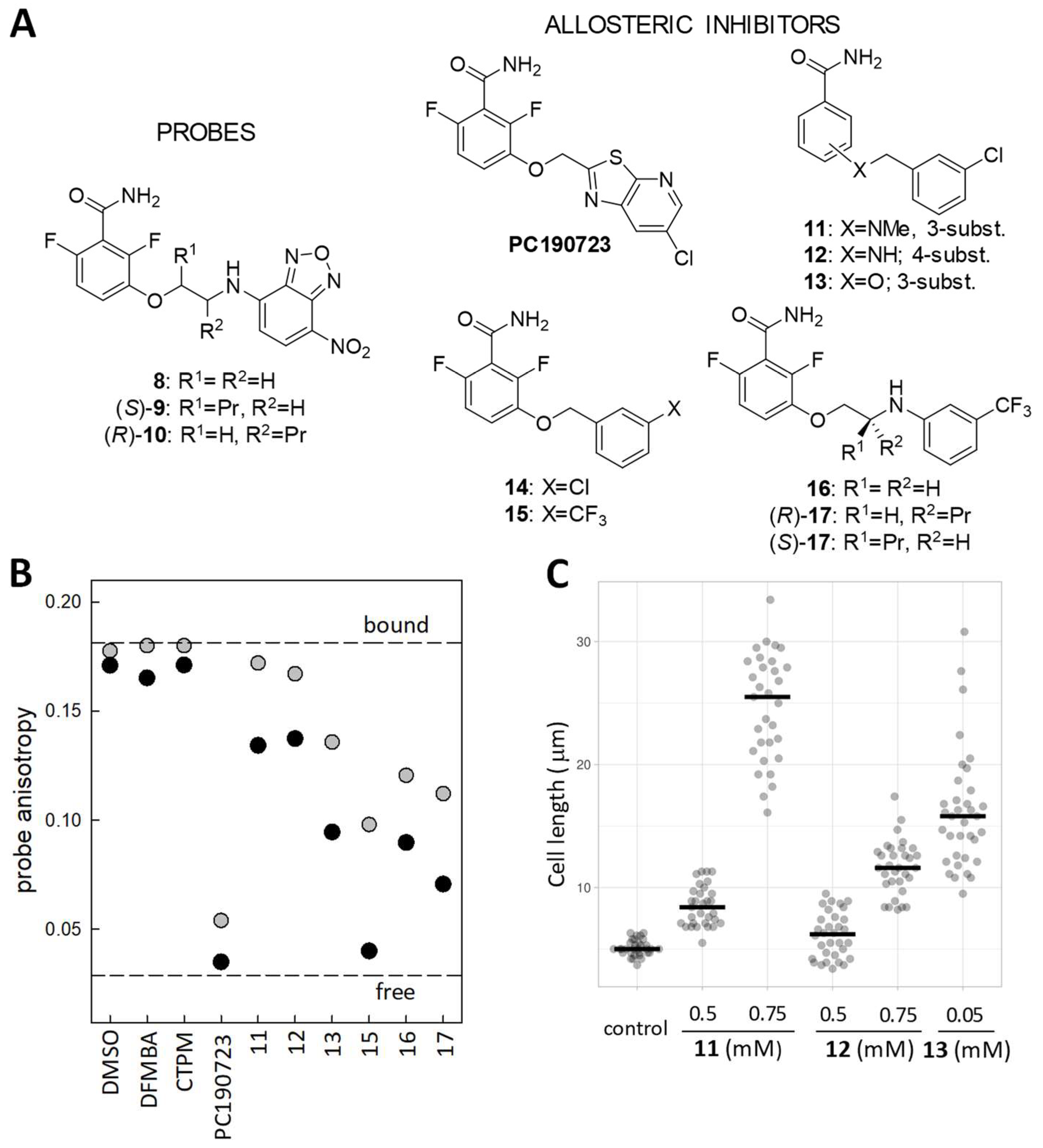
4.2. Fluorescent Probes for the FtsZ Interdomain Cleft and the Assembly Switch
4.3. Fluorescence Polarization Assay for Allosteric FtsZ Inhibitors
4.4. Enhancing the Affinity of Simplified Benzamide Inhibitors
4.5. Structural Insights into Allosteric Inhibitor Binding to S. aureus FtsZ
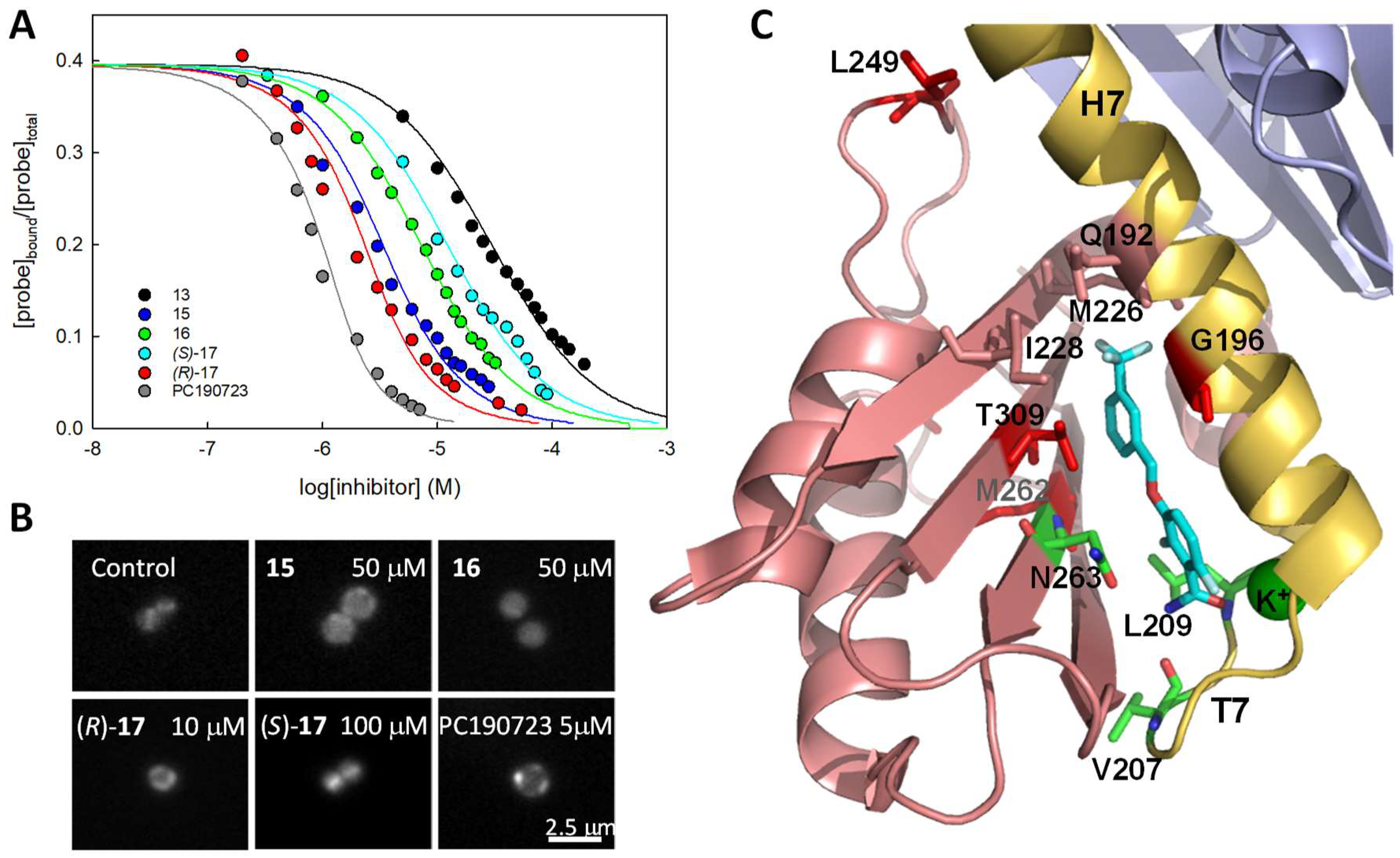
4.6. Antibacterial Activity of Benzamide-Based Allosteric FtsZ Inhibitors
5. Proposed Experimental Strategies for FtsZ Inhibitor Screening and Characterization
- (A)
- To screen for FtsZ inhibitors, we currently favor the following workflow:
- (1)
- Phenotypic screening putative FtsZ inhibitors (1–102 compounds) of B. subtilis and E. coli (cell filamentation), with the determination of the division inhibitory concentration (MDC) and membrane integrity.
- For HTS of large compound libraries, robotized phenotypic screening of B. subtilis or E. coli plate cultures, based on automatic image analysis of bacterial filamentation [79] may be implemented, if available.
- (2)
- FP determination of binding affinity (KD) to the nucleotide and allosteric binding sites of FtsZ and compound solubility limit determination.
- (3)
- Antibacterial activity (MIC) on pathogens and frequency of resistance (FOR) mutations of susceptible pathogens.
- (B)
- As an alternate HTS route, we suggest a binding site-directed approach starting from a robotized FP plate screen of compound binding to the FtsZ nucleotide or allosteric sites, and compound solubility limit determination, followed by phenotypic analysis on B. subtilis and E. coli (filamentation), the determination of MDC and membrane integrity, and a final assessment of MIC and FOR in pathogens.
