Pathophysiological Response to SARS-CoV-2 Infection Detected by Infrared Spectroscopy Enables Rapid and Robust Saliva Screening for COVID-19
Abstract
:1. Introduction
2. Materials and Methods
2.1. SARS-CoV-2 Virus
2.2. In Vitro Cell Infection Model
2.3. Nucleic Acid Extraction and RT–qPCR
2.4. Mouse Model
2.5. Mouse Oral Lavage Proteomics
2.6. Human Cohort Study
2.7. ATR-FTIR Spectra Acquisition and Processing
2.8. Statistical Analysis
3. Results
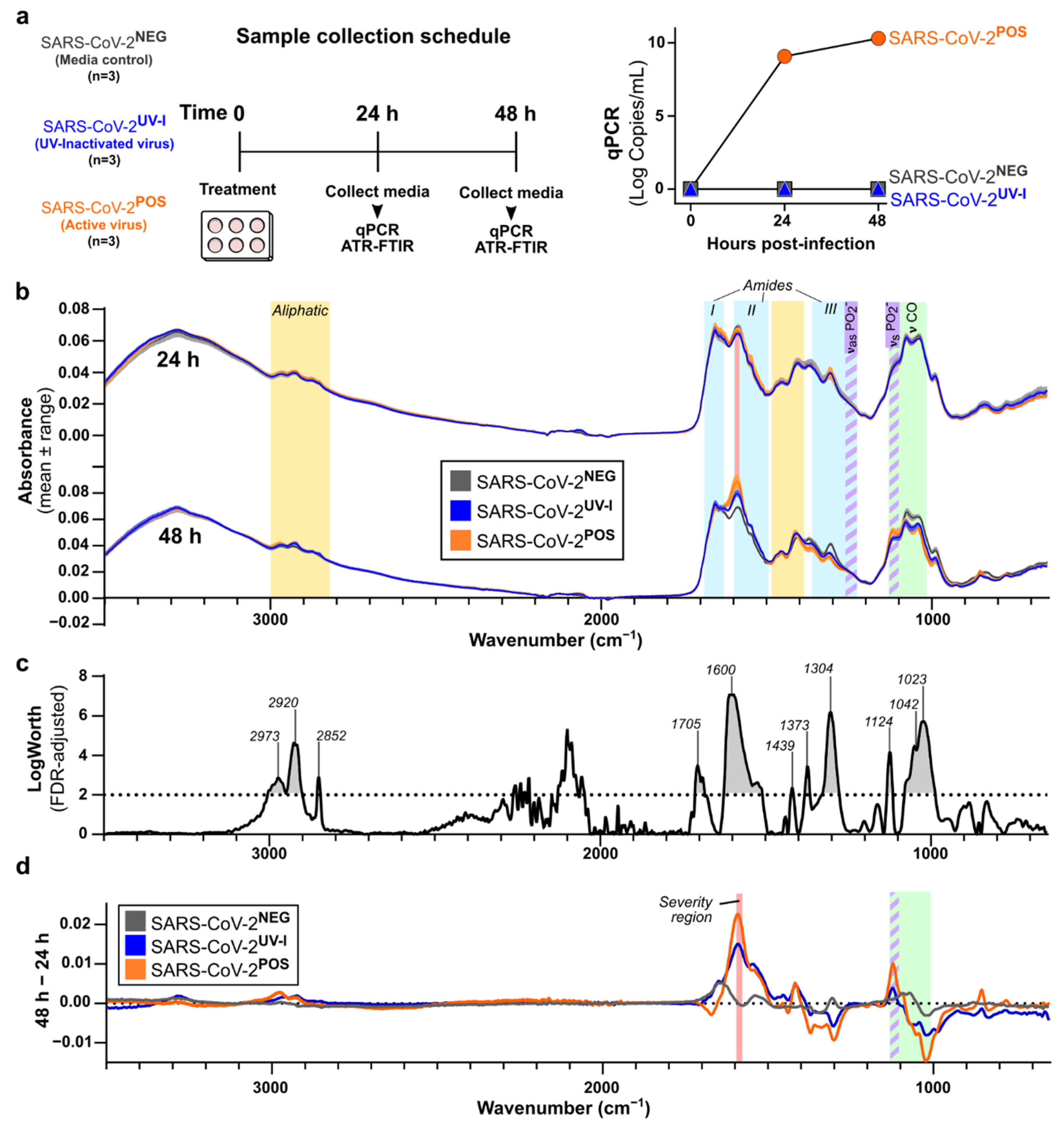
3.1. Characterisation of In Vitro SARS-CoV-2 Infection-Induced Secretome ATR-FTIR Spectra
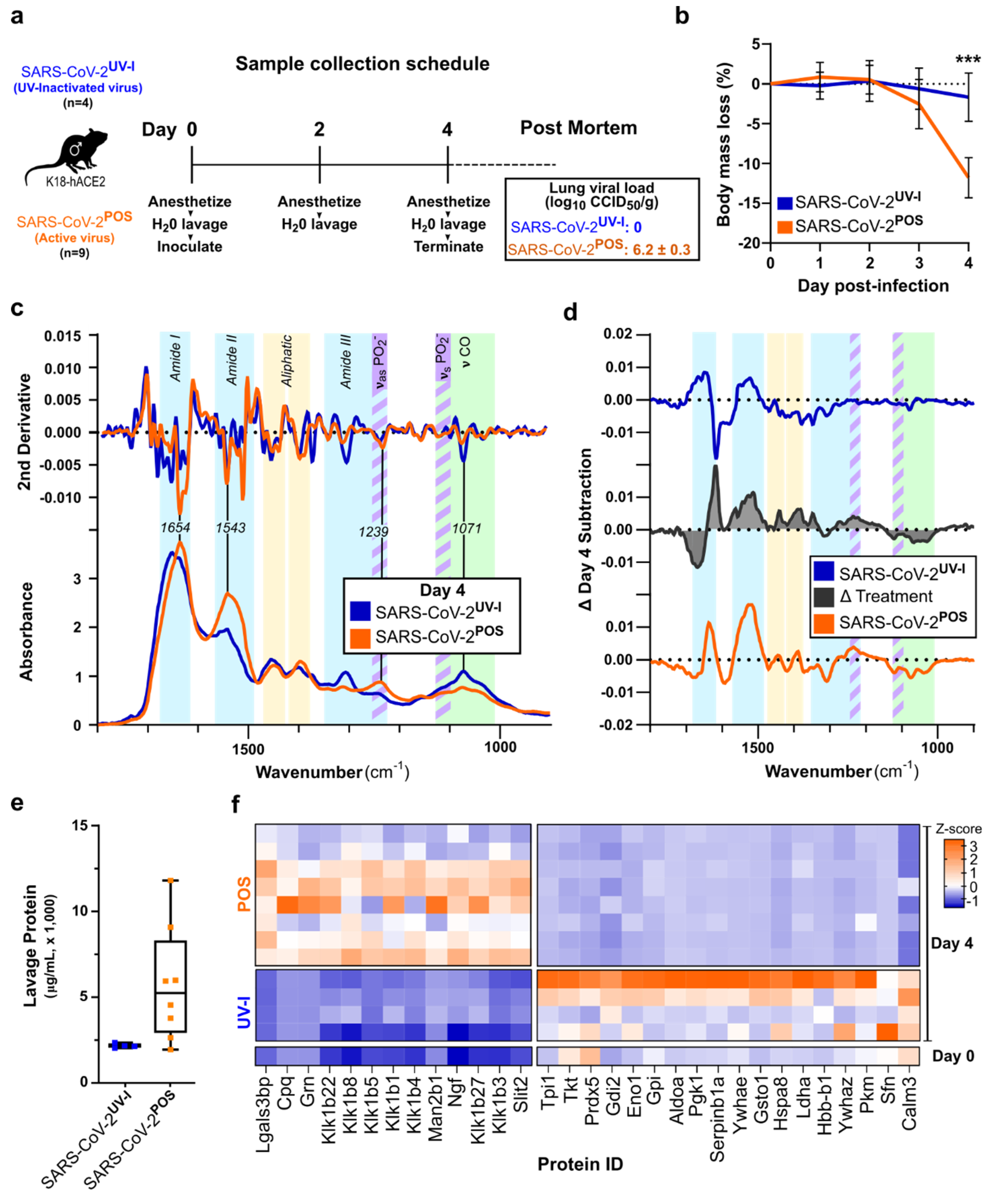
3.2. ATR-FTIR Spectra of Oral Lavage from Respiratory SARS-CoV-2-Infected Mouse Model
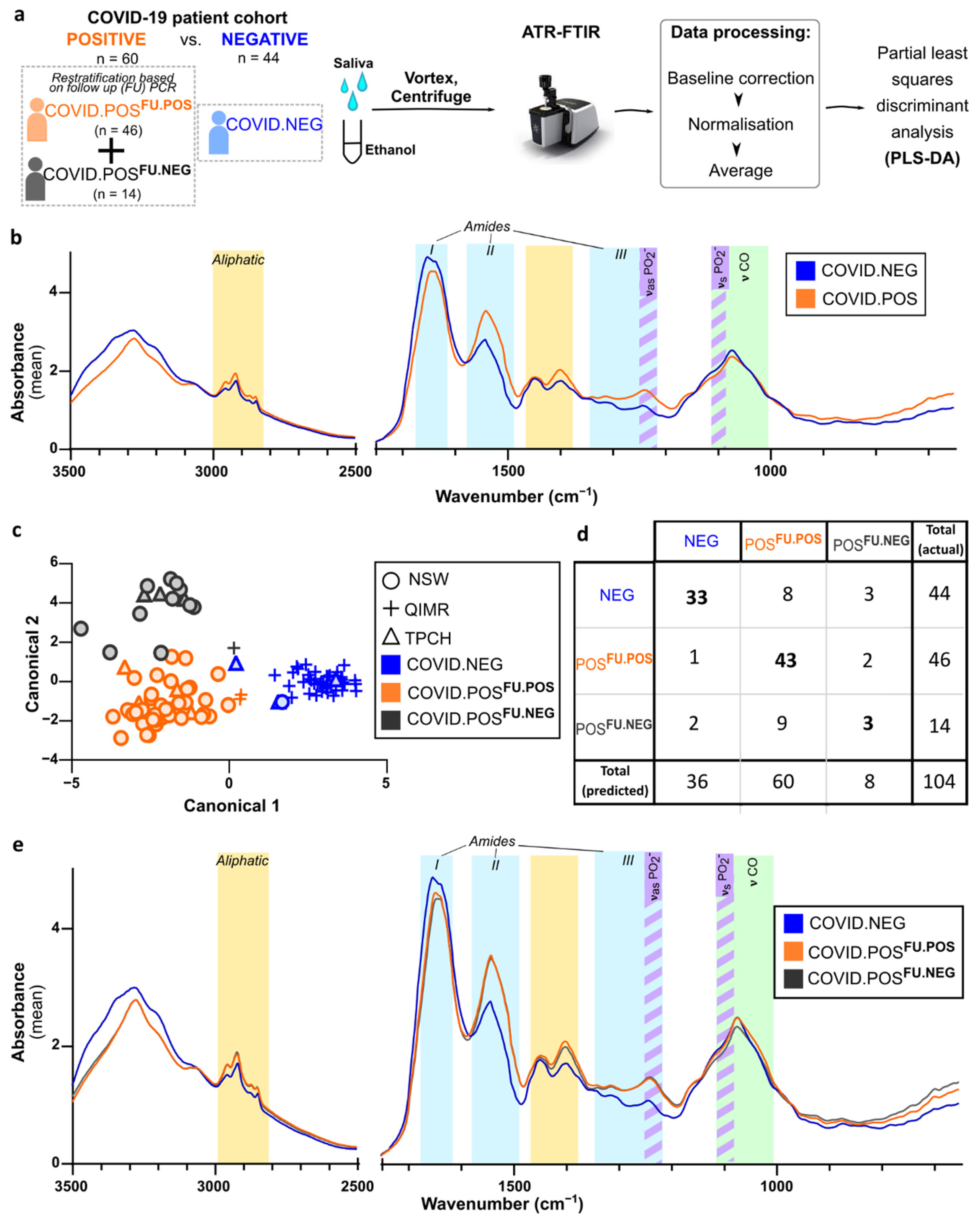
3.3. ATR-FTIR Spectra of Human Saliva Distinguishes SARS-CoV-2 Infection Status
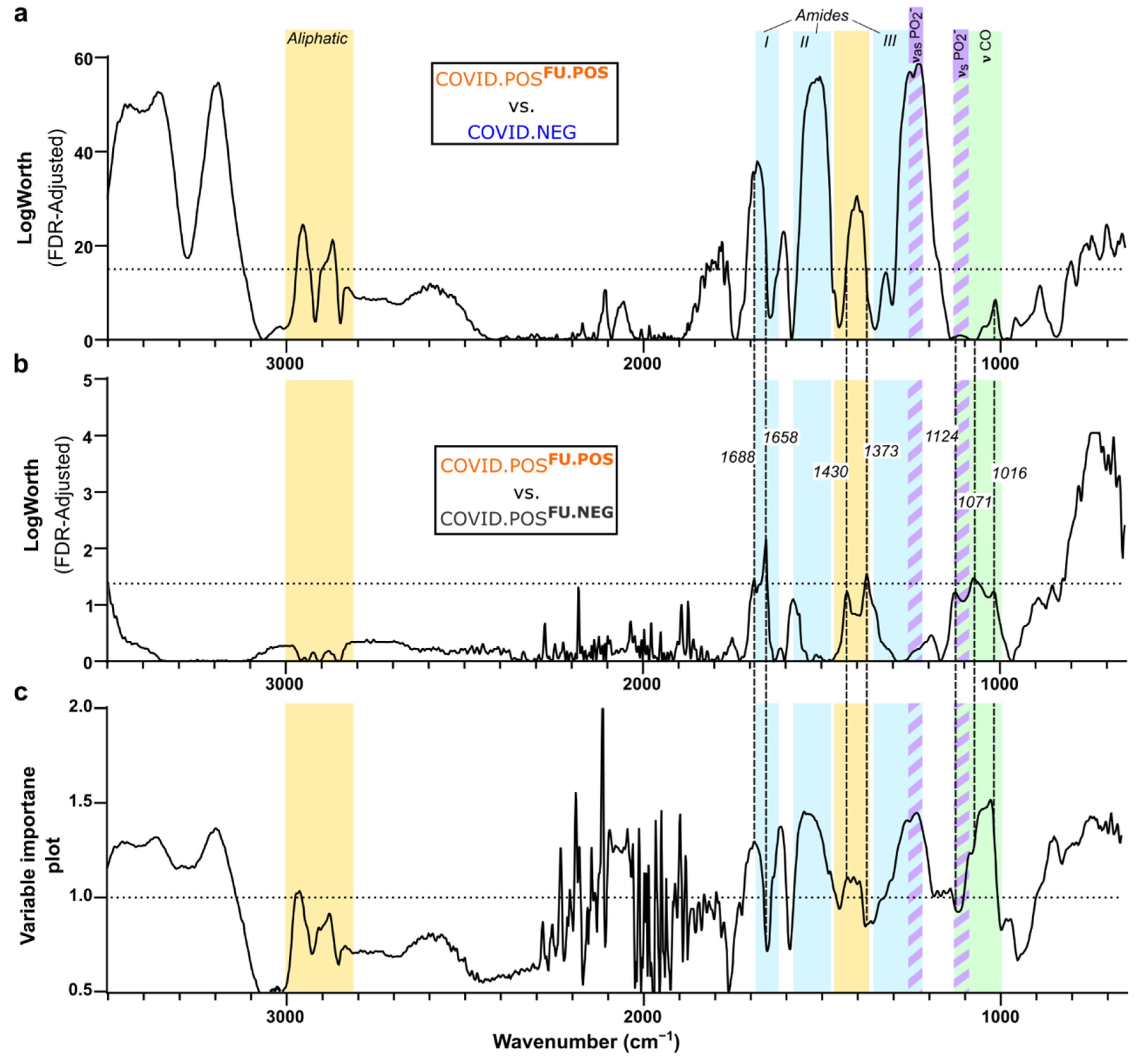
3.4. Delineation of Spectral Signature for COVID.POSFU.POS Saliva
3.5. Comparison of COVID-19 Spectral Signature across Diverse Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [CrossRef]
- Sohrabi, C.; Alsafi, Z.; O’Neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, R. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 9 January 2022).
- Guo, J.; Ge, J.; Guo, Y. Recent advances in methods for the diagnosis of Corona Virus Disease 2019. J. Clin. Lab. Anal. 2021, 36, e24178. [Google Scholar] [CrossRef] [PubMed]
- Fathi Karkan, S.; Maleki Baladi, R.; Shahgolzari, M.; Gholizadeh, M.; Shayegh, F.; Arashkia, A. The evolving direct and indirect platforms for the detection of SARS-CoV-2. J. Virol. Methods 2022, 300, 114381. [Google Scholar] [CrossRef] [PubMed]
- Toropov, N.; Osborne, E.; Joshi, L.T.; Davidson, J.; Morgan, C.; Page, J.; Pepperell, J.; Vollmer, F. SARS-CoV-2 Tests: Bridging the Gap between Laboratory Sensors and Clinical Applications. ACS Sens. 2021, 6, 2815–2837. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Berhane, S.; Taylor, M.; Adriano, A.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2021, 3, CD013705. [Google Scholar] [CrossRef]
