Abstract
Tau proteins are known to be mainly involved in regulation of microtubule dynamics. Besides this function, which is critical for axonal transport and signal transduction, tau proteins also have other roles in neurons. Moreover, tau proteins are turned into aggregates and consequently trigger many neurodegenerative diseases termed tauopathies, of which Alzheimer’s disease (AD) is the figurehead. Such pathological aggregation processes are critical for the onset of these diseases. Among the various causes of tau protein pathogenicity, abnormal tau mRNA metabolism, expression and dysregulation of tau post-translational modifications are critical steps. Moreover, the relevance of tau function to general mRNA metabolism has been highlighted recently in tauopathies. In this review, we mainly focus on how mRNA metabolism impacts the onset and development of tauopathies. Thus, we intend to portray how mRNA metabolism of, or mediated by, tau is associated with neurodegenerative diseases.
1. Tau Protein and Associated Pathologies
In 1975, the tau protein was first isolated and recognized as a microtubule-associated protein (MAP) [1]. In fact, tau is a MAP that is highly expressed in neurons, predominantly in the axons [1,2,3,4]. Furthermore, tau binds to the outer microtubule surface tethering together tubulin dimers (α and β), stabilizing polymerization by a kiss-and-hop mechanism. Moreover, this mechanism was found to be essential for axonal transport in neurons [5,6,7].
Besides its primary function as microtubule dynamics regulator, tau also has roles in chromatin structure, signal transduction, synaptic plasticity, neuronal stress response and nucleic acid protection [8,9,10,11,12]. Considering this wide range of biological functions, it was surprising that pioneer reports on tau knockout mice (Mapt-/-) presented ‘physiological’ phenotypes with normal cognitive function in spatial learning tasks and presynaptic-mediated transmission [13,14]. However, more recently, Mapt-/- mice exhibited age-dependent short-term memory deficits, hyperactivity, and synaptic plasticity defects. Thus, although tau has a role in normal neuronal function, its absence or reduction can be surpassed under certain conditions [15]. Nowadays, it is well established that tau KO animals display abnormal phenotypes [16]. In humans, the high relevance of tau goes beyond its physiological function. Indeed, abnormal tau aggregates are the hallmarks of several diseases/disorders classified under the general term of tauopathies.
Tauopathies comprise a group of proteinopathies of the human nervous system characterized by the assembly into filaments of tau proteins, alone or in association with other proteins. Among tauopathies are pathologies such as Alzheimer’s disease (AD), Pick’s disease (PiD), chronic traumatic encephalopathies (CTE), progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), argyrophilic grain disease (AGD), frontotemporal dementia and Parkinsonism linked to chromosome 17 (FTDP-17) and several other, rarer diseases [17,18]. Moreover, tau aggregates display distinct morphological and biochemical features between tauopathies. In the brains of AD patients, tau aggregates are named neurofibrillary tangles (NFTs). NFTs are composed of two distinguishable intracellular assemblies, the paired helical filaments (PHFs) and the straight filaments (SFs) [18]. Besides affecting microtubule dynamics, pathogenic forms of tau are reported to affect nuclear pore localization, function and nucleocytoplasmic trafficking [19,20], induce heterochromatin relaxation [21], promote DNA damage [22,23], alter RNA stability and RNA export [24,25,26], promote ribosome instability [27] and activate transposable elements [28,29].
2. Tau mRNA Metabolism
In humans, tau is encoded by a single gene named MAPT (Microtubule Associated Protein Tau) located at human chromosome 17q21.31. Owing to an inversion polymorphism, the MAPT locus is found as two major haplotypes, named H1 and H2. The haplotype effect is related to changes in tau mRNA expression and splicing [30]. The MAPT gene consists of 16 exons numbered from 0 to 14. In the adult human brain, six major tau isoforms that arise from the alternative mRNA splicing of exons 2, 3 and 10 are expressed. The MAPT gene codes for tau isoforms that all consist of four domains, the N-terminal domain, a proline-rich domain (PRD), a microtubule binding domain (MTBD) that contains microtubule binding repeats, and a C-terminal domain that is involved in the regulation of microtubule polymerization [17,31].
2.1. Splicing Variants
Considering its six isoforms in the human adult brain, tau protein can vary between 352 and 441 amino acids. The alternatively spliced exons 2 and 3 code for 29 amino acids each, located in the N-terminal part of the protein (Figure 1A). The presence of exon 2 is mandatory for the inclusion of exon 3. Regarding exon 2 and 3 splicing, isoforms are named 0N (absence of both), 1N (exon 2 inclusion) and 2N (inclusion of both). Additionally, it has been reported that in the human brain, the 0N, 1N and 2N isoforms are differentially expressed, comprising 37%, 54% and 9%, respectively [32]. Furthermore, the alternative splicing of exons 2 and 3 can also affect tau protein interactions, as exon 3 inclusion is associated with decreased tau assembly into filaments [33,34]. Accordingly, the MAPT H1 haplotype that is known to be associated with tauopathies decreases exon 3 inclusion [35].
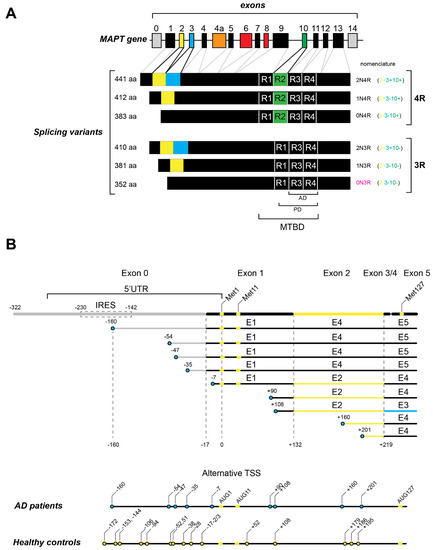
Figure 1.
Tau mRNA transcripts. (A) Tau mRNA transcripts are generated from the single MAPT gene and consist of 16 exons (numbered 0 to 14). The coding sequence starts in exon 1. In the adult human brain, six splicing variants are generated by alternative splicing of exon 2 (light green), exon 3 (blue) and exon 10 (green). All of these variants contain three (3R) or four (4R) microtubule binding repeats (R). Exon 4a (orange) is only transcribed in peripheral nervous system. Exons 6 and 8 (red) are not transcribed in human brain. The microtubule binding repeats constitute the microtubule -binding domain (MTBD). The regions of Tau filaments that have been observed by cryo-EM in Alzheimer’s disease patients (G273/305 to E380) and Pick’s disease patients (K254 to F378 in 3R Tau) are indicated as (AD) and (PD), respectively. (B) The 5′ ends of tau mRNA transcripts in Alzheimer’s disease patients determined by 5′ RACE experiments (Huin et al., 2017). The position of the IRES in the 5′ UTR is boxed by a dashed line. The positions of methionine codons 1, 11 and 127 are shown in light green. The positions of the 5′ ends are represented by blue circles. The numbers of nucleotides upstream of the AUG start are indicated by numbers prefixed with the minus sign (−). The numbers of nucleotides downstream of the AUG start are indicated by numbers prefixed with the positive sign (+). The deduced alternative transcription start sites (TSS) found in AD patients are summarized in the bottom of the panel and compared with the TSS found in a normal brain (light green circles).
Alternative splicing of tau mRNA can also vary in its MTBD constitution, by either inclusion or exclusion of exon 10. Exon 10 encodes a 30-amino acid microtubule binding repeat. Consequently, the MTBD of tau can be composed of three or four microtubule binding repeats (3R or 4R). Given that 4R tau isoforms display increased MT-binding affinity and MT assembly when compared to 3R tau isoforms, the exon 10 alternative splicing has been reported to be crucial for the onset of neurodegenerative diseases [17,36,37]. Furthermore, it is known that the 3R/4R isoform ratio is a key factor for several tauopathies. Indeed, tau-mediated aggregates are heterogeneous and neurodegenerative diseases that can be divided into 3R, 4R or 3R + 4R tauopathies according to the biochemical composition of tau inclusions. Moreover, several mutations in and around exon 10 were found to affect its splicing and consequently linked to tau pathogenesis (detailed below) [17,18]. In this sense, AD and CTE are classified as 3R + 4R tauopathies; PiD and some types of FTDP-17 are 3R tauopathies; the 4R tauopathies include AGD, CBD and PSP [17,18,38]. Recently, Bachmann and colleagues (2021) showed that in human SH-SY5Y neuroblastoma cells, 2N tau isoforms diminished axonal enrichment while 4R isoforms increased microtubule count. These results point to the different but equally important consequences of alternative splicing of exons 2, 3 and 10 [39]. Thus, the alternative splicing of tau mRNA was proven to be crucial for tau-mediated pathology. Together with splicing deregulation, tau-mediated pathology is also dependent on the total amount of the protein [40,41]. Additionally, tau mRNA variants can arise from transcription initiation from different promoters and likely contribute to pathological processes.
2.2. Tau Transcription and Variable 5′ UTR
A promoter is a genomic sequence located upstream of the transcriptional start site (TSS) that defines where the transcription starts. Consequently, a promoter determines the 5′ end of an mRNA. The existence of multiple transcripts with different 5′ ends originated from the same gene reflects the presence of alternative promoters in the genome. Moreover, the promoter usage may change according to tissues or cell types [42]. The alternative promoter usage (APU) contributes to transcriptome diversity and flexibility and the regulation of gene expression. Indeed, APU has been described to affect alternative splicing, tissue and/or subcellular specificity and gene expression during development [43,44,45,46].
For several years, only one promoter, located upstream of exon 0, was described for the MAPT gene. Interestingly, a hypomethylated CpG island (a feature associated with promoter regions) was found 13 kb upstream of exon 1. In addition, this region harbors binding sites for a transcription factor and a histone marker. These features suggest the existence of an alternative promoter in that region, a hypothesis corroborated by reports of shorter MAPT mRNAs starting on exon 1 in genome databases [47,48,49]. Indeed, Huin and colleagues (2017) reported a functional alternative promoter located in exon 1 of the MAPT gene that originates 5′ truncated transcripts in human brain (Figure 1B). These 5′ truncated transcripts could be translated into normal or N-terminal-truncated tau proteins, which could lead to changes in protein localization and function. In the brain, up to 60% of the genes are transcribed by APU [44,50]. Moreover, the usage of different promoters has been described for genes involved in Alzheimer’s disease, such as apolipoprotein E gene (APOE) and the presenilin-2 gene (PSEN2) [50,51,52]. Even though the activity of the MAPT promoter located in exon 1 is weak, it was found to be relevant for tissue-specific expression [53,54]. Interestingly, the 5′ end truncated variants of tau mRNAs were found to be overexpressed in AD and PSP brains [48]. Notably, as depicted in Figure 1B, several 5′ end truncated mRNAs were found in AD patient brains (−160, −54, −47, −35, −7, +90, +160 and +201) and absent from healthy control brains. Once the 5′ end truncated transcripts can lead to 5′ terminal truncated proteins, this can be relevant for the onset of neurodegenerative diseases. Tau truncated protein expression was associated with neurodegenerative diseases in mice and humans [55,56,57,58,59]. Among the 5′ truncated tau mRNAs were particularly interesting transcripts with the first AUG codon located in the 48th position of the coding sequence (Met11 in the protein) or in exon 5 (Met127 in the protein) (Figure 1B). Interestingly, these N-terminally truncated tau proteins starting from Met11 and Met127 had already been detected in human brain tissue using a proteomic approach [60]. In the long 5′ UTR of tau mRNA, the existence of an internal ribosome entry site (IRES) was unveiled [61,62]. Initially characterized in viral genomes, an IRES is a sequence of mRNA in the 5′ UTR that can initiate translation without the need for all translation initiation factors [63]. In fact, the IRESs fold into a secondary structure that is able to directly recruit the 40S ribosome in a cap-independent manner. Cellular IRESs were shown to work as a by-pass mechanism when canonical translation is inhibited [64]. The IRES in tau mRNA is defined by two main structural domains and is very sensitive to small nucleotide substitutions. Additionally, the tau IRES located between 230 and 142 nt upstream of the canonical AUG was able to directly recruit the 40S ribosome (Figure 1B) [61,62]. Interestingly, the IRES is present only in tau mRNAs with a long 5′ UTR, thereby raising questions about the translation initiation mechanism used for tau mRNA variants with short 5′ UTRs.