6. Outlook for Other Methods, VS, and Machine Learning Approaches for Selecting Potential Antibacterial FtsZ Inhibitors
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwon, J.H.; Powderly, W.G. The post-antibiotic era is here. Science 2021, 373, 471. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- den Blaauwen, T.; Andreu, J.M.; Monasterio, O. Bacterial cell division proteins as antibiotic targets. Bioorganic Chem. 2014, 55, 27–38. [Google Scholar] [CrossRef]
- Bi, E.; Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 1991, 354, 161–164. [Google Scholar] [CrossRef] [PubMed]
- McQuillen, R.; Xiao, J. Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu. Rev. Biophys. 2020, 49, 309–341. [Google Scholar] [CrossRef]
- Barrows, J.M.; Goley, E.D. FtsZ dynamics in bacterial division: What, how, and why? Curr. Opin. Cell Biol. 2021, 68, 163–172. [Google Scholar] [CrossRef]
- Levin, P.A.; Janakiraman, A.; Slauch, J.M. Localization, assembly, and activation of the Escherichia coli cell division machinery. EcoSal Plus 2021, 9, eESP-0022. [Google Scholar] [CrossRef] [PubMed]
- Nogales, E.; Downing, K.H.; Amos, L.A.; Lowe, J. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 1998, 5, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, M.O.; Prota, A.E. Microtubule-targeting agents: Strategies to hijack the cytoskeleton. Trends Cell Biol. 2018, 28, 776–792. [Google Scholar] [CrossRef]
- Läppchen, T.; Pinas, V.A.; Hartog, A.F.; Koomen, G.J.; Schaffner-Barbero, C.; Andreu, J.M.; Trambaiolo, D.; Lowe, J.; Juhem, A.; Popov, A.V.; et al. Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem. Biol. 2008, 15, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.M.; Therien, A.G.; Lu, J.; Lee, S.H.; Caron, A.; Gill, C.J.; Lebeau-Jacob, C.; Benton-Perdomo, L.; Monteiro, J.M.; Pereira, P.M.; et al. Restoring methicillin-resistant Staphylococcus aureus susceptibility to beta-lactam antibiotics. Sci. Transl. Med. 2012, 4, 126ra135. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Yamane, J.; Mogi, N.; Yamaguchi, H.; Takemoto, H.; Yao, M.; Tanaka, I. Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus. Acta Cryst. Sect. D-Biol. Crystallogr. 2012, 68, 1175–1188. [Google Scholar] [CrossRef]
- Fujita, J.; Maeda, Y.; Mizohata, E.; Inoue, T.; Kaul, M.; Parhi, A.K.; LaVoie, E.J.; Pilch, D.S.; Matsumura, H. Structural flexibility of an inhibitor overcomes drug resistance mutations in Staphylococcus aureus FtsZ. ACS Chem. Biol. 2017, 12, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-González, E.; Fujita, J.; Yoshizawa, T.; Nelson, J.M.; Pilch, A.J.; Hillman, E.; Ozawa, M.; Kuroda, N.; Al-Tameemi, H.M.; Boyd, J.M.; et al. Structure-guided design of a fluorescent probe for the visualization of FtsZ in clinically important gram-positive and gram-negative bacterial pathogens. Sci. Rep. 2019, 9, 20092. [Google Scholar] [CrossRef] [PubMed]
- Alnami, A.; Norton, R.S.; Pena, H.P.; Haider, S.; Kozielski, F. Conformational flexibility of a highly conserved helix controls cryptic pocket formation in FtsZ. J. Mol. Biol. 2021, 433, 167061. [Google Scholar] [CrossRef] [PubMed]
- Huecas, S.; Araújo-Bazán, L.; Ruiz, F.M.; Ruiz-Ávila, L.B.; Martínez, R.F.; Escobar-Peña, A.; Artola, M.; Vázquez-Villa, H.; Martín-Fontecha, M.; Fernández-Tornero, C.; et al. Targeting the FtsZ allosteric binding site with a novel fluorescence polarization screen, cytological and structural approaches for antibacterial discovery. J. Med. Chem. 2021, 64, 5730–5745. [Google Scholar] [CrossRef] [PubMed]
- Bisson-Filho Alexandre, W.; Hsu, Y.-P.; Squyres Georgia, R.; Kuru, E.; Wu, F.; Jukes, C.; Sun, Y.; Dekker, C.; Holden, S.; VanNieuwenhze Michael, S.; et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 2017, 355, 739–743. [Google Scholar] [CrossRef]