- Barauna, V.G.; Singh, M.N.; Barbosa, L.L.; Marcarini, W.D.; Vassallo, P.F.; Mill, J.G.; Ribeiro-Rodrigues, R.; Campos, L.C.G.; Warnke, P.H.; Martin, F.L. Ultrarapid On-Site Detection of SARS-CoV-2 Infection Using Simple ATR-FTIR Spectroscopy and an Analysis Algorithm: High Sensitivity and Specificity. Anal. Chem. 2021, 93, 2950–2958. [Google Scholar] [CrossRef]
- Martinez-Cuazitl, A.; Vazquez-Zapien, G.J.; Sanchez-Brito, M.; Limon-Pacheco, J.H.; Guerrero-Ruiz, M.; Garibay-Gonzalez, F.; Delgado-Macuil, R.J.; de Jesus, M.G.G.; Corona-Perezgrovas, M.A.; Pereyra-Talamantes, A.; et al. ATR-FTIR spectrum analysis of saliva samples from COVID-19 positive patients. Sci. Rep. 2021, 11, 19980. [Google Scholar] [CrossRef]
- Wood, B.R.; Kochan, K.; Bedolla, D.E.; Salazar-Quiroz, N.; Grimley, S.L.; Perez-Guaita, D.; Baker, M.J.; Vongsvivut, J.; Tobin, M.J.; Bambery, K.R.; et al. Infrared Based Saliva Screening Test for COVID-19. Angew. Chem. Int. Ed. Engl. 2021, 60, 17102–17107. [Google Scholar] [CrossRef]
- Naseer, K.; Ali, S.; Qazi, J. ATR-FTIR spectroscopy as the future of diagnostics: A systematic review of the approach using bio-fluids. Appl. Spectrosc. Rev. 2021, 56, 85–97. [Google Scholar] [CrossRef]
- Banerjee, A.; Gokhale, A.; Bankar, R.; Palanivel, V.; Salkar, A.; Robinson, H.; Shastri, J.S.; Agrawal, S.; Hartel, G.; Hill, M.M.; et al. Rapid Classification of COVID-19 Severity by ATR-FTIR Spectroscopy of Plasma Samples. Anal. Chem. 2021, 93, 10391–10396. [Google Scholar] [CrossRef] [PubMed]
- Amarilla, A.A.; Sng, J.D.J.; Parry, R.; Deerain, J.M.; Potter, J.R.; Setoh, Y.X.; Rawle, D.J.; Le, T.T.; Modhiran, N.; Wang, X.; et al. A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat. Commun. 2021, 12, 3431. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.J.; Le, T.T.; Dobbin, C.A.; Banovic, T.; Howard, C.B.; Flores Fde, M.; Vanags, D.; Naylor, D.J.; Hill, G.R.; Suhrbier, A. Heat shock protein 10 inhibits lipopolysaccharide-induced inflammatory mediator production. J. Biol. Chem. 2005, 280, 4037–4047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, R.J.; Humphrey, S.J.; Fortuna, P.R.J.; Lor, M.; Foster, S.R.; Quaife-Ryan, G.A.; Johnston, R.L.; Dumenil, T.; Bishop, C.; Rudraraju, R.; et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell 2021, 184, 2167–2182.e2122. [Google Scholar] [CrossRef]