In fact, APU diversifies the transcriptional profile not only by the different 5′ ends but also by, among others, modulating alternative splicing, tissue specificity and/or expression levels [43,65,66,67]. In addition, the existence of APU allows the promoter switching that could lead to a harmful process or to better responses to diverse stimuli. As previously described, these 5′ end truncated transcripts of tau could also drive different splicing isoforms. Even though the tau second promoter function has yet to be fully described, it may alter the 3R/4R tau protein ratio and possibly drive tau pathology [48]. This report suggests that beyond the protein truncation driven by post-translational proteolysis, APU can also generate N-terminally truncated tau variants. Therefore, APU can represent an underestimated source of tau protein variants that could have an impact in tauopathies.
2.3. The 3′ UTR Is Important for Tau mRNA Trafficking
Despite being controversial, it was reported that 3′ UTR of tau mRNA could be essential for its axonal localization and, consequently, tau function [68]. In their report, Aronov and colleagues stated that absence or mutation in what they called 3′ UTR axonal targeting region of tau mRNA resulted in mislocalization of tau mRNA and protein in neuronal cells. Furthermore, swapping the tau 3′ UTR with the dendrite targeting 3′ UTR of MAP 2 (microtubule-associated protein 2 that localizes to cell body and dendrites) caused tau mRNA and protein to fail to localize to axons [68]. On the other hand, dendritic microinjections of tau protein or neuron transfection of tau cDNA without tau 3′ UTR also localized in axons [69,70]. More recently, tau proper localization was linked to autophagy and proteasomal pathways rather than mRNA localization [71]. Moreover, there are additional hypotheses for the regulation of tau protein sorting such as annexin, which interacts with tau exon 1 and thereby contributes to tau axonal localization [72]. In addition, it has been reported that miR-132 microRNA directly targets tau 3′ UTR [73,74]. Remarkably, the activity of this miRNA can impact tau aggregation, cognitive function, and tau metabolism [74]. The link and relevance of noncoding RNAs to tau post-transcriptional and post-translational regulation will be explored below.
2.4. Noncoding RNAs: From microRNAs to Long Noncoding RNAs
Noncoding RNAs (ncRNAs) are a class of RNAs that are transcribed like mRNAs but do not code for functional proteins. Instead, ncRNAs are known to play crucial regulatory roles in mRNA and protein metabolism. The ncRNA class includes long noncoding RNAs (lncRNAs), piwi-interacting RNAs (piRNAs) and microRNAs (miRNAs) [75,76].
MicroRNAs (miRNAs) are small RNAs (~22 nts long) that regulate the expression of complementary messenger RNAs and, therefore, play important posttranscriptional regulatory roles in eukaryotic cells [77]. In humans and other mammals, these small RNAs help modulate the expression of most mRNAs shaping the transcriptome [77]. In brief, miRNAs are transcribed by RNA polymerase II as primary miRNAs (pri-miRNAs). The pri-miRNAs are processed by Drosha/DGRC8 complex in the nucleus before being exported to cytoplasm as a hairpin-shaped precursor miRNA (pre-miRNA). Once in the cytoplasm, Dicer generates a ~22 nt long miRNA duplex [77]. Then, one strand of the miRNA duplex (the mature miRNA) in association with the RNA-silencing complex (RISC) will encounter the complementary target mRNA, leading to its translational repression or degradation. The miRNA-mediated regulation is very versatile, since a single miRNA can target several mRNAs and a single mRNA can be under the control of different miRNAs [77]. Due to the versatile nature of miRNAs, it is not surprising that miRNAs are involved in several brain functions such as neurogenesis, memory, synaptic function, and neuronal survival [78,79]. In this context, several miRNAs were indicated as good biomarkers for some neurodegenerative diseases, specifically AD. In fact, several miRNAs were found to be differentially expressed in the brain, serum and plasma of AD patients [73,80,81]. Interestingly, several miRNAs were directly or indirectly involved in tau pathology. Among them, miR-125b, which is up-regulated in AD, promotes tau hyper-phosphorylation in neuronal cells. This indirect effect arises most likely through down-regulation of the phosphatases DUSP6 and PPP1CA, which are considerably reduced in AD brains [82]. Corroborating this hypothesis, suppression of miR-125b in neurons reduces tau phosphorylation and kinase expression/activity. Conversely, miR-125b overexpression impairs associative learning and downregulates DUSP6 and PPP1CA, resulting in enhanced tau phosphorylation in mice. These results show that miR-125b has a role in the tau pathology in AD [82]. Additionally, dysregulation of miR-146a was also reported to induce tau hyperphosphorylation and AD pathogenesis via ROCK1-PTEN-tau pathway [83]. Interestingly, deficiency in the miR-132/212 cluster, which is downregulated in tauopathies, leads to tau overexpression, phosphorylation, and aggregation. The miR-132 is very versatile and, besides directly targeting tau mRNA in its 3′ UTR, is predicted to target several other proteins relevant to neuronal survival and synaptic function [73,74]. Furthermore, miR-132/212 deletion induced tau aggregation in mice expressing endogenous or mutant tau. Treatment of AD mice with miR-132 mimics partially restored memory function and tau metabolism. Interestingly, miR-132 and miR-212 levels correlate with insoluble tau filaments and cognitive impairment in humans [74]. Moreover, miR-132 was also implicated in the regulation of tau alternative splicing, which is dysregulated in PSP [84]. However, miR-132 is not the only miRNA modulating tau alternative splicing. MiR-124, miR-9, miR-137, and miR-153 were also reported to affect the alternative spicing of tau exon 10 in neuronal cells by targeting splicing factors such as poly-pyrimidine tract-binding proteins 1 and 2 (PTBP1/2) [74,85,86]. The miR-132/232, among other miRNAs, such as mir-9 and miR-181c, was also reported to directly inhibit the deacetylase SIRT1 [87]. In AD brains, reduced SIRT1 levels increased acetylated tau levels and pathological phosphorylated tau in vivo [88]. Interestingly, miR-132-3p and miR-22-3p shared 48 target genes, such as PTEN and SIRT1, which directly or indirectly impact mechanisms associated with tau pathology [73].
Besides microRNAs, another noncoding class of RNAs comes into play in the regulation of tau—the long noncoding RNAs (lncRNAs). LncRNAs are commonly defined as transcripts longer than 200 nucleotides that are not translated into proteins [75,76]. Lately, an increasing number of reports have described and characterized the function of lncRNAs. Broadly, lncRNAs can be categorized as cis regulators and trans regulators. The former regulate chromatin structure and/or gene expression near the transcription site. The latter perform cellular functions throughout the cell far from the transcription site. Furthermore, lncRNAs can be generated by pervasive transcription mostly in the opposite direction of the transcript that they regulate (cis regulation) [76]. Of note in the scope of this review, an lncRNA was described and characterized for an MAPT locus, the MAPT-AS1. MAPT-AS1 is an 840 nt lncRNA transcribed from the anti-sense strand of the MAPT promoter region [89,90]. Moreover, MAPT-AS1 expression was reported to be reduced in brains of patients with Parkinson’s disease [89,90]. MAPT-AS1 is upregulated during neuronal differentiation [91]. Additionally, while overexpression of MAPT-AS1 leads to a decrease in MAPT promoter activity, MAPT-AS1 knockdown increased methylation and 4R isoform expression (but not of total tau transcripts) in cellular assays. This suggests that MAPT-AS1 has a role in both gene transcription and alternative splicing of tau [89].
More recently, MAPT-AS1 was further characterized as a functional expression regulator of neuronal tau [92]. Indeed, this report showed that the 75-nt overlap with the tau mRNA in its 5′ UTR modulates neuronal tau protein levels and tau pathology in human brain [92]. Moreover, MAPT-AS1 as well as tau is enriched in the brain, being found in both nucleus and cytoplasm. Further, it was proposed that MAPT-AS1 mediates repression of tau translation by competing for ribosomal-RNA pairing with tau mRNA IRES [62,92]. Altogether, these results define a role of MAPT-AS1 in regulating tau translation as an important player in the prevention of tau-mediated pathology.
3. Tau Protein Metabolism
All Tau isoforms comprises four domains: N-terminal domain (aa 1–150), proline-rich domain (PRD), microtubule binding domain (MTBD) and C-terminal domain (aa 369–441) (Figure 2) [18]. The tau N-terminal domain, also known as projection domain, plays a role in signaling and protein interactions, acting as function regulator and contributing to tau’s physiological paperclip conformation by interacting with the C-terminal domain. The so-called projection domain projects away from microtubules when tau binds to microtubules [31,93]. Tau N-terminal domain contributes to its axonal localization and has been proposed to mediate interactions with neuronal proteins. Further, N-terminally truncated tau showed increased association with microtubules [72,94].
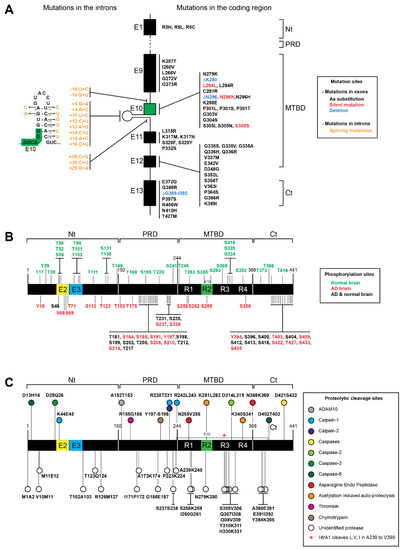
Figure 2.