- Yang, X.; Lyu, Z.; Miguel, A.; McQuillen, R.; Huang, K.C.; Xiao, J. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 2017, 355, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.M.; Pereira, A.R.; Reichmann, N.T.; Saraiva, B.M.; Fernandes, P.B.; Veiga, H.; Tavares, A.C.; Santos, M.; Ferreira, M.T.; Macario, V.; et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 2018, 554, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Squyres, G.R.; Holmes, M.J.; Barger, S.R.; Pennycook, B.R.; Ryan, J.; Yan, V.T.; Garner, E.C. Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis. Nat. Microbiol. 2021, 6, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, F.M.; Huecas, S.; Santos-Aledo, A.; Prim, E.A.; Andreu, J.M.; Fernández-Tornero, C. FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics. PLoS Biol. 2022, 20, e3001497. [Google Scholar] [CrossRef] [PubMed]
- Huecas, S.; Llorca, O.; Boskovic, J.; Martin-Benito, J.; Valpuesta, J.M.; Andreu, J.M. Energetics and geometry of FtsZ polymers: Nucleated self-assembly of single protofilaments. Biophys. J. 2008, 94, 1796–1806. [Google Scholar] [CrossRef]
- Miraldi, E.R.; Thomas, P.J.; Romberg, L. Allosteric models for cooperative polymerization of linear polymers. Biophys. J. 2008, 95, 2470–2486. [Google Scholar] [CrossRef] [PubMed]
- Fujita, J.; Harada, R.; Maeda, Y.; Saito, Y.; Mizohata, E.; Inoue, T.; Shigeta, Y.; Matsumura, H. Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus. J. Struct. Biol. 2017, 198, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Artola, M.; Ruiz-Avila, L.B.; Ramirez-Aportela, E.; Martinez, R.F.; Araujo-Bazán, L.; Vázquez-Villa, H.; Martín-Fontecha, M.; Oliva, M.A.; Martín-Galiano, A.J.; Chacón, P.; et al. The structural assembly switch of cell division protein FtsZ probed with fluorescent allosteric inhibitors. Chem. Sci. 2017, 8, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, J.M.; Tsim, M.; Oliva, M.A.; García-Sanchez, A.; Kureisaite-Ciziene, D.; Andreu, J.M.; Löwe, J. A polymerisation-associated conformational switch in FtsZ. mBio 2017, 8, e00254. [Google Scholar]
- Du, S.; Pichoff, S.; Kruse, K.; Lutkenhaus, J. FtsZ filaments have the opposite kinetic polarity of microtubules. Proc. Natl. Acad. Sci. USA 2018, 115, 10768–10773. [Google Scholar] [CrossRef]
- Zorrilla, S.; Monterroso, B.; Robles-Ramos, M.-Á.; Margolin, W.; Rivas, G. FtsZ interactions and biomolecular condensates as potential targets for new antibiotics. Antibiotics 2021, 10, 254. [Google Scholar] [CrossRef] [PubMed]
- Andreu, J.M.; Oliva, M.A.; Monasterio, O. Reversible unfolding of FtsZ cell division proteins from archaea and bacteria. Comparison with eukaryotic tubulin folding and assembly. J. Biol. Chem. 2002, 277, 43262–43270. [Google Scholar] [CrossRef]
- Huecas, S.; Canosa-Valls, A.J.; Araújo-Bazán, L.; Ruiz, F.M.; Laurents, D.V.; Fernández-Tornero, C.; Andreu, J.M. Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus. FEBS J. 2020, 287, 4048–4067. [Google Scholar] [CrossRef] [PubMed]
- Silber, N.; Pan, S.; Schäkermann, S.; Mayer, C.; Brötz-Oesterhelt, H.; Sass, P.; Søgaard-Andersen, L. Cell division protein FtsZ is unfolded for N-terminal degradation by antibiotic-activated ClpP. mBio 2020, 11, e01006–e01020. [Google Scholar] [CrossRef]
- Cordell, S.C.; Robinson, E.J.; Lowe, J. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl. Acad. Sci. USA 2003, 100, 7889–7894. [Google Scholar] [CrossRef] [PubMed]
- Bisson-Filho, A.W.; Discola, K.F.; Castellen, P.; Blasios, V.; Martins, A.; Sforca, M.L.; Garcia, W.; Zeri, A.C.; Erickson, H.P.; Dessen, A.; et al. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc. Natl. Acad. Sci. USA 2015, 112, E2130–E2138. [Google Scholar] [CrossRef] [PubMed]