- Simonova, D.; Karamancheva, I. Application of Fourier Transform Infrared Spectroscopy for Tumor Diagnosis. Biotechnol. Biotec. Eq. 2013, 27, 4200–4207. [Google Scholar] [CrossRef]
- Mateus, T.; Almeida, I.; Costa, A.; Viegas, D.; Magalhaes, S.; Martins, F.; Herdeiro, M.T.; Silva, O.A.B.D.E.; Fraga, C.; Alves, I.; et al. Fourier-Transform Infrared Spectroscopy as a Discriminatory Tool for Myotonic Dystrophy Type 1 Metabolism: A Pilot Study. Int. J. Environ. Res. Pub. Health 2021, 18, 3800. [Google Scholar] [CrossRef]
- Yinda, C.K.; Port, J.R.; Bushmaker, T.; Offei Owusu, I.; Purushotham, J.N.; Avanzato, V.A.; Fischer, R.J.; Schulz, J.E.; Holbrook, M.G.; Hebner, M.J.; et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021, 17, e1009195. [Google Scholar] [CrossRef]
- Arce, V.M.; Costoya, J.A. SARS-CoV-2 infection in K18-ACE2 transgenic mice replicates human pulmonary disease in COVID-19. Cell Mol. Immunol. 2021, 18, 513–514. [Google Scholar] [CrossRef]
- Rosenfeld, R.; Noy-Porat, T.; Mechaly, A.; Makdasi, E.; Levy, Y.; Alcalay, R.; Falach, R.; Aftalion, M.; Epstein, E.; Gur, D.; et al. Post-exposure protection of SARS-CoV-2 lethal infected K18-hACE2 transgenic mice by neutralizing human monoclonal antibody. Nat. Commun. 2021, 12, 944. [Google Scholar] [CrossRef]
- Zheng, J.; Wong, L.R.; Li, K.; Verma, A.K.; Ortiz, M.E.; Wohlford-Lenane, C.; Leidinger, M.R.; Knudson, C.M.; Meyerholz, D.K.; McCray, P.B., Jr.; et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 2021, 589, 603–607. [Google Scholar] [CrossRef]
- Garcia-Arriaza, J.; Garaigorta, U.; Perez, P.; Lazaro-Frias, A.; Zamora, C.; Gastaminza, P.; Del Fresno, C.; Casasnovas, J.M.; Sorzano, C.O.S.; Sancho, D.; et al. COVID-19 vaccine candidates based on modified vaccinia virus Ankara expressing the SARS-CoV-2 spike induce robust T- and B-cell immune responses and full efficacy in mice. J. Virol. 2021, 95, e02260-20. [Google Scholar] [CrossRef] [PubMed]
- Alsoussi, W.B.; Turner, J.S.; Case, J.B.; Zhao, H.; Schmitz, A.J.; Zhou, J.Q.; Chen, R.E.; Lei, T.; Rizk, A.A.; McIntire, K.M.; et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020, 205, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.O.; Case, J.B.; Winkler, E.S.; Thackray, L.B.; Kafai, N.M.; Bailey, A.L.; McCune, B.T.; Fox, J.M.; Chen, R.E.; Alsoussi, W.B.; et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 2020, 182, 744–753.e4. [Google Scholar] [CrossRef] [PubMed]