Tau proteins, mutations, phosphorylation and truncation sites. (A) Mutations found in tau associated with neurodegenerative diseases. The most frequent mutations are found in the microtubule binding domain (MTBD) and in the C-terminal domain (Ct). Single amino acid substitutions are shown in black, deletions are shown in blue, and silent mutations are shown in red. Several mutations (shown in orange) that affect splicing are also found in the introns flanking exon 10 (shown in green). The mutations are also shown on the intron hairpin that is located close to exon 10 (B) The phosphorylation sites that have been found only in normal brain are shown in green above the graphic representing tau protein; phosphorylation sites found only in Alzheimer’s disease (AD) patients are shown in red under the graphic, and phosphorylation sites found in both AD and normal brain are shown in black. (C) Cleavage sites by identified proteases are shown above the graphic of tau protein. The color code of the corresponding proteases is indicated in the figure. Other cleavage sites that have been generated by yet unidentified proteases are shown under the graphic and represented by white circles.
The PRD (aa 151–243) is responsible for interaction with proteins containing SH3 domains and plays an important role in tau protein folding. Furthermore, the PRD plays a role in cell signaling and interacts with protein kinases. Consequently, the PRD impacts not only microtubule binding and assembly, but also tau phosphorylation sites [31,93,95].
The MTDB domain (aa 244–368) mediates the tau binding to microtubules and is responsible for microtubule stabilization [17,31].
The C-terminal part (aa 369–441), in addition to contributing to microtubule binding, interacts with the N-terminal part conferring the paperclip conformation to tau protein [17,31].
3.1. MAPT Mutations of Tau Proteins
To date, more than 100 MAPT mutations have been associated with neurodegenerative diseases [96] (https://www.alzforum.org/mutations/mapt) (accessed on 17 January 2022). These mutations can be divided into two groups, missense mutations or splicing mutations. The primary effect of the missense mutations is at the protein level, while splicing mutations modulate the alternative splicing of tau exon 10. Mutations that introduce amino acid substitution have an impact by altering the tau interactions with microtubules promoting tau assembly into filaments [97,98,99].
Besides affecting the mRNA splicing of MAPT exon 10, these mutations can disrupt tau protein folding and/or protein–protein interactions, enhancing tau aggregation. Actually, most of the mutations are located between exon 9 and exon 12, which encodes for the MTBRs (Figure 2A). Thus, these mutations can affect the tau interaction with microtubules and promote tau assembly into filaments [18,96,97,98,99,100,101,102]. Remarkably, a direct correlation between MAPT mutations and post-translational modifications of tau has not been established [102].
As depicted in Figure 2, most of the MAPT missense mutations cluster in or near the MTBR domain of tau as well as the splicing mutations. The mutations that have an impact on splicing of exon 10 can be either intronic or exonic. These mutations usually result in overproduction of 4R tau and its assembly into filaments [17,103]. Although less common, it has been shown in several reports that mutations can also increase 3R tau expression. Indeed, mutations in exons 9–13 (K257T, L266V, G272V, G273R, S305N, L315R, S320F, S320Y, P332S, Q336H, Q336R, K369I, E372G and G389) were associated with 3R tauopathies, such as PiD [17,104] (https://www.alzforum.org/mutations/mapt) (accessed on 17 January 2022). Recently, a deletion from G389 to I392 was also described as predominant in 3R tauopathy [105]. However, the most characteristic MAPT mutations affect the alternative splicing by inclusion of exon 10, resulting in the overproduction of 4R Tau. In fact, several intronic mutations (I9-10 G > T, I9-15 > C, I10+3 G > A, I10+4 A > C, I10+11 T > C, I10+12 C > T, I10+13 A > G, I10+14 C > T, I10+15 A > C, I10+16 C > T, I10+19 C > G, I10+25 C > T, I10+29 G > A) and exonic mutations (N279K, ΔK280, L284L, L284R, C291R, ΔN296, N296D, N296H, N296N, K298E, G303V, G304S, S305I, S305N, S305S and V363I) were associated with neurodegenerative diseases, often associated with 4R overproduction [17,103,104,106,107,108]. In addition, the P301 residue, present only in 4R tau isoforms, is of major importance for regulation of tau aggregation [103]. Indeed, mutations on P301 are highly relevant for studying tauopathies mainly because P301L is the most common tau mutation. Furthermore, in transgenic mice, the P301L mutation induces aberrant tau hyperphosphorylation and aggregation [20,109]. Additionally, there are three mutations on this residue (P301L, P301S, P301T) associated with tau pathology [110]. Indeed, it was shown that P301L, P301T, and P301S recombinant proteins are prone to induce more oligomers when compared with wild-type recombinant tau [110]. In addition, a PS19 mouse model that bears a P301S tau mutation spontaneously develops tau pathology [111]. In fact, tau transgenic mice expressing P301L or P301S human tau develop tau-mediated pathology [111,112,113,114]. Additionally, the combination of P301L or P301S tau with S320F generated aggressive models of tauopathy without exogenous seeding [115]. Furthermore, THY-Tau 22 mice (bearing two mutations on tau G272V and P301S) show hyperphosphorylation of tau, NFT-like inclusions and cognitive impairment [116]. However, MAPT mutations other than P301 also enable the generation of in vivo models of tau pathology.
Mutations outside exon 10, such as I260V in exon 9, K317M and K317N in exon 11, E342V in exon 12 and N410H in exon 13, were also able to alter the 3R/4R ratio [17]. Nevertheless, not all mutations located in exons 11 and 12 affect exon 10 splicing like G335S, G335V, G335A and V337M. Instead, they reduce the ability of tau to promote microtubule assembly [17,117]. Conversely, Q336H and Q336R mutations increased tau-mediated microtubule assembly [17,118]. Despite not altering the 3R/4R ratio or having the ability to bind MTs, the D348G tau mutation was associated with early amyotrophic lateral sclerosis (ALS) onset. For the D348G mutation, ALS seems to be caused by tau accumulation due to evasion of proteasomal degradation [119,120]. The S352L, S356T, P364S, G366R, P397S, R406W and T427M mutations were found in FTLD, FTD or PD patients [121,122,123,124,125,126]. Notably, the P364S tau showed a special propensity for aggregation, even higher than that of P301L tau. Moreover, P364 (in R4) corresponds to the residue P301 in R2 and to P322 in R3, whose mutations were strongly associated with different tauopathies [123]. It is also worth noting that more C-terminal mutations (P397S, R406W and T427M) were associated with later age of onset and/or longer disease duration, which suggests that the C-terminal part of the protein is less critical for tau-mediated pathology [124,125,126].
Interestingly, there is only one residue (R5) located in exon 1 in the N-terminal of the tau protein that is associated with tau-mediated pathology. The R5H, R5L and R5C mutations were associated with AD, PSP and Parkinson’s disease, respectively [127,128,129]. The R5H and R5L mutations reduce the ability of tau to promote microtubule assembly and promote tau fibril formation [128,130]. Furthermore, the R5L mutation was shown to affect tau polymerization and MT assembly in 0N4R isoforms [131].
3.2. Post-Translational Modifications of Tau Protein
Tau pathology is often explained by changes in its post-translational state, regarding for example, its phosphorylation and truncation pattern [132,133]. Indeed, tau protein can be subjected to several post-translational modifications, such as phosphorylation, acetylation, methylation, glycation, ubiquitination and truncation [132,133,134,135,136,137]. Tau post-translational modifications are tightly regulated and essential for its proper function and metabolism as well as its relevance for neurodegenerative disorders [135,137].
Tau phosphorylation is a normal event, occurring on serine, threonine and tyrosine residues. On average, in the normal adult brain, tau has 2–3 phosphorylated residues, whereas in the AD brain the phosphorylation is at least 3-fold greater. Interestingly, depending on the isoform, tau can have up to 85 putative phosphorylation sites. However, not all of the putative phosphorylation sites were described and/or characterized (Figure 2B) [93,135,138]. Usually, hyperphosphorylation impairs the ability of tau to interact with MTs and can induce tau missorting. Consequently, tau hyperphosphorylation can drastically alter the function of the protein and induce tau pathology [93,139]. In addition, the hyperphosphorylation of tau has been associated with aggregate formation and microtubule assembly disruption [31,103,140,141]. Thus, there are some therapeutic approaches focusing on serine/threonine dephosphorylation [93]. Although hyperphosphorylation is strongly associated with aggregate formation, there are phosphorylation sites that can have a protective effect against aggregation. In fact, several phosphorylation sites are shared by normal and AD brains, and some phosphorylation sites are exclusively found in normal brains (Figure 2B) [93,135]. For example, on one hand, the phosphorylation on S262 and S356 residues is AD-specific and decreases the binding affinity of tau to microtubules. On the other hand, S293 and S305 residues were found phosphorylated only in normal brains, which suggests that they might have a protective effect against aggregation (Figure 2B) [132].
Of note, it has been shown that phosphorylation at AT8 sites in tau (S199E+S202E+T205E) promotes the split of the N-terminal and C-terminal domains [135,142]. However, controversial data showing that this AT8 epitope actually corresponds to phosphorylated residues S202, T205 and S208 have also been presented [143]. In turn, phosphorylation at the PHF1 epitope (S396E+S404E) moves the C-terminal domain away from the MTBR domain. Interestingly, combining both phosphorylations leads to compaction of the paperclip conformation, which leads to the tau aggregation characteristic of tau in AD [142]. Furthermore, there is growing evidence that acetylation is the second most important PTM for the function of tauopathies after phosphorylation. Since the longest tau isoform (2N4R) has 44 lysine residues, up to 10 percent of the protein has the potential to be acetylated [137]. In fact, more than 20 lysine residues of tau are subjected to acetylation [88]. Interestingly, tau acetylation blocks its own degradation, inhibits microtubule binding and promotes aggregation [88,144,145,146]. Moreover, tau acetylation affects tau phosphorylation and ubiquitination [88,147]. Additionally, as mentioned above, the deacetylase SIRT1 directly targets tau, reducing tau acetylation and, consequently, tau accumulation [148]. Furthermore, the strong relevance of tau acetylation might be linked to the prevention of other modifications at tau lysine residues, including methylation, glycation and ubiquitination [137]. In addition to acetylation, lysine residues of tau can also be ubiquitinated. Ubiquitination is the most common PTM in mammalian cells and can affect the activity, localization or stability of the targeted proteins [149]. Strikingly, tau ubiquitination was reported to increase both tau clearance and tau aggregation [137,150]. Besides acetylation and ubiquitination, tau lysine residues can also undergo methylation and glycation. Both methylation and glycosylation can impair tau microtubule-binding activity, proper clearance and aggregation [137,151]. Thus, tau methylation and glycation also contribute to neurodegenerative disease pathogenesis [137]. Taken together, PTM combinations, competition and crosstalk add a new layer of complexity to the regulation of tau function. Notably, this seems particularly relevant at tau lysine residues. Besides these usual PTMs, tau can be subjected to proteolytic truncation (tau proteolysis). The resulting tau fragments are also recognized to drive tau pathology and contribute to dysregulation of tau function, as described in more detail below (Figure 2C) [72,152,153]
3.3. Tau Proteolysis
For several years, it has been suggested that PTMs of tau induce protein structural changes that disrupt the physiological MT-binding paperclip conformation [93]. The MT release and consequent cytoplasmic accumulation allows tau proteolysis that ultimately can lead to tau aggregates [93,152]. Along with this process, tau proteolysis, also known as tau truncation, was found to promote tau aggregation [59]. Moreover, a wide range of tau fragments already have been found to be neurotoxic, contributing to the onset of tauopathies [140,152].