- Araujo-Bazan, L.; Huecas, S.; Valle, J.; Andreu, D.; Andreu, J.M. Synthetic developmental regulator MciZ targets FtsZ across Bacillus species and inhibits bacterial division. Mol. Microbiol. 2019, 111, 965–980. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W. The prokaryotic cytoskeleton: A putative target for inhibitors and antibiotics? Appl. Microbiol. Biotechnol. 2006, 73, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Lock, R.L.; Harry, E.J. Cell-division inhibitors: New insights for future antibiotics. Nat. Rev. Drug. Discov. 2008, 7, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Haydon, D.J.; Stokes, N.R.; Ure, R.; Galbraith, G.; Bennett, J.M.; Brown, D.R.; Baker, P.J.; Barynin, V.V.; Rice, D.W.; Sedelnikova, S.E.; et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 2008, 321, 1673–1675. [Google Scholar] [CrossRef]
- Schaffner-Barbero, C.; Martin-Fontecha, M.; Chacon, P.; Andreu, J.M. Targeting the assembly of bacterial cell division protein FtsZ with small molecules. ACS Chem. Biol. 2012, 7, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Kusuma, K.D.; Payne, M.; Ung, A.T.; Bottomley, A.L.; Harry, E.J. FtsZ as an Antibacterial Target: Status and Guidelines for Progressing This Avenue. ACS Infect. Dis. 2019, 5, 1279–1294. [Google Scholar] [CrossRef] [PubMed]
- Silber, N.; Matos de Opitz, C.L.; Mayer, C.; Sass, P. Cell division protein FtsZ: From structure and mechanism to antibiotic target. Future Microbiol. 2020, 15, 801–831. [Google Scholar] [CrossRef] [PubMed]
- Casiraghi, A.; Suigo, L.; Valoti, E.; Straniero, V. Targeting bacterial cell division: A binding site-centered approach to the most promising inhibitors of the essential protein FtsZ. Antibiotics 2020, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wang, Z.; Li, T.; Teng, D.; Mao, R.; Hao, Y.; Yang, N.; Wang, X.; Wang, J. Recent progress of bacterial FtsZ inhibitors with a focus on peptides. FEBS J. 2021, 288, 1091–1106. [Google Scholar] [CrossRef]
- Pradhan, P.; Margolin, W.; Beuria, T.K. Targeting the achilles heel of FtsZ: The interdomain cleft. Front. Microbiol. 2021, 12, 732796. [Google Scholar] [CrossRef] [PubMed]
- Lehar, S.M.; Pillow, T.; Xu, M.; Staben, L.; Kajihara, K.K.; Vandlen, R.; DePalatis, L.; Raab, H.; Hazenbos, W.L.; Hiroshi Morisaki, J.; et al. Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature 2015, 527, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Nejad, A.J.; Shahrokhi, N.; Nielsen, P.E. Targeting of the essential acpP, ftsZ, and rne genes in carbapenem-resistant Acinetobacter baumannii by antisense PNA precision antibacterials. Biomedicines 2021, 9, 429. [Google Scholar] [CrossRef] [PubMed]
- Mückl, A.; Schwarz-Schilling, M.; Fischer, K.; Simmel, F.C. Filamentation and restoration of normal growth in Escherichia coli using a combined CRISPRi sgRNA/antisense RNA approach. PLoS ONE 2018, 13, e0198058. [Google Scholar] [CrossRef] [PubMed]
- Sass, P.; Josten, M.; Famulla, K.; Schiffer, G.; Sahl, H.-G.; Hamoen, L.; Brötz-Oesterhelt, H. Antibiotic acyldepsipeptides activate ClpP peptidase to degrade the cell division protein FtsZ. Proc. Natl. Acad. Sci. USA 2011, 108, 17474–17479. [Google Scholar] [CrossRef] [PubMed]
- Marcelo, F.; Huecas, S.; Ruiz-Avila, L.B.; Canada, F.J.; Perona, A.; Poveda, A.; Martin-Santamaria, S.; Morreale, A.; Jimenez-Barbero, J.; Andreu, J.M. Interactions of bacterial cell division protein FtsZ with C8-substituted guanine nucleotide inhibitors. A combined NMR, biochemical and molecular modeling perspective. J. Am. Chem. Soc. 2013, 135, 16418–16428. [Google Scholar] [CrossRef]
- Schrodinger, LLC. The PyMOL Molecular Graphics System; Version 1.8; Schrodinger, LCC.: New York, NY, USA, 2015. [Google Scholar]
- Lappchen, T. Synthesis of GTP Analogues and Evaluation of Their Effect on the Antibiotic Target FtsZ and Its Eukaryotic Homologue Tubulin. Ph.D. Thesis, University of Amsterdam, Amsterdam, The Netherlands, 2007. Available online: https://hdl.handle.net/11245/1.270147 (accessed on 27 May 2022).