- Zucchiatti, P.; Mitri, E.; Kenig, S.; Bille, F.; Kourousias, G.; Bedolla, D.E.; Vaccari, L. Contribution of Ribonucleic Acid (RNA) to the Fourier Transform Infrared (FTIR) Spectrum of Eukaryotic Cells. Anal. Chem. 2016, 88, 12090–12098. [Google Scholar] [CrossRef] [PubMed]
- Ami, D.; Lavatelli, F.; Rognoni, P.; Palladini, G.; Raimondi, S.; Giorgetti, S.; Monti, L.; Doglia, S.M.; Natalello, A.; Merlini, G. In situ characterization of protein aggregates in human tissues affected by light chain amyloidosis: A FTIR microspectroscopy study. Sci. Rep. 2016, 6, 29096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milosevic, J.; Prodanovic, R.; Polovic, N. On the Protein Fibrillation Pathway: Oligomer Intermediates Detection Using ATR-FTIR Spectroscopy. Molecules 2021, 26, 970. [Google Scholar] [CrossRef]
- Wood, B.R. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem. Soc. Rev. 2016, 45, 1980–1998. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Geinguenaud, F.; Militello, V.; Arluison, V. Application of FTIR Spectroscopy to Analyze RNA Structure. Methods Mol. Biol. 2020, 2113, 119–133. [Google Scholar] [CrossRef]
- Zhuang, J.S.; Li, M.; Pu, Y.Q.; Ragauskas, A.J.; Yoo, C.G. Observation of Potential Contaminants in Processed Biomass Using Fourier Transform Infrared Spectroscopy. Appl. Sci. 2020, 10, 4345. [Google Scholar] [CrossRef]
- Banyay, M.; Sarkar, M.; Graslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef]
- Mehrotra, R. Infrared Spectroscopy, Gas Chromatography/Infrared in Food Analysis. In Encyclopedia of Analytical Chemistry; Wiley: Hoboken, NJ, USA, 2006. [Google Scholar] [CrossRef]
- Williams, E.; Bond, K.; Zhang, B.W.; Putland, M.; Williamson, D.A. Saliva as a Noninvasive Specimen for Detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e00776-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manabe, Y.C.; Reuland, C.; Yu, T.; Azamfirei, R.; Hardick, J.P.; Church, T.; Brown, D.M.; Sewell, T.T.; Antar, A.; Blair, P.W.; et al. Self-Collected Oral Fluid Saliva Is Insensitive Compared with Nasal-Oropharyngeal Swabs in the Detection of Severe Acute Respiratory Syndrome Coronavirus 2 in Outpatients. Open Forum Infect. Dis. 2021, 8, ofaa648. [Google Scholar] [CrossRef]