Tau protein cleavage can be classified by the cleavage site and responsible protease, as depicted in Figure 2C [140,152]. Several tau fragments were already attributed to several proteases. However, several proteases responsible for many of the tau fragments are still not characterized (Figure 2C) [140,152]. The relevance of proteases and the different tau fragments to tau pathology has been shown in several reports. For example, blocking caspase-2-mediated cleavage rescues memory deficits in P301L tau transgenic mice [154]. Furthermore, deletion of the first 150 or 230 amino acids of tau enhances tau self-aggregation and seeding. Conversely, loss of the 50 or 20 C-terminal amino acids does not produce significant effects [155]. Moreover, Gu and colleagues (2020) showed that the tau fragment from residues 151 to 391 has the highest pathological activity in vitro. Thus, these data indicate that some sections of tau can prevent tau aggregation and consequently protect tau from pathological features [155].
Tau truncation can be mediated by a wide variety of proteins. Indeed, several reports show that tau can be cleaved by ADAM10, calpains (calpain-1 and calpain-2), caspases (caspase-2, -3 and -6), asparagine endo peptidase, thrombin, chymotrypsin and cathepsins (cathepsin B, D and L). Besides this, tau can also be cleaved by acetylation-induced auto-proteolysis.
Amongst the caspases, caspase-2 was shown to produce the fragment 1-314, and this C-terminally truncated tau fragment promotes tau aggregation in dendritic spines [154]. Moreover, caspase-3 cleavage between D421-S422 was associated with AD and PSP. Additionally, caspase-6 produces a 1-402 tau fragment used as biomarker for AD [140,152].
The calpains that mediate tau proteolysis are calpain-1 and calpain-2. Calpain-1 has been associated with several neurodegenerative diseases such as Parkinson’s disease and Huntington’s disease. Moreover, a fragment generated by calpain-1, tau 232-441, was considered cytotoxic and associated with aging [140,152].
In conclusion, there are many different PTMs that affect tau and is functions. Ultimately, these PTMs, such as proteolysis, can induce tau aggregation and, consequently, tau pathology. Thus, the better our understanding of how tau PTMs are regulated, the sooner we will have in our hands tools to decipher the disease mechanisms of tauopathies and consequently design new therapeutic strategies.
4. Tau Protein and Cellular mRNA Metabolism
The longstanding relation between tau and RNA was established by the ability of RNA to act as a cofactor for tau aggregation [156,157]. The negatively charged nature of RNA neutralizes the positively charged residues of tau [156,157]. Interestingly, an increasing number of reports have shown that mutations in genes that encode RNA binding proteins cause neurodegenerative diseases such as ALS, FTLD, AD and spinal muscular atrophy (SMA) [158,159,160]. Interestingly, some interactome studies unveiled that tau protein interacts with RNA binding proteins such as TIA1, TDP-43, DDX6, PABP and RPL7 [160,161,162,163].
The link between the RBP T-cell intracellular antigen 1 (TIA1) and tau appears to be of major importance, since TIA1 is a core component of stress granules (SGs). SGs are cytoplasmic membraneless ribonucleoprotein complexes that under stressful conditions reversibly aggregate in the cytoplasm. SGs can arise in response to diverse cellular stresses such as nutritional stress, heat or osmotic shock, DNA damage or proteostasis dysfunction [164,165,166,167,168,169,170]. Indeed, under different stresses, the nuclear TIA1 is translocated to the cytoplasm, where it becomes one of the main building blocks of SGs [166]. Additionally, SGs are known to be constituted by mRNAs, RNA binding proteins, the small 40S ribosome subunit and translation factors [170,171,172]. Moreover, SGs can regulate mRNA metabolism and protein translation [173]. Concerning tauopathies, TIA1 reduction was able to prolong the lifetimes and increased neuronal survival in transgenic P301S tau mice [162]. Interestingly, under stress triggered by arsenite, glucocorticoids, Aβ and tau oligomers, the TIA1 and hyperphosphorylated tau co-localize in the SGs [174,175,176,177]. Even though SGs are not homogeneous, hyperphosphorylated tau inclusions strongly colocalize with TIA1-containing granules in the neuronal SH-SY5Y cell line [175,178]. Furthermore, in tau KO neurons, TIA1 is strictly nuclear, which hints that tau may have a role in TIA1 translocation from the nucleus [177]. Moreover, Tau/TIA1 co-localization was correlated with larger tau aggregates [176]. More recently, it was shown that reducing TIA1 in vivo rescued the tau-associated phenotype in P301S tau mice [162,179]. Even upon tau overexpression, reducing TIA1 levels not only decreased the number and size of cytoplasmic TIA1-containing granules, but also rescued behavior, neuronal degeneration and survival [162,179]. Similarly, TIA reduction decreased the hyperphosphorylated tau accumulation in 3-month-old mice [162,179]. In accordance with this observation, it has been shown in vitro that TIA1 knockdown also reduces tau pathology and provides neuroprotection [177]. Recently, Ash and colleagues showed that tau oligomerization in association with TIA1 alone is more toxic than tau aggregates generated by incubation with RNA alone or artificial crowding agent polyethylene glycol (PEG) [12].
Besides TIA1, pathological tau (hyperphosphorylated or misfolded) was found to co-localize with multiple RNA binding proteins [161,162,175,176,177,179]. These studies raise the hypothesis that oligomeric or misfolded tau is cytotoxic and able to drive cognitive decline independently of NFT formation [180,181]. Moreover, it was reported that RNA splicing is dysregulated in AD brains and in a PS19 mouse model (P301S tau). In the PS19 brain, transcripts encoding spliceosomal and mRNA binding proteins are reduced, favoring transcripts that encode synaptic proteins. Interestingly, reducing TIA1 expression in PS19 mice partially rescued the “physiological” RNA splicing [179].
Even though the tau association with SGs has not been thoroughly described, the link between tau and RNA metabolism is emphasized by the direct interaction of phosphorylated tau with TDP-43 [159,160]. TDP-43 is an RNA and DNA binding protein mainly associated with mRNA metabolism [182,183,184]. TDP-43 is mainly nuclear but can also be found in cytoplasm [185,186], where it is known to incorporate SGs [186,187]. As part of its nuclear function, TDP-43 is involved in transcription, translation, mRNA transport, microRNA biogenesis and long non-coding RNA processing [186,188]. Interestingly, it was shown that tau/TDP-43 interactions might be involved in several tauopathies by affecting each other’s functions. On one hand, TDP-43 oligomers may serve as nucleation sites for tau aggregates; on the other hand, tau oligomers modulate the TDP-43 aggregation state and cellular localization [160].
Interestingly, TDP-43 also directly interacts with TIA1 and is detected in SGs of FTD and ALS patients [160,189,190]. Furthermore, TDP-43 has been implicated in several neurodegenerative diseases, such as FTLD, ALS, FTD and AD [160,191,192]. It has been suggested that tau association with TDP-43 oligomers might play a role in AD, ALS and FTD [160]. Although TDP-43 aggregates may modulate AD pathology, there are AD cases without TDP-43 pathology [193,194]. Even though not strictly required, these reports suggest that TDP-43/tau interaction in SGs might be critical for several tauopathies and neuronal function.
Still in the context of SGs, tau also interacts with the splicing factor proline and glutamine rich (SFPQ) protein. SFPQ is an abundant nuclear RBP, being a key component of neuronal transport granules. SFPQ is involved in various processes such as alternative splicing, DNA repair, transcriptional regulation and RNA processing and transport [195,196]. SFPQ is dysregulated in association with tau and TIA1 proteins in patients with rapidly progressive AD. This dysregulation has been associated with the nuclear depletion and cytoplasmic co-localization with TIA1 and tau proteins [197].
More recently, Jiang and colleagues (2021) showed that hnRNPA2B1 protein is required for tau-mediated neurodegeneration in vitro and in vivo [198]. The hnRNPA2B1 protein is a reader of m6A in the nucleus, where it regulates the chromatin state and transcription [199,200,201]. Moreover, the oligomeric tau (oTau) complexes with hnRNPA2B1 and m6A-containing RNA seem to regulate RNA translation stress response and promote SG formation. It was suggested that hnRNPA2B1 functions as an m6A reader bridging oTau to m6A labeled transcripts. Interestingly, hnRNPA2B1 knockdown reduces the toxicity induced by pathological tau and dampens the stress response in neurons [198].
Beyond its involvement in SGs, the link between tau and RNA translation regulation is strengthened by tau acting as a negative regulator for protein synthesis by binding to ribosomes [27]. Indeed, tau can function as translation inhibitor by sequestering ribosomal protein S6 (rpS6), a component of the 40S ribosome complex, which is essential for canonical translation [202]. Moreover, human AD brains showed decreased rpS6-dependent translation, establishing tau as a selective driver for global mRNA translation [202]. This tau–ribosome interaction could help to explain the link between tauopathies and cognitive impairment [202,203].
Moreover, tau oligomers co-localize with Musashi proteins (MSI1 and MSI2) in AD, ALS and FTD brains [204]. MSI proteins are RNA-binding proteins that regulate mRNA translation during neuronal development in Drosophila melanogaster [205]. MSI1 can be found in both cytoplasm and nucleus, while MSI2 was found in the cytoplasm and associated with polysomes [206]. Tau and MSI proteins co-localize in the cytoplasm and nuclei of mouse neurons [204]. Tau regulates MSI function by modulating MSI levels and controlling their cellular localization and interactome [204].
Recently, an additional, direct link between tau and mRNA translation inhibition was found. Two reports revealed a direct interaction between tau and DDX6 [163,207]. DDX6 is a DEAD-box RNA helicase known as mediator of mRNA decay and miRNA biogenesis [208]. The interaction between tau and DDX6 increased the silencing activity of let-7a, miR-21 and miR-124 miRNAs. Indeed, it was shown that the tau/DDX6 interaction potentiates let-7 miRNA silencing activity rather than affecting the translational repression mediated by DDX6. Furthermore, disease-associated mutations of tau P301S and P301L disrupted tau/DDX6 interaction and consequently impaired let-7 mi-RISC-mediated gene silencing. Altogether, these data reveal a new role for tau in regulating miRNA activity [163]. Furthermore, DDX6 accumulates in AD and CBD human brains, where it co-localizes with hyperphosphorylated tau [163].