- Shahsavari, N.; Wang, B.; Imai, Y.; Mori, M.; Son, S.; Liang, L.; Böhringer, N.; Manuse, S.; Gates Michael, F.; Morrissette, M.; et al. A silent operon of photorhabdus luminescens encodes a prodrug mimic of GTP. mBio 2022, 0, e00700–e00722. [Google Scholar] [CrossRef] [PubMed]
- Huecas, S.; Schaffner-Barbero, C.; Garcia, W.; Yebenes, H.; Palacios, J.M.; Diaz, J.F.; Menendez, M.; Andreu, J.M. The interactions of cell division protein FtsZ with guanine nucleotides. J. Biol. Chem. 2007, 282, 37515–37528. [Google Scholar] [CrossRef] [PubMed]
- Huecas, S.; Marcelo, F.; Perona, A.; Ruiz-Avila, L.B.; Morreale, A.; Canada, F.J.; Jimenez-Barbero, J.; Andreu, J.M. Beyond a Fluorescent Probe: Inhibition of Cell Division Protein FtsZ by mant-GTP Elucidated by NMR and Biochemical Approaches. ACS Chem. Biol. 2015, 10, 2382–2392. [Google Scholar] [CrossRef] [PubMed]
- Schaffner-Barbero, C.; Gil-Redondo, R.; Ruiz-Avila, L.B.; Huecas, S.; Lappchen, T.; den Blaauwen, T.; Diaz, J.F.; Morreale, A.; Andreu, J.M. Insights into nucleotide recognition by cell division protein FtsZ from a mant-GTP competition assay and molecular dynamics. Biochemistry 2010, 49, 10458–10472. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruiz-Avila, L.B.; Huecas, S.; Artola, M.; Vergonos, A.; Ramirez-Aportela, E.; Cercenado, E.; Barasoain, I.; Vazquez-Villa, H.; Martin-Fontecha, M.; Chacon, P.; et al. Synthetic Inhibitors of Bacterial Cell Division Targeting the GTP-Binding Site of FtsZ. ACS Chem. Biol. 2013, 8, 2072–2083. [Google Scholar] [CrossRef]
- Artola, M.; Ruiz-Avila, L.B.; Vergoñós, A.; Huecas, S.; Araujo-Bazán, L.; Martín-Fontecha, M.; Vázquez-Villa, H.; Turrado, C.; Ramírez-Aportela, E.; Hoegl, A.; et al. Effective GTP-replacing FtsZ inhibitors and antibacterial mechanism of action. ACS Chem. Biol. 2015, 10, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.E.; Kim, M.B.; Moore, J.T.; O’Brien, T.E.; Sorto, N.A.; Grove, C.I.; Lackner, L.L.; Ames, J.B.; Shaw, J.T. Comparison of small molecule inhibitors of the bacterial cell division protein FtsZ and identification of a reliable cross-species inhibitor. ACS Chem. Biol. 2012, 7, 1918–1928. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Plaza, A.; Keffer, J.L.; Bifulco, G.; Lloyd, J.R.; Bewley, C.A. Chrysophaentins A-H, antibacterial bisdiarylbutene macrocycles that inhibit the bacterial cell division protein FtsZ. J. Am. Chem. Soc. 2010, 132, 9069–9077. [Google Scholar] [CrossRef] [PubMed]
- Keffer, J.L.; Huecas, S.; Hammill, J.T.; Wipf, P.; Andreu, J.M.; Bewley, C.A. Chrysophaentins are competitive inhibitors of FtsZ and inhibit Z-ring formation in live bacteria. Bioorg. Med. Chem. 2013, 21, 5673–5678. [Google Scholar] [CrossRef] [PubMed]
- Matthew, S.; Chen, Q.-Y.; Ratnayake, R.; Fermaintt Charles, S.; Lucena-Agell, D.; Bonato, F.; Prota Andrea, E.; Lim Seok, T.; Wang, X.; Díaz, J.F.; et al. Gatorbulin-1, a distinct cyclodepsipeptide chemotype, targets a seventh tubulin pharmacological site. Proc. Natl. Acad. Sci. USA 2021, 118, e2021847118. [Google Scholar] [CrossRef] [PubMed]
- Mühlethaler, T.; Gioia, D.; Prota, A.E.; Sharpe, M.E.; Cavalli, A.; Steinmetz, M.O. Comprehensive analysis of binding sites in tubulin. Angew. Chem. Int. Ed. 2021, 60, 13331–13342. [Google Scholar] [CrossRef] [PubMed]
- Lutkenhaus, J.F.; Wolf-Watz, H.; Donachie, W.D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J. Bacteriol. 1980, 142, 615–620. [Google Scholar] [CrossRef]
- Strahl, H.; Hamoen, L.W. Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. USA 2010, 107, 12281–12286. [Google Scholar] [CrossRef] [PubMed]
- Foss, M.H.; Eun, Y.J.; Grove, C.I.; Pauw, D.A.; Sorto, N.A.; Rensvold, J.W.; Pagliarini, D.J.; Shaw, J.T.; Weibel, D.B. Inhibitors of bacterial tubulin target bacterial membranes. MedChemComm 2013, 4, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Araujo-Bazan, L.; Ruiz-Avila, L.B.; Andreu, D.; Huecas, S.; Andreu, J.M. Cytological profile of antibacterial FtsZ inhibitors and synthetic peptide MciZ. Front. Microbiol. 2016, 7, 1558. [Google Scholar] [CrossRef] [PubMed]