- Butler-Laporte, G.; Lawandi, A.; Schiller, I. Comparison of Saliva and Nasopharyngeal Swab Nucleic Acid Amplification Testing for Detection of SARS-CoV-2: A Systematic Review and Meta-analysis. JAMA Intern. Med. 2021, 181, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Yang, H.; Zeng, K.; Zhang, J.; Zhong, N.; Wang, Y.; Ye, J.; Tu, P.; Liu, Z. EGCG Inhibited Lipofuscin Formation Based on Intercepting Amyloidogenic beta-Sheet-Rich Structure Conversion. PLoS ONE 2016, 11, e0152064. [Google Scholar] [CrossRef]
- Sinha, N.; Thakur, A.K. Likelihood of amyloid formation in COVID-19-induced ARDS. Trends Microbiol. 2021, 29, 967–969. [Google Scholar] [CrossRef] [PubMed]
- Jana, A.K.; Greenwood, A.B.; Hansmann, U.H.E. Presence of a SARS-CoV-2 Protein Enhances Amyloid Formation of Serum Amyloid A. J. Phys. Chem. B 2021, 125, 9155–9167. [Google Scholar] [CrossRef] [PubMed]
- Zinellu, A.; Paliogiannis, P.; Carru, C.; Mangoni, A.A. Serum amyloid A concentrations, COVID-19 severity and mortality: An updated systematic review and meta-analysis. Int. J. Infect. Dis. 2021, 105, 668–674. [Google Scholar] [CrossRef]
- Tavassoly, O.; Safavi, F.; Tavassoly, I. Seeding Brain Protein Aggregation by SARS-CoV-2 as a Possible Long-Term Complication of COVID-19 Infection. ACS Chem. Neurosci. 2020, 11, 3704–3706. [Google Scholar] [CrossRef]
- Idrees, D.; Kumar, V. SARS-CoV-2 spike protein interactions with amyloidogenic proteins: Potential clues to neurodegeneration. Biochem. Biophys. Res. Commun. 2021, 554, 94–98. [Google Scholar] [CrossRef]
- Geng, H.; Subramanian, S.; Wu, L.; Bu, H.F.; Wang, X.; Du, C.; De Plaen, I.G.; Tan, X.D. SARS-CoV-2 ORF8 Forms Intracellular Aggregates and Inhibits IFNgamma-Induced Antiviral Gene Expression in Human Lung Epithelial Cells. Front. Immunol. 2021, 12, 679482. [Google Scholar] [CrossRef] [PubMed]
- Savitt, A.G.; Manimala, S.; White, T.; Fandaros, M.; Yin, W.; Duan, H.; Xu, X.; Geisbrecht, B.V.; Rubenstein, D.A.; Kaplan, A.P.; et al. SARS-CoV-2 Exacerbates COVID-19 Pathology Through Activation of the Complement and Kinin Systems. Front. Immunol. 2021, 12, 767347. [Google Scholar] [CrossRef] [PubMed]