Interestingly, it was described that tau also has a nuclear role, being crucial for DNA and RNA integrity in vivo. Furthermore, tau oligomerization triggers the loss of the nucleic acid protective effect of monomeric tau [10,23]. Besides that, tau has a major impact on transcription and post-transcription regulation. Indeed, tau nuclear levels and phosphorylation state reduced global synthesis of RNA (including rRNA) and was suggested to be relevant to ribosomal biogenesis and function [209,210]. Moreover, tau was shown to be crucial for ribosomal DNA transcriptional repression through its involvement in heterochromatin and DNA methylation [210]. In addition, tau seems to have a role in translocating nuclear RBPs to cytoplasm [176,177]. This might be due to tau interaction with nucleoporins impairing protein transport between the nucleus and the cytoplasm [20]. Considering that under basal conditions most RBPs are nuclear, under stress, they are translocated to the cytoplasm, where they may trigger SG formation [161,211]. Although the precise mechanism by which tau modulates transcription is unknown, tau involvement in heterochromatin and DNA methylation is a factor to take into consideration. Moreover, tau-mediated translocation of known transcription regulators such as SFPQ and TDP-43 to the cytoplasm might be of utmost importance. Thus, tau aggregation into SGs may also be a key factor for tau transcription modulation. The relevance of tau in this process grants tau a pivotal role in global mRNA metabolism. Altogether, SG formation and protein aggregation have a huge impact on tauopathies due to their role in transcription regulation, RNA integrity, ribosomal function and subcellular compartmentalization. Finally, these studies show that tau-mediated RNA metabolism can be a powerful driver of neurodegenerative diseases. However, whether tau-mediated RNA metabolism alone can drive tau pathology is still an open question.
5. Conclusions
The molecular mechanisms underlying the onset and development of tauopathies are very complex. The undeniable role of tau post-translational modifications has been extensively studied. In this review, we highlighted the interest in investigating not only the role of tau protein in general mRNA metabolism but also the role of tau mRNA metabolism per se in the development of tauopathies. The diversity of tau mRNA variants impacts tau protein properties and its physiological and pathological functions. This diversity is mainly related to alternative splicing, but other variables such as the alternative promoter usage (APU) or the involvement in the general mRNA metabolism (SG formation) may also contribute and need to be further investigated. Likewise, APU could have an impact on tau translation and the production of some N-terminal truncated tau species whose role in tauopathies is well-known. Therefore, some aspects of tau mRNA metabolism are likely among the mechanisms underlying translation deregulation and able to play an instrumental role in the physiopathology of tauopathies.
Funding
Our research is funded by ‘Centre National de la Recherche Scientifique’, by ‘Fondation pour la Recherche Médicale’ and ‘Fondation Alzheimer’ (Grant number: ALZ201912009641) and by ‘Agence Nationale pour la Recherche’ (Grant number: ANR-21-CE12-0028).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Weingarten, M.D.; Lockwood, A.H.; Hwo, S.Y.; Kirschner, M.W. A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cleveland, D.; Hwo, S.-Y.; Kirschner, M.W. Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J. Mol. Biol. 1977, 116, 227–247. [Google Scholar] [CrossRef]
- Binder, L.I.; Frankfurter, A.; Rebhun, L.I. The distribution of tau in the mammalian central nervous central nervous. J. Cell Biol. 1985, 101, 1371–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goedert, M.; Crowther, R.A.; Garner, C.C. Molecular characterization of microtubule-associated proteins tau and map2. Trends Neurosci. 1991, 14, 193–199. [Google Scholar] [CrossRef]
- Janning, D.; Igaev, M.; Sündermann, F.; Brühmann, J.; Beutel, O.; Heinisch, J.J.; Bakota, L.; Piehler, J.; Junge, W.; Brandt, R. Single-molecule tracking of tau reveals fast kiss-and-hop interaction with microtubules in living neurons. Mol. Biol. Cell 2014, 25, 3541–3551. [Google Scholar] [CrossRef]
- Kellogg, E.H.; Hejab, N.M.A.; Poepsel, S.; Downing, K.H.; DiMaio, F.; Nogales, E. Near-atomic model of microtubule-tau interactions. Science 2018, 360, 1242–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbier, P.; Zejneli, O.; Martinho, M.; Lasorsa, A.; Belle, V.; Smet-Nocca, C.; Tsvetkov, P.O.; Devred, F.; Landrieu, I. Role of Tau as a Microtubule-Associated Protein: Structural and Functional Aspects. Front. Aging Neurosci. 2019, 11, 204. [Google Scholar] [CrossRef] [Green Version]
- Sotiropoulos, I.; Galas, M.-C.; Silva, J.; Skoulakis, E.; Wegmann, S.; Maina, M.B.; Blum, D.; Sayas, C.L.; Mandelkow, E.-M.; Mandelkow, E.; et al. Atypical, non-standard functions of the microtubule associated Tau protein. Acta Neuropathol. Commun. 2017, 5, 91. [Google Scholar] [CrossRef] [Green Version]
- Marciniak, E.; Leboucher, A.; Caron, E.; Ahmed, T.; Tailleux, A.; Dumont, J.; Issad, T.; Gerhardt, E.; Pagesy, P.; Vileno, M.; et al. Tau deletion promotes brain insulin resistance. J. Exp. Med. 2017, 214, 2257–2269. [Google Scholar] [CrossRef]
- Violet, M.; Chauderlier, A.; Delattre, L.; Tardivel, M.; Chouala, M.S.; Sultan, A.; Marciniak, E.; Humez, S.; Binder, L.; Kayed, R.; et al. Prefibrillar Tau oligomers alter the nucleic acid protective function of Tau in hippocampal neurons in vivo. Neurobiol. Dis. 2015, 82, 540–551. [Google Scholar] [CrossRef]
- Ahmed, T.; Van der Jeugd, A.; Blum, D.; Galas, M.-C.; D’Hooge, R.; Buee, L.; Balschun, D. Cognition and hippocampal synaptic plasticity in mice with a homozygous tau deletion. Neurobiol. Aging 2014, 35, 2474–2478. [Google Scholar] [CrossRef] [PubMed]
- Ash, P.E.A.; Lei, S.; Shattuck, J.; Boudeau, S.; Carlomagno, Y.; Medalla, M.; Mashimo, B.L.; Socorro, G.; Al-Mohanna, L.F.A.; Jiang, L.; et al. TIA1 potentiates tau phase separation and promotes generation of toxic oligomeric tau. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.D.; Suchowerska, A.; Van Der Hoven, J.; De Silva, D.M.; Wu, C.W.; van Eersel, J.; Ittner, A.; Ittner, L. Lessons from Tau-Deficient Mice. Int. J. Alzheimer’s Dis. 2012, 2012, 873270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, T.; Whitcomb, D.; Eunjoon, K.; Regan, P.; Piers, T.; Heo, S.; Brown, C.; Hashikawa, T.; Murayama, M.; Seok, H.; et al. Microtubule-associated protein tau is essential for long-term depression in the hippocampus. Philos. Trans. R. Soc. B: Biol. Sci. 2014, 369, 20130144. [Google Scholar] [CrossRef] [Green Version]
- Biundo, F.; Del Prete, D.; Zhang, H.; Arancio, O.; D’Adamio, L. A role for tau in learning, memory and synaptic plasticity. Sci. Rep. 2018, 8, 3184. [Google Scholar] [CrossRef] [Green Version]
- Brandt, R.; Trushina, N.I.; Bakota, L. Much More Than a Cytoskeletal Protein: Physiological and Pathological Functions of the Non-microtubule Binding Region of Tau. Front. Neurol. 2020, 11, 590059. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G. Ordered Assembly of Tau Protein and Neurodegeneration. Adv. Exp. Med. Biol. 2019, 1184, 3–21. [Google Scholar]
- Sexton, C.; Snyder, H.; Beher, D.; Boxer, A.L.; Brannelly, P.; Brion, J.P.; Buée, L.; Cacace, A.M.; Chételat, G.; Citron, M. Current directions in tau research: Highlights from Tau. Alzheimer’s Dement. 2021, 1–20. [Google Scholar] [CrossRef]
- Paonessa, F.; Evans, L.D.; Solanki, R.; Larrieu, D.; Wray, S.; Hardy, J.; Jackson, S.P.; Livesey, F.J. Microtubules Deform the Nuclear Membrane and Disrupt Nucleocytoplasmic Transport in Tau-Mediated Frontotemporal Dementia. Cell Rep. 2019, 26, 582–593.e5. [Google Scholar] [CrossRef] [Green Version]
- Eftekharzadeh, B.; Daigle, J.G.; Kapinos, L.E.; Coyne, A.; Schiantarelli, J.; Carlomagno, Y.; Cook, C.; Miller, S.J.; Dujardin, S.; Amaral, A.S.; et al. Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron 2018, 99, 925–940. [Google Scholar] [CrossRef] [Green Version]
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014, 17, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Khurana, V.; Merlo, P.; DuBoff, B.; Fulga, T.A.; Sharp, K.A.; Campbell, S.D.; Götz, J.; Feany, M.B. A neuroprotective role for the DNA damage checkpoint in tauopathy. Aging Cell 2012, 11, 360–362. [Google Scholar] [CrossRef]
- Violet, M.; Delattre, L.; Tardivel, M.; Sultan, A.; Chauderlier, A.; Caillierez, R.; Talahari, S.; Nesslany, F.; Lefebvre, B.; Bonnefoy, E.; et al. A major role for Tau in neuronal DNA and RNA protection in vivo under physiological and hyperthermic conditions. Front. Cell. Neurosci. 2014, 8, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelison, G.L.; Levy, S.A.; Jenson, T.; Frost, B. Tau-induced nuclear envelope invagination causes a toxic accumulation of mRNA in Drosophila. Aging Cell 2018, 18, e12847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, Y.-C.; Guo, C.; Yalamanchili, H.K.; Abreha, M.; Al-Ouran, R.; Li, Y.; Dammer, E.B.; Lah, J.J.; Levey, A.I.; Bennett, D.A.; et al. Tau-Mediated Disruption of the Spliceosome Triggers Cryptic RNA Splicing and Neurodegeneration in Alzheimer’s Disease. Cell Rep. 2019, 29, 301–316.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bou Samra, E.; Buhagiar-Labarchède, G.; Machon, C.; Guitton, J.; Onclercq-Delic, R.; Green, M.R.; Alibert, O.; Gazin, C.; Veaute, X.; Amor-Guéret, M. A role for Tau protein in maintaining ribosomal DNA stability and cytidine deaminase-deficient cell survival. Nat. Commun. 2017, 8, 693. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Bell, M.; Lyons, D.N.; Rodriguez-Rivera, J.; Ingram, A.; Fontaine, S.N.; Mechas, E.; Chen, J.; Wolozin, B.; LeVine, H.; et al. Pathological Tau Promotes Neuronal Damage by Impairing Ribosomal Function and Decreasing Protein Synthesis. J. Neurosci. 2016, 36, 1001–1007. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Jeong, H.-H.; Hsieh, Y.-C.; Klein, H.-U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Samimi, H.; Gamez, M.; Zare, H.; Frost, B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci. 2018, 21, 1038–1048. [Google Scholar] [CrossRef]