- Nonejuie, P.; Burkart, M.; Pogliano, K.; Pogliano, J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. USA 2013, 110, 16169–16174. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Chijiiwa, Y.; Suzuki, K.; Takahashi, K.; Nanamiya, H.; Sato, T.; Hosoya, Y.; Ochi, K.; Kawamura, F. The lethal effect of a benzamide derivative, 3-methoxybenzamide, can be suppressed by mutations within a cell division gene, ftsZ, in Bacillus subtilis. J. Bacteriol. 1999, 181, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Haydon, D.J.; Bennett, J.M.; Brown, D.; Collins, I.; Galbraith, G.; Lancett, P.; Macdonald, R.; Stokes, N.R.; Chauhan, P.K.; Sutariya, J.K.; et al. Creating an Antibacterial with in Vivo Efficacy: Synthesis and Characterization of Potent Inhibitors of the Bacterial Cell Division Protein FtsZ with Improved Pharmaceutical Properties. J. Med. Chem. 2010, 53, 3927–3936. [Google Scholar] [CrossRef]
- Andreu, J.M.; Schaffner-Barbero, C.; Huecas, S.; Alonso, D.; Lopez-Rodriguez, M.L.; Ruiz-Avila, L.B.; Nunez-Ramirez, R.; Llorca, O.; Martin-Galiano, A.J. The antibacterial cell division inhibitor PC190723 is an FtsZ polymer-stabilizing agent that induces filament assembly and condensation. J. Biol. Chem. 2010, 285, 14239–14246. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.W.; Wu, L.J.; Czaplewski, L.G.; Errington, J. Multiple effects of benzamide antibiotics on FtsZ function. Mol. Microbiol. 2011, 80, 68–84. [Google Scholar] [CrossRef]
- Elsen, N.L.; Lu, J.; Parthasarathy, G.; Reid, J.C.; Sharma, S.; Soisson, S.M.; Lumb, K.J. Mechanism of action of the cell-division inhibitor PC190723: Modulation of FtsZ assembly cooperativity. J. Am. Chem. Soc. 2012, 134, 12342–12345. [Google Scholar] [CrossRef]
- Stokes, N.R.; Baker, N.; Bennett, J.M.; Berry, J.; Collins, I.; Czaplewski, L.G.; Logan, A.; Macdonald, R.; Macleod, L.; Peasley, H.; et al. An improved small-molecule inhibitor of FtsZ with superior in vitro potency, drug-like properties, and in vivo efficacy. Antimicrob. Agents Chemother. 2013, 57, 317–325. [Google Scholar] [CrossRef]
- Kaul, M.; Mark, L.; Parhi, A.K.; LaVoie, E.J.; Pilch, D.S. Combining the FtsZ-targeting prodrug TXA709 and the cephalosporin cefdinir confers synergy and reduces the frequency of resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2016, 60, 4290–4296. [Google Scholar] [CrossRef]
- Kellogg, E.H.; Hejab, N.M.A.; Howes, S.; Northcote, P.; Miller, J.H.; Díaz, J.F.; Downing, K.H.; Nogales, E. Insights into the distinct mechanisms of action of taxane and non-taxane microtubule stabilizers from cryo-EM structures. J. Mol. Biol. 2017, 429, 633–646. [Google Scholar] [CrossRef]
- Sharma, A.K.; Poddar, S.M.; Chakraborty, J.; Nayak, B.S.; Kalathil, S.; Mitra, N.; Pananghat, G.; Srinivasan, R. A salt bridge mediated resistance mechanism to FtsZ inhibitor PC190723 revealed by a single step cell-based screen. bioRxiv 2022. [Google Scholar] [CrossRef]
- Richter, M.F.; Drown, B.S.; Riley, A.P.; Garcia, A.; Shirai, T.; Svec, R.L.; Hergenrother, P.J. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 2017, 545, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Rosado-Lugo, J.D.; Sun, Y.; Banerjee, A.; Cao, Y.; Datta, P.; Zhang, Y.; Yuan, Y.; Parhi, A.K. Evaluation of 2,6-difluoro-3-(oxazol-2-ylmethoxy)benzamide chemotypes as Gram-negative FtsZ inhibitors. J. Antibiot. 2022, 75, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Straniero, V.; Sebastián-Pérez, V.; Suigo, L.; Margolin, W.; Casiraghi, A.; Hrast, M.; Zanotto, C.; Zdovc, I.; Radaelli, A.; Valoti, E. Computational design and development of benzodioxane-benzamides as potent inhibitors of FtsZ by exploring the hydrophobic subpocket. Antibiotics 2021, 10, 442. [Google Scholar] [CrossRef]
- Fredborg, M.; Rosenvinge, F.S.; Spillum, E.; Kroghsbo, S.; Wang, M.; Sondergaard, T.E. Automated image analysis for quantification of filamentous bacteria. BMC Microbiol. 2015, 15, 255. [Google Scholar] [CrossRef]
- Wang, J.; Galgoci, A.; Kodali, S.; Herath, K.B.; Jayasuriya, H.; Dorso, K.; Vicente, F.; González, A.; Cully, D.; Bramhill, D.; et al. Discovery of a small molecule that inhibits cell division by blocking FtsZ, a novel therapeutic target of tntibiotics. J. Biol. Chem. 2003, 278, 44424–44428. [Google Scholar] [CrossRef]