- Renne, T.; Stavrou, E.X. Roles of Factor XII in Innate Immunity. Front. Immunol. 2019, 10, 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, W.E.G.; Neufurth, M.; Wang, S.F.; Tan, R.W.; Schroder, H.C.; Wang, X.H. Morphogenetic (Mucin Expression) as Well as Potential Anti-Corona Viral Activity of the Marine Secondary Metabolite Polyphosphate on A549 Cells. Mar. Drugs 2020, 18, 639. [Google Scholar] [CrossRef]
- Maas, C.; Renne, T. Coagulation factor XII in thrombosis and inflammation. Blood 2018, 131, 1903–1909. [Google Scholar] [CrossRef] [Green Version]
- Muller, F.; Mutch, N.J.; Schenk, W.A.; Smith, S.A.; Esterl, L.; Spronk, H.M.; Schmidbauer, S.; Gahl, W.A.; Morrissey, J.H.; Renne, T. Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell 2009, 139, 1143–1156. [Google Scholar] [CrossRef] [Green Version]
- Mailer, R.K.; Hanel, L.; Allende, M.; Renne, T. Polyphosphate as a Target for Interference with Inflammation and Thrombosis. Front Med. 2019, 6, 76. [Google Scholar] [CrossRef] [Green Version]
- Laubli, H.; Alisson-Silva, F.; Stanczak, M.A.; Siddiqui, S.S.; Deng, L.W.; Verhagen, A.; Varki, N.; Varki, A. Lectin Galactoside-binding Soluble 3 Binding Protein (LGALS3BP) Is a Tumor-associated Immunomodulatory Ligand for CD33-related Siglecs. J. Biol. Chem. 2014, 289, 33481–33491. [Google Scholar] [CrossRef] [Green Version]
- Montefusco, L.; Ben Nasr, M.; D’Addio, F.; Loretelli, C.; Rossi, A.; Pastore, I.; Daniele, G.; Abdelsalam, A.; Maestroni, A.; Dell’Acqua, M.; et al. Acute and long-term disruption of glycometabolic control after SARS-CoV-2 infection. Nat. Metab. 2021, 3, 774–785. [Google Scholar] [CrossRef]
- Reiterer, M.; Rajan, M.; Gomez-Banoy, N.; Lau, J.D.; Gomez-Escobar, L.G.; Ma, L.; Gilani, A.; Alvarez-Mulett, S.; Sholle, E.T.; Chandar, V.; et al. Hyperglycemia in acute COVID-19 is characterized by insulin resistance and adipose tissue infectivity by SARS-CoV-2. Cell Metab. 2021, 33, 2174–2188.e5. [Google Scholar] [CrossRef]