- Trabzuni, D.; Wray, S.; Vandrovcova, J.; Ramasamy, A.; Walker, R.; Smith, C.; Luk, C.; Gibbs, J.R.; Dillman, A.; Hernandez, D.G.; et al. MAPT expression and splicing is differentially regulated by brain region: Relation to genotype and implication for tauopathies. Hum. Mol. Genet. 2012, 21, 4094–4103. [Google Scholar] [CrossRef] [Green Version]
- Sinsky, J.; Pichlerova, K.; Hanes, J. Tau Protein Interaction Partners and Their Roles in Alzheimer’s Disease and Other Tauopathies. Int. J. Mol. Sci. 2021, 22, 9207. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Jakes, R. Expression of separate isoforms of human tau protein: Correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J 1990, 9, 4225–4230. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Song, X.; Nisbet, R.; Götz, J. Co-immunoprecipitation with Tau Isoform-specific Antibodies Reveals Distinct Protein Interactions and Highlights a Putative Role for 2N Tau in Disease. J. Biol. Chem. 2016, 291, 8173–8188. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Q.; Congdon, E.; Nagaraja, H.N.; Kuret, J. Tau Isoform Composition Influences Rate and Extent of Filament Formation. J. Biol. Chem. 2012, 287, 20711–20719. [Google Scholar] [CrossRef] [Green Version]
- Caffrey, T.M.; Joachim, C.; Wade-Martins, R. Haplotype-specific expression of the N-terminal exons 2 and 3 at the human MAPT locus. Neurobiol. Aging 2008, 29, 1923–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Kosik, K.S. Competition for microtubule-binding with dual expression of tau missense and splice isoforms. Mol. Biol. Cell 2001, 12, 171–184. [Google Scholar] [CrossRef] [Green Version]
- Goode, B.L.; Chau, M.; Denis, P.E.; Feinstein, S.C. Structural and functional differences between 3-repeat and 4-repeat tau isoforms: Implications for normal tau function and the onset of neurodegenerative disease. J. Biol. Chem. 2000, 275, 38182–38189. [Google Scholar] [CrossRef] [Green Version]
- Buée, L.; Delacourte, A. Comparative biochemistry of tau in progressive supranuclear palsy, corticobasal degenera-tion, FTDP-17 and Pick’s disease. Brain Pathol. 1999, 9, 681–693. [Google Scholar] [CrossRef]
- Bachmann, S.; Bell, M.; Klimek, J.; Zempel, H. Differential Effects of the Six Human TAU Isoforms: Somatic Retention of 2N-TAU and Increased Microtubule Number Induced by 4R-TAU. Front. Neurosci. 2021, 15, 643115. [Google Scholar] [CrossRef]
- DeVos, S.L.; Miller, R.L.; Schoch, K.M.; Holmes, B.B.; Kebodeaux, C.S.; Wegener, A.J.; Chen, G.; Shen, T.; Tran, H.; Nichols, B.; et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Lasagna-Reeves, C.A.; de Haro, M.; Hao, S.; Park, J.; Rousseaux, M.W.C.; Al-Ramahi, I.; Jafar-Nejad, P.; Vilanova-Velez, L.; See, L.; De Maio, A.; et al. Reduction of Nuak Decreases Tau and Reverses Phenotypes in a Tauopathy Mouse Model. Neuron 2016, 92, 407–418. [Google Scholar] [CrossRef] [Green Version]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.; Taylor, M.; Engström, P.; Frith, M.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Golovine, K.; Schwerin, M.; Vanselow, J. Three Different Promoters Control Expression of the Aromatase Cytochrome PGene (Cyp19) in Mouse Gonads and Brain1. Biol. Reprod. 2003, 68, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Pardo, L.M.; Rizzu, P.; Francescatto, M.; Vitezic, M.; Leday, G.G.; Sanchez, J.S.; Khamis, A.; Takahashi, H.; van de Berg, W.D.; Medvedeva, Y.A.; et al. Regional differences in gene expression and promoter usage in aged human brains. Neurobiol. Aging 2013, 34, 1825–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.H.; Liu, T.Y.C.; Li, H.; Choo, K.B. Use of a common promoter by two juxtaposed and intronless mouse early embryonic genes, Rnf33 and Rnf35: Implications in zygotic gene expression. Genomics 2002, 80, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Makrigiannis, A.P.; Hodge, D.L.; Anderson, S.K. Identification of a novel Ly49 promoter that is active in bone marrow and fetal thymus. J. Immunol. 2002, 168, 5163–5169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caillet-Boudin, M.-L.; Buee, L.; Sergeant, N.; Lefebvre, B. Regulation of human MAPT gene expression. Mol. Neurodegener. 2015, 10, 28. [Google Scholar] [CrossRef] [Green Version]
- Huin, V.; Buée, L.; Behal, H.; Labreuche, J.; Sablonniere, B.; Dhaenens, C.-M. Alternative promoter usage generates novel shorter MAPT mRNA transcripts in Alzheimer’s disease and progressive supranuclear palsy brains. Sci. Rep. 2017, 7, 12589. [Google Scholar] [CrossRef] [Green Version]
- Huin, V.; Deramecourt, V.; Caparros-Lefebvre, D.; Maurage, C.A.; Duyckaerts, C.; Kovari, E.; Pasquier, F.; Buée-Scherrer, V.; Labreuche, J.; Behal, H.; et al. The MAPT gene is differentially methylated in the pro-gressive supranuclear palsy brain. Mov. Disord. 2016, 31, 1883–1890. [Google Scholar] [CrossRef]
- Renbaum, P.; Beeri, R.; Gabai, E.; Amiel, M.; Gal, M.; Ehrengruber, M.U.; Levy-Lahad, E. Egr-1 upregulates the Alzheimer’s disease presenilin-2 gene in neuronal cells. Gene 2003, 318, 113–124. [Google Scholar] [CrossRef]
- Twine, N.A.; Janitz, K.; Wilkins, M.R.; Janitz, M. Whole Transcriptome Sequencing Reveals Gene Expression and Splicing Differences in Brain Regions Affected by Alzheimer’s Disease. PLoS ONE 2011, 6, e16266. [Google Scholar] [CrossRef] [PubMed]
- Ounallah-Saad, H.; Beeri, R.; Goshen, I.; Yirmiya, R.; Renbaum, P.; Levy-Lahad, E. Transcriptional regulation of the murine Presenilin-2 gene reveals similarities and differences to its human orthologue. Gene 2009, 446, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Borowsky, B.; Hoffman, B.J. Analysis of a gene encoding two glycine transporter variants reveals alternative promoter usage and a novel gene structure. J. Biol. Chem. 1998, 273, 29077–29085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udono, T.; Yasumoto, K.-I.; Takeda, K.; Amae, S.; Watanabe, K.; Saito, H.; Fuse, N.; Tachibana, M.; Takahashi, K.; Tamai, M.; et al. Structural organization of the human microphthalmia-associated transcription factor gene containing four alternative promoters. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2000, 1491, 205–219. [Google Scholar] [CrossRef]
- Rissman, R.A.; Poon, W.W.; Blurton-Jones, M.; Oddo, S.; Torp, R.; Vitek, M.P.; LaFerla, F.M.; Rohn, T.T.; Cotman, C.W. Caspa-se-cleavage of tau is an early event in Alzheimer disease tangle pathology. J. Clin. Investig. 2004, 114, 121–130. [Google Scholar] [CrossRef] [Green Version]
- De Calignon, A.; Fox, L.M.; Pitstick, R.; Carlson, G.A.; Bacskai, B.J.; Spires-Jones, T.L.; Hyman, B.T. Caspase activation precedes and leads to tangles. Nature 2010, 464, 1201–1204. [Google Scholar] [CrossRef]
- Matsumoto, S.-E.; Motoi, Y.; Ishiguro, K.; Tabira, T.; Kametani, F.; Hasegawa, M.; Hattori, N. The twenty-four KDa C-terminal tau fragment increases with aging in tauopathy mice: Implications of prion-like properties. Hum. Mol. Genet. 2015, 24, 6403–6416. [Google Scholar] [CrossRef] [Green Version]
- Zilka, N.; Filipcik, P.; Koson, P.; Fialova, L.; Skrabana, R.; Zilkova, M.; Rolkova, G.P.; Kontsekova, E.; Novak, M. Truncated tau from sporadic Alzheimer’s disease suffices to drive neurofibrillary degeneration in vivo. FEBS Lett. 2006, 580, 3582–3588. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.P.; Biernat, J.; Pickhardt, M.; Mandelkow, E. Stepwise proteolysis liberates tau fragments that nucleate the Alzheimer-like aggregation of full-length tau in a neuronal cell model. Proc. Natl. Acad. Sci. USA 2007, 104, 10252–10257. [Google Scholar] [CrossRef] [Green Version]
- Derisbourg, M.; Leghay, C.; Chiappetta, G.; Fernandez-Gomez, F.J.; Laurent, C.; Demeyer, D.; Carrier, S.; Buée-Scherrer, V.; Blum, D.; Vinh, J.; et al. Role of the Tau N-terminal region in microtubule stabilization revealed by new endogenous truncated forms. Sci. Rep. 2015, 5, 9659. [Google Scholar] [CrossRef] [Green Version]
- Veo, B.L.; Krushel, L.A. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J. Alzheimer’s Dis. 2009, 16, 271–275. [Google Scholar] [CrossRef]
- Veo, B.L.; Krushel, L.A. Secondary RNA structure and nucleotide specificity contribute to internal initiation mediated by the human tau 5’ leader. RNA Biol. 2012, 9, 1344–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mailliot, J.; Martin, F. Viral internal ribosomal entry sites: Four classes for one goal. Wiley Interdiscip. Rev. RNA 2018, 9, 1–34. [Google Scholar] [CrossRef]
- Lacerda, R.; Menezes, J.; Romão, L. More than just scanning: The importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell Mol. Life Sci. 2016, 74, 1659–1680. [Google Scholar] [CrossRef] [PubMed]
- Logette, E.; Wotawa, A.; Solier, S.; Desoche, L.; Solary, E.; Corcos, L. The human caspase-2 gene: Alternative promoters, pre-mRNA splicing and AUG usage direct isoform-specific expression. Oncogene 2003, 22, 935–946. [Google Scholar] [CrossRef] [Green Version]
- Kadener, S.; Fededa, J.P.; Rosbash, M.; Kornblihtt, A. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. USA 2002, 99, 8185–8190. [Google Scholar] [CrossRef] [Green Version]
- Cramer, P.; Cáceres, J.F.; Cazalla, D.; Kadener, S.; Muro, A.F.; Baralle, F.E.; Kornblihtt, A.R. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 1999, 4, 251–258. [Google Scholar] [CrossRef]