- Stokes, N.R.; Sievers, J.; Barker, S.; Bennett, J.M.; Brown, D.R.; Collins, I.; Errington, V.M.; Foulger, D.; Hall, M.; Halsey, R.; et al. Novel inhibitors of bacterial cytokinesis identified by a cell-based antibiotic screening assay. J. Biol. Chem. 2005, 280, 39709–39715. [Google Scholar] [CrossRef]
- Charoensutthivarakul, S.; Thomas, S.E.; Curran, A.; Brown, K.P.; Belardinelli, J.M.; Whitehouse, A.J.; Acebrón-García-de-Eulate, M.; Sangan, J.; Gramani, S.G.; Jackson, M.; et al. Development of inhibitors of SAICAR synthetase (PurC) from Mycobacterium abscessus using a fragment-based approach. ACS Infect. Dis. 2022, 8, 296–309. [Google Scholar] [CrossRef]
- Dalvit, C.; Vulpetti, A. Ligand-based fluorine NMR screening: Principles and applications in drug discovery projects. J. Med. Chem. 2019, 62, 2218–2244. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.-Y.; Sun, N.; Neves, M.A.C.; Lam, P.C.-H.; Chung, W.-H.; Wong, L.-K.; Chow, H.-Y.; Ma, D.-L.; Chan, P.-H.; Leung, Y.-C.; et al. Identification of a new class of FtsZ inhibitors by structure-based design and in vitro screening. J. Chem. Inf. Model. 2013, 53, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Chan, F.-Y.; Lu, Y.-J.; Neves, M.A.C.; Lui, H.-K.; Wang, Y.; Chow, K.-Y.; Chan, K.-F.; Yan, S.-C.; Leung, Y.-C.; et al. Rational design of berberine-based FtsZ inhibitors with broad-spectrum antibacterial activity. PLoS ONE 2014, 9, e97514. [Google Scholar] [CrossRef] [PubMed]
- Totrov, M.; Abagyan, R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins 1997, 29 (Suppl. 1), 215–220. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. ZINC − A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- ZINC15. Available online: https://zinc.docking.org (accessed on 27 May 2022).
- Vergoñós-Tomás, A. Inhibidores de la Proteína de División Celular Bacteriana FtsZ Dirigidos al Sitio de Unión del Nucleótido. Ph.D. Thesis, Autonomous University of Madrid, Madrid, Spain, 2017. Available online: https://repositorio.uam.es/handle/10486/680161?show=full&locale-attribute=en (accessed on 27 May 2022).
- Ramirez-Aportela, E. Dinámica de los Filamentos de FtsZ y Búsqueda Racional de Inhibidores Sintéticos con Actividad Antibacteriana. Ph.D. Thesis, Autonomous University of Madrid, Madrid, Spain, 2017. Available online: https://repositorio.uam.es/handle/10486/677469 (accessed on 27 May 2022).
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A deep learning approach to antibiotic discovery. Cell 2020, 180, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Jayatunga, M.K.P.; Xie, W.; Ruder, L.; Schulze, U.; Meier, C. AI in small-molecule drug discovery: A coming wave? Nat. Rev. Drug Discov. 2022, 21, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Gentile, F.; Yaacoub, J.C.; Gleave, J.; Fernandez, M.; Ton, A.-T.; Ban, F.; Stern, A.; Cherkasov, A. Artificial intelligence–enabled virtual screening of ultra-large chemical libraries with deep docking. Nat. Protoc. 2022, 17, 672–697. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreu, J.M.; Huecas, S.; Araújo-Bazán, L.; Vázquez-Villa, H.; Martín-Fontecha, M. The Search for Antibacterial Inhibitors Targeting Cell Division Protein FtsZ at Its Nucleotide and Allosteric Binding Sites. Biomedicines 2022, 10, 1825. https://doi.org/10.3390/biomedicines10081825
Andreu JM, Huecas S, Araújo-Bazán L, Vázquez-Villa H, Martín-Fontecha M. The Search for Antibacterial Inhibitors Targeting Cell Division Protein FtsZ at Its Nucleotide and Allosteric Binding Sites. Biomedicines. 2022; 10(8):1825. https://doi.org/10.3390/biomedicines10081825
Chicago/Turabian StyleAndreu, José M., Sonia Huecas, Lidia Araújo-Bazán, Henar Vázquez-Villa, and Mar Martín-Fontecha. 2022. "The Search for Antibacterial Inhibitors Targeting Cell Division Protein FtsZ at Its Nucleotide and Allosteric Binding Sites" Biomedicines 10, no. 8: 1825. https://doi.org/10.3390/biomedicines10081825
APA StyleAndreu, J. M., Huecas, S., Araújo-Bazán, L., Vázquez-Villa, H., & Martín-Fontecha, M. (2022). The Search for Antibacterial Inhibitors Targeting Cell Division Protein FtsZ at Its Nucleotide and Allosteric Binding Sites. Biomedicines, 10(8), 1825. https://doi.org/10.3390/biomedicines10081825