- Kitane, D.L.; Loukman, S.; Marchoudi, N.; Fernandez-Galiana, A.; El Ansari, F.Z.; Jouali, F.; Badir, J.; Gala, J.L.; Bertsimas, D.; Azami, N.; et al. A simple and fast spectroscopy-based technique for COVID-19 diagnosis. Sci. Rep. 2021, 11, 16740. [Google Scholar] [CrossRef] [PubMed]
- Lucas, C.; Klein, J.; Sundaram, M.E.; Liu, F.; Wong, P.; Silva, J.; Mao, T.; Oh, J.E.; Mohanty, S.; Huang, J.; et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat. Med. 2021, 27, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Heymann, D.L.; Teo, Y.Y.; Garcia, P.J. Diagnostics for COVID-19: Moving from pandemic response to control. Lancet 2021. [Google Scholar] [CrossRef]
| Band Designation | In Vitro a | Mouse a | Human a | Chemical Components [26,28,29,30,31,32,33] | Barauna [8] | Wood [10] | Martinez- Cuazitl c [9] |
|---|---|---|---|---|---|---|---|
| Amide A | 3246 | 3358 | N-H, O-H stretching | X | |||
| 3518–3280 b | |||||||
| Amide B | 3067 | 3190 | Amide II overtone, aromatic amino acids | X | |||
| 3248–3110 b | |||||||
| Aliphatic | 2973 | 2931–2880 | 2954 | -CH3/-CH2; C-H symmetric (νs) & asymmetric (νas) stretching | X | ||
| 2858 | 2870 | ||||||
| 2837 | 2968–2944 b | ||||||
| Fatty Acids | 1705 | 1702 | 1722–1704 | -COOH, C = O ν; and ketones | X | O | |
| 1714–1690 b | |||||||
| * Amide I | 1690 | 1680 b | Protein β-sheets; C = O guanine | ||||
| 1638 | Protein β-sheets | ||||||
| 1600 | 1625–1594 | Protein aggregates; amyloid fibrils | X | ||||
| 1632–1585 b | |||||||
| Amide II | 1524 | 1578–31 | 1572–1470 | N-H; primarily β-sheet | X | X | |
| Aliphatic Fingerprint | 1468 | 1464 | -CH2 δ, bending vibrations | X | |||
| 1439 | 1431 | 1416 | -CH2 δ, symmetric stretching band of carboxyl group, CH2 ω, wagging; RNA | X | X | X | |
| 1420b | |||||||
| 1402 | C–H deformation; CH2 ω; C–N stretching; In-plane C2′OH in RNA | X | X | ||||
| 1400b | |||||||
| 1373 | 1370 | 1375 | -CH3 δ, C-H ν; methyl bending/stretching | X | X | ||
| 1388–1376b | |||||||
| Amide III | 1302 | 1302 | 1319 | Amino acid side-chains; terminal oxygen (PO3-) | X | X | |
| 1378–1354 | 1340–1285 | -CH2 ω; -CH3 δ, amyloid contribution | X | X | |||
| 1335–1280 | 1330–1177 b | ||||||
| 1250 | PO2 νas; C-N ν | X | X | ||||
| 1250 b | |||||||
| 1243–1218 | PO2 νas; amyloid fibrils | X | X | ||||