- Aronov, S.L.; Aranda, G.; Behar, L.; Ginzburg, I. Axonal Tau mRNA Localization Coincides with Tau Protein in Living Neuronal Cells and Depends on Axonal Targeting Signal. J. Neurosci. 2001, 21, 6577–6587. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, N.; Funakoshi, T.; Sato-Harada, R.; Kanai, Y. Selective stabilization of tau in axons and microtubule-associated protein 2C in cell bodies and dendrites contributes to polarized localization of cytoskeletal proteins in mature neurons. J. Cell Biol. 1996, 132, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Zempel, H.; Dennissen, F.J.A.; Kumar, Y.; Luedtke, J.; Biernat, J.; Mandelkow, E.-M.; Mandelkow, E. Axodendritic sorting and pathological missorting of Tau are isoform-specific and determined by axon initial segment architecture. J. Biol. Chem. 2017, 292, 12192–12207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balaji, V.; Kaniyappan, S.; Mandelkow, E.; Wang, Y.; Mandelkow, E.M. Pathological missorting of endogenous MAPT/Tau in neurons caused by failure of protein degradation systems. Autophagy 2018, 14, 2139–2154. [Google Scholar] [CrossRef] [PubMed]
- Gauthier-Kemper, A.; Alonso, M.S.; Sündermann, F.; Niewidok, B.; Fernandez, M.-P.; Bakota, L.; Heinisch, J.J.; Brandt, R. Annexins A2 and A6 interact with the extreme N terminus of tau and thereby contribute to tau’s axonal localization. J. Biol. Chem. 2018, 293, 8065–8076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauretti, E.; Dincer, O.; Praticò, D. Regional and temporal miRNAs expression profile in a transgenic mouse model of tauopathy: Implication for its pathogenesis. Mol. Psychiatry 2020, 26, 7020–7028. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.Y.; Hernandez-Rapp, J.; Jolivette, F.; Lecours, C.; Bisht, K.; Goupil, C.; Dorval, V.; Parsi, S.; Morin, F.; Planel, E.; et al. MiR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 2015, 24, 6721–6735. [Google Scholar] [CrossRef] [Green Version]
- Lekka, E.; Hall, J. Noncoding RNAs in disease. FEBS Lett. 2018, 592, 2884–2900. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Reddy, P.H. MicroRNA-455-3p as a potential biomarker for Alzheimer’s disease: An update. Front. Aging Neurosci. 2018, 10, 41. [Google Scholar] [CrossRef] [Green Version]
- Cosín-Tomàs, M.; Antonell, A.; Lladó, A.; Alcolea, D.; Fortea, J.; Ezquerra, M.; Lleó, A.; Martí, M.J.; Pallàs, M.; Sanchez-Valle, R.; et al. Plasma miR-34a-5p and miR-545-3p as Early Biomarkers of Alzheimer’s Disease: Potential and Limitations. Mol. Neurobiol. 2017, 54, 5550–5562. [Google Scholar] [CrossRef] [PubMed]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. Micro RNA -125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Huang, Y.; Wang, L.-L.; Zhang, Y.-F.; Xu, J.; Zhou, Y.; Lourenco, G.; Zhang, B.; Wang, Y.; Ren, R.-J.; et al. MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer’s disease. Sci. Rep. 2016, 6, 26697. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.Y.; Delay, C.; Girard, J.; Papon, M.-A.; Planel, E.; Sergeant, N.; Buée, L.; Hébert, S.S. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum. Mol. Genet. 2011, 20, 4016–4024. [Google Scholar] [CrossRef] [Green Version]
- Bazrgar, M.; Khodabakhsh, P.; Mohagheghi, F.; Prudencio, M.; Ahmadiani, A. Brain microRNAs dysregulation: Implication for missplicing and abnormal post-translational modifications of tau protein in Alzheimer’s disease and related tauopathies. Pharmacol. Res. 2020, 155, 104729. [Google Scholar] [CrossRef]
- Praticò, D. The Functional Role of microRNAs in the Pathogenesis of Tauopathy. Cells 2020, 9, 2262. [Google Scholar] [CrossRef]
- Zhao, J.; Yue, D.; Zhou, Y.; Jia, L.; Wang, H.; Guo, M.; Xu, H.; Chen, C.; Zhang, J.; Xu, L. The role of MicroRNAs in Aβ deposition and Tau phosphorylation in Alzheimer’s disease. Front. Neurol. 2017, 8, 342. [Google Scholar] [CrossRef] [Green Version]
- Min, S.-W.; Cho, S.-H.; Zhou, Y.; Schroeder, S.; Haroutunian, V.; Seeley, W.W.; Huang, E.; Shen, Y.; Masliah, E.; Mukherjee, C.; et al. Acetylation of Tau Inhibits Its Degradation and Contributes to Tauopathy. Neuron 2010, 67, 953–966. [Google Scholar] [CrossRef] [Green Version]
- Coupland, K.; Kim, W.S.; Halliday, G.; Hallupp, M.; Dobson-Stone, C.; Kwok, J.B.J. Role of the Long Non-Coding RNA MAPT-AS1 in Regulation of Microtubule Associated Protein Tau (MAPT) Expression in Parkinson’s Disease. PLoS ONE 2016, 11, e0157924. [Google Scholar] [CrossRef] [Green Version]
- Speir, M.L.; Zweig, A.S.; Rosenbloom, K.R.; Raney, B.J.; Paten, B.; Nejad, P.; Lee, B.T.; Learned, K.; Karolchik, D.; Hinrichs, A.; et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016, 44, D717–D725. [Google Scholar] [CrossRef]
- Elkouris, M.; Kouroupi, G.; Vourvoukelis, A.; Papagiannakis, N.; Kaltezioti, V.; Matsas, R.; Stefanis, L.; Xilouri, M.; Politis, P.K. Long Non-coding RNAs Associated With Neurodegeneration-Linked Genes Are Reduced in Parkinson’s Disease Patients. Front. Cell. Neurosci. 2019, 13, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simone, R.; Javad, F.; Emmett, W.; Wilkins, O.G.; Almeida, F.L.; Barahona-Torres, N.; Zareba-Paslawska, J.; Ehteramyan, M.; Zuccotti, P.; Modelska, A.; et al. MIR-NATs repress MAPT translation and aid proteostasis in neurodegeneration. Nature 2021, 594, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016, 17, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Stefanoska, K.; Volkerling, A.; Bertz, J.; Poljak, A.; Ke, Y.; Ittner, L.; Ittner, A. An N-terminal motif unique to primate tau enables differential protein–protein interactions. J. Biol. Chem. 2018, 293, 3710–3719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goode, B.L.; Denis, P.E.; Panda, D.; Radeke, M.J.; Miller, H.P.; Wilson, L.; Feinstein, S.C. Functional interactions between the proline-rich and repeat regions of tau enhance microtubule binding and assembly. Mol. Biol. Cell 1997, 8, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Z.; Ikezu, T. Tau Secretion. Adv. Exp. Med. Biol. 2019, 1184, 123–134. [Google Scholar]
- Hasegawa, M.; Smith, M.J.; Goedert, M. Tau proteins with FTDP-17 mutations have a reduced ability to promote microtubule assembly. FEBS Lett. 1998, 437, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Goedert, M.; Jakes, R.; Crowther, R. Effects of frontotemporal dementia FTDP-17 mutations on heparin-induced assembly of tau filaments. FEBS Lett. 1999, 450, 306–311. [Google Scholar] [CrossRef] [Green Version]
- Nacharaju, P.; Lewis, J.; Easson, C.; Yen, S.; Hackett, J.; Hutton, M. Accelerated filament formation from Tau protein with SPECIFIC FTDP-17 missense mutations. J. Neuropathol. Exp. Neurol. 1999, 58, 545. [Google Scholar] [CrossRef]
- Hutton, M.; Lendon, C.L.; Rizzu, P.; Baker, M.; Froelich, S.; Houlden, H.H.; Pickering-Brown, S.; Chakraverty, S.; Isaacs, A.; Grover, A.; et al. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998, 393, 702–704. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Murrell, J.R.; Goedert, M.; Farlow, M.R.; Klug, A.; Ghetti, B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. USA 1998, 95, 7737–7741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goedert, M.; Yamaguchi, Y.; Mishra, S.K.; Higuchi, M.; Sahara, N. Tau Filaments and the Development of Positron Emission Tomography Tracers. Front. Neurol. 2018, 9, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strang, K.H.; Golde, T.E.; Giasson, B.I. MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab. Investig. 2019, 99, 912–928. [Google Scholar] [CrossRef] [PubMed]
- Ghetti, B.; Oblak, A.L.; Boeve, B.F.; Johnson, K.A.; Dickerson, B.C.; Goedert, M. Invited review: Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: A chameleon for neuropathology and neuroimaging. Neuropathol. Appl. Neurobiol. 2014, 41, 24–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafei, R.; Woollacott, I.O.; Mummery, C.J.; Bocchetta, M.; Guerreiro, R.; Bras, J.; Warren, J.; Lashley, T.; Jaunmuktane, Z.; Rohrer, J.D. Two pathologically confirmed cases of novel mutations in the MAPT gene causing frontotemporal dementia. Neurobiol. Aging 2020, 87, 141.e15–141.e20. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Tagliavini, F. Frontotemporal lobar degeneration: Old knowledge and new insight into the pathogenetic mechanisms of tau mutations. Front. Aging Neurosci. 2015, 7, 192. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Gong, C.X. Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener. 2008, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.; Fairen, M.D.; Sabir, M.S.; Pastor, P.; Ding, J.; Ispierto, L.; Butala, A.; Morris, C.M.; Schulte, C.; Gasser, T.; et al. MAPT p.V363I mutation A rare cause of corticobasal degeneration. Neurol. Genet. 2019, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Sotiropoulos, I.; Silva, J.; Kimura, T.; Rodrigues, A.J.; Costa, P.; Almeida, O.F.X.; Sousa, N.; Takashima, A. Female hippo-campus vulnerability to environmental stress, a precipitating factor in tau aggregation pathology. J Alzheimer’s Dis. 2015, 43, 763–774. [Google Scholar] [CrossRef] [Green Version]
- Maeda, S.; Sato, Y.; Takashima, A. Frontotemporal dementia with Parkinsonism linked to chromosome-17 mutations enhance tau oligomer formation. Neurobiol. Aging 2018, 69, 26–32. [Google Scholar] [CrossRef]