| 1226b | |||||||
| RNA | 1124 | 1129 b | PO2 νs, phosphodiester stretching | X | |||
| Saccharide | 1094 | -C-O-C, ether linkages; -O-Ca2+ c | X | ||||
| 1072 | 1064 | 1077 b | PO2− νs, symmetric and C-O ν | X | X | X | |
| 1050 | -C-O ν, C-OH group; C-C ν (sugars) | X | X | ||||
| 1023 | 1034–1003 | C-O ν; P-O ν; C-OH δ | X | ||||
| 1095–997 b | |||||||
| 1008 | 1012 | 1012 | C4-OH, Glucose | X | X | ||
| 988 | 988–974 | PO2− νs; -C-O-, ribose | X | ||||
| 940 | P-O ν, phosphorylation; -C-C- ν | X | |||||
| 887–866 | 889 | P-O-P νas; -C-C- ν; aromatics | O | X | |||
| 830 | 836 b | P-O-C ν; = C-H δ; aromatics | O | X |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazmer, S.T.; Hartel, G.; Robinson, H.; Richards, R.S.; Yan, K.; van Hal, S.J.; Chan, R.; Hind, A.; Bradley, D.; Zieschang, F.; et al. Pathophysiological Response to SARS-CoV-2 Infection Detected by Infrared Spectroscopy Enables Rapid and Robust Saliva Screening for COVID-19. Biomedicines 2022, 10, 351. https://doi.org/10.3390/biomedicines10020351
Kazmer ST, Hartel G, Robinson H, Richards RS, Yan K, van Hal SJ, Chan R, Hind A, Bradley D, Zieschang F, et al. Pathophysiological Response to SARS-CoV-2 Infection Detected by Infrared Spectroscopy Enables Rapid and Robust Saliva Screening for COVID-19. Biomedicines. 2022; 10(2):351. https://doi.org/10.3390/biomedicines10020351
Chicago/Turabian StyleKazmer, Seth T., Gunter Hartel, Harley Robinson, Renee S. Richards, Kexin Yan, Sebastiaan J. van Hal, Raymond Chan, Andrew Hind, David Bradley, Fabian Zieschang, and et al. 2022. "Pathophysiological Response to SARS-CoV-2 Infection Detected by Infrared Spectroscopy Enables Rapid and Robust Saliva Screening for COVID-19" Biomedicines 10, no. 2: 351. https://doi.org/10.3390/biomedicines10020351
APA StyleKazmer, S. T., Hartel, G., Robinson, H., Richards, R. S., Yan, K., van Hal, S. J., Chan, R., Hind, A., Bradley, D., Zieschang, F., Rawle, D. J., Le, T. T., Reid, D. W., Suhrbier, A., & Hill, M. M. (2022). Pathophysiological Response to SARS-CoV-2 Infection Detected by Infrared Spectroscopy Enables Rapid and Robust Saliva Screening for COVID-19. Biomedicines, 10(2), 351. https://doi.org/10.3390/biomedicines10020351