- Iba, M.; Guo, J.L.; McBride, J.D.; Zhang, B.; Trojanowski, J.Q.; Lee, V.M.-Y. Synthetic Tau Fibrils Mediate Transmission of Neurofibrillary Tangles in a Transgenic Mouse Model of Alzheimer’s-Like Tauopathy. J. Neurosci. 2013, 33, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- Peeraer, E.; Bottelbergs, A.; Van Kolen, K.; Stancu, I.-C.; Vasconcelos, B.; Mahieu, M.; Duytschaever, H.; Donck, L.V.; Torremans, A.; Sluydts, E.; et al. Intracerebral injection of preformed synthetic tau fibrils initiates widespread tauopathy and neuronal loss in the brains of tau transgenic mice. Neurobiol. Dis. 2015, 73, 83–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.J.; Kerridge, C.; Cooper, J.; Cavallini, A.; Falcon, B.; Cella, C.V.; Landi, A.; Szekeres, P.G.; Murray, T.K.; Ahmed, Z.; et al. Short Fibrils Constitute the Major Species of Seed-Competent Tau in the Brains of Mice Transgenic for Human P301S Tau. J. Neurosci. 2016, 36, 762–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iba, M.; McBride, J.D.; Guo, J.L.; Zhang, B.; Trojanowski, J.Q.; Lee, V.M.-Y. Tau pathology spread in PS19 tau transgenic mice following locus coeruleus (LC) injections of synthetic tau fibrils is determined by the LC’s afferent and efferent connections. Acta Neuropathol. 2015, 130, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Strang, K.H.; Croft, C.L.; Sorrentino, Z.A.; Chakrabarty, P.; Golde, T.E.; Giasson, B.I. Distinct differences in prion-like seeding and aggregation between Tau protein variants provide mechanistic insights into tauopathies. J. Biol. Chem. 2018, 293, 2408–2421. [Google Scholar] [CrossRef] [Green Version]
- Schindowski, K.; Bretteville, A.; Leroy, K.; Bégard, S.; Brion, J.-P.; Hamdane, M.; Buee, L. Alzheimer’s Disease-Like Tau Neuropathology Leads to Memory Deficits and Loss of Functional Synapses in a Novel Mutated Tau Transgenic Mouse without Any Motor Deficits. Am. J. Pathol. 2006, 169, 599–616. [Google Scholar] [CrossRef] [Green Version]
- Neumann, M.; Diekmann, S.; Bertsch, U.; Vanmassenhove, B.; Bogerts, B.; Kretzschmar, H.A. Novel G335V mutation in the tau gene associated with early onset familial frontotemporal dementia. Neurogenetics 2005, 6, 91–95. [Google Scholar] [CrossRef]
- Ando, K.; Ferlini, L.; Suain, V.; Yilmaz, Z.; Mansour, S.; Le Ber, I.; Bouchard, C.; Leroy, K.; Durr, A.; Clot, F.; et al. De novo MAPT mutation G335A causes severe brain atrophy, 3R and 4R PHF-tau pathology and early onset frontotemporal dementia. Acta Neuropathol. Commun. 2020, 8, 94. [Google Scholar] [CrossRef]
- Origone, P.; Geroldi, A.; Lamp, M.; Sanguineri, F.; Caponnetto, C.; Cabona, C.; Gotta, F.; Trevisan, L.; Bellone, E.; Manganelli, F.; et al. Role of MAPT in Pure Motor Neuron Disease: Report of a Recurrent Mutation in Italian Patients. Neurodegener. Dis. 2019, 18, 310–314. [Google Scholar] [CrossRef]
- Di Fonzo, A.; Ronchi, D.; Gallia, F.; Cribiù, F.M.; Trezzi, I.; Vetro, A.; Mina, E.; Della Limongelli, I.; Bellazzi, R.; Ricca, I.; et al. Lower motor neuron disease with respiratory failure caused by a novel MAPT mutation. Neurology 2014, 82, 1990–1998. [Google Scholar] [CrossRef]
- Momeni, P.; Wickremaratchi, M.M.; Bell, J.; Arnold, R.; Beer, R.; Hardy, J.; Revesz, T.; Neal, J.W.; Morris, H.R. Familial early onset frontotemporal dementia caused by a novel S356T MAPT mutation, initially diagnosed as schizophrenia. Clin. Neurol. Neurosurg. 2010, 112, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Nicholl, D.J.; Greenstone, M.A.; Clarke, C.E.; Rizzu, P.; Crooks, D.; Crowe, A.; Trojanowski, J.Q.; Lee, V.M.-Y.; Heutink, P. An English kindred with a novel recessive tauopathy and respiratory failure. Ann. Neurol. 2003, 54, 682–686. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Bastone, A.; Piccoli, E.; Mazzoleni, G.; Morbin, M.; Uggetti, A.; Giaccone, G.; Sperber, S.; Beeg, M.; Salmona, M.; et al. New mutations in MAPT gene causing frontotemporal lobar degeneration: Biochemical and structural characteriza-tion. Neurobiol. Aging 2012, 33, 834.e1–834.e6. [Google Scholar]
- Borrego-Écija, S.; Antonell, A.; Puig-Butillé, J.A.; Pericot, I.; Prat-Bravo, C.; Abellan-Vidal, M.T.; Mallada, J.; Olives, J.; Falgàs, N.; Oliva, R.; et al. Novel P397S MAPT variant associated with late onset and slow progressive fronto-temporal dementia. Ann. Clin. Transl. Neurol. 2019, 6, 1559–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostojic, J.; Elfgren, C.; Passant, U.; Fabre, S.F.; Nilsson, K.; Gustafson, L.; Lannfelt, L. The Tau R406W Mutation Causes Progressive Presenile Dementia with Bitemporal Atrophy. Dement. Geriatr. Cogn. Disord. 2004, 17, 298–301. [Google Scholar] [CrossRef]
- Giaccone, G.; Rossi, G.; Farina, L.; Marcon, G.; Di Fede, G.; Catania, M.; Morbin, M.; Sacco, L.; Bugiani, O.; Tagliavini, F. Familial frontotemporal dementia associated with the novel MAPT mutation T427M. J. Neurol. 2005, 252, 1543–1545. [Google Scholar] [CrossRef]
- Cruchaga, C.; Chakraverty, S.; Mayo, K.; Vallania, F.L.M.; Mitra, R.D.; Faber, K.; Williamson, J.; Bird, T.; Diaz-Arrastia, R.; Foroud, T.M.; et al. Goate AM & for the NIA-LOAD—NCRAD Family Study Consortium (2012) Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PLoS ONE 2012, 7, e31039. [Google Scholar]
- Poorkaj, P.; Muma, N.A.; Zhukareva, V.; Cochran, E.J.; Shannon, K.M.; Hurtig, H.; Koller, W.C.; Bird, T.D.; Trojanowski, J.Q.; Lee, V.M.-Y.; et al. An R5L τ mutation in a subject with a progressive supranuclear palsy phenotype. Ann. Neurol. 2002, 52, 511–516. [Google Scholar] [CrossRef]
- Schulte, E.C.; Fukumori, A.; Mollenhauer, B.; Hor, H.; Arzberger, T.; Perneczky, R.; Kurz, A.; Diehl-Schmid, J.; Hull, M.; Lichtner, P.; et al. Rare variants in β-Amyloid precursor protein (APP) and Parkinson’s disease. Eur. J. Hum. Genet. 2015, 23, 1328–1333. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, S.; Toyoshima, Y.; Hasegawa, M.; Umeda, Y.; Wakabayashi, K.; Tokiguchi, S.; Iwatsubo, T.; Takahashi, H. Late-onset frontotemporal dementia with a novel exon 1 (Arg5His) tau gene mutation. Ann. Neurol. 2002, 51, 525–530. [Google Scholar] [CrossRef]
- Mutreja, Y.; Combs, B.; Gamblin, T.C. FTDP-17 mutations alter the aggregation and microtubule stabilization propensity of tau in an isoform-specific fashion. Biochemistry 2019, 58, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Šimić, G.; Babić Leko, M.; Wray, S.; Harrington, C.; Delalle, I.; Jovanov-Milošević, N.; Bažadona, D.; Buée, L.; de Silva, R.; Giovanni, G.D.; et al. Tau protein hyperphosphorylation and aggregation in alzheimer’s disease and other tauopathies, and possible neuroprotective strategies. Biomolecules 2016, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fichou, Y.; Al-Hilaly, Y.K.; Devred, F.; Smet-Nocca, C.; Tsvetkov, P.O.; Verelst, J.; Winderickx, J.; Geukens, N.; Vanmechelen, E.; Perrotin, A.; et al. The elusive tau molecular structures: Can we translate the recent breakthroughs into new targets for intervention? Acta Neuropathol. Commun. 2019, 7, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goedert, M.; Eisenberg, D.S.; Crowther, R.A. Propagation of Tau Aggregates and Neurodegeneration. Annu. Rev. Neurosci. 2017, 40, 189–210. [Google Scholar] [CrossRef] [PubMed]
- Mair, W.; Muntel, J.; Tepper, K.; Tang, S.; Biernat, J.; Seeley, W.W.; Kosik, K.S.; Mandelkow, E.; Steen, H.; Steen, J.A. FLEXITau: Quantifying Post-translational Modifications of Tau Protein in Vitro and in Human Disease. Anal. Chem. 2016, 88, 3704–3714. [Google Scholar] [CrossRef] [Green Version]
- Morris, M.; Knudsen, G.M.; Maeda, S.; Trinidad, J.C.; Ioanoviciu, A.; Burlingame, A.L.; Mucke, L. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nat. Neurosci. 2015, 18, 1183–1189. [Google Scholar] [CrossRef]
- Alquezar, C.; Arya, S.; Kao, A.W. Tau Post-translational Modifications: Dynamic Transformers of Tau Function, Degradation, and Aggregation. Front. Neurol. 2020, 11, 595532. [Google Scholar] [CrossRef]
- Wang, J.-Z.; Xia, Y.-Y.; Grundke-Iqbal, I.; Iqbal, K. Abnormal Hyperphosphorylation of Tau: Sites, Regulation, and Molecular Mechanism of Neurofibrillary Degeneration. J. Alzheimer’s Dis. 2013, 33 (Suppl. 1), S123–S139. [Google Scholar] [CrossRef]
- Thies, E.; Mandelkow, E.M. Missorting of tau in neurons causes degeneration of synapses that can be rescued by the kinase MARK2/Par-1. J. Neurosci. 2007, 27, 2896–2907. [Google Scholar] [CrossRef] [Green Version]
- Boyarko, B.; Hook, V. Human Tau Isoforms and Proteolysis for Production of Toxic Tau Fragments in Neuro-degeneration. Front. Neurosci. 2021, 15, S123–S139. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeganathan, S.; Hascher, A.; Chinnathambi, S.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. Proline-directed pseudo-phosphorylation at AT8 and PHF1 epitopes induces a compaction of the paperclip folding of tau and generates a patho-logical (MC-1) conformation. J. Biol. Chem. 2008, 283, 32066–32076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malia, T.J.; Teplyakov, A.; Ernst, R.; Wu, S.J.; Lacy, E.R.; Liu, X.; Vandermeeren, M.; Mercken, M.; Luo, J.; Sweet, R.W.; et al. Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8. Proteins Struct. Funct. Bioinform. 2016, 84, 427–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohn, P.D.; Tracy, T.E.; Son, H.I.; Zhou, Y.; Leite, R.E.P.; Miller, B.L.; Seeley, W.W.; Grinberg, L.T.; Gan, L. Acetylated tau destabilizes the cytoskeleton in the axon initial segment and is mislocalized to the somatodendritic compartment. Mol. Neurodegener. 2016, 11, 47. [Google Scholar] [CrossRef] [Green Version]
- Min, S.-W.; Chen, X.; Tracy, T.E.; Li, Y.; Zhou, Y.; Wang, C.; Shirakawa, K.; Minami, S.S.; Defensor, E.; Mok, S.-A.; et al. Critical role of acetylation in tau-mediated neurodegeneration and cognitive deficits. Nat. Med. 2015, 21, 1154–1162. [Google Scholar] [CrossRef] [Green Version]
- Cohen, T.J.; Guo, J.L.; Hurtado, D.E.; Kwong, L.K.; Mills, I.P.; Trojanowski, J.Q.; Lee, V.M.Y. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2011, 2, 252. [Google Scholar] [CrossRef] [Green Version]
- Cook, C.; Carlomagno, Y.; Gendron, T.F.; Dunmore, J.; Scheffel, K.; Stetler, C.; Davis, M.; Dickson, D.W.; Jarpe, M.; DeTure, M.; et al. Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 2014, 23, 104–116. [Google Scholar] [CrossRef]
- Min, S.W.; Sohn, P.D.; Li, Y.; Devidze, N.; Johnson, J.R.; Krogan, N.J.; Masliah, E.; Mok, S.A.; Gestwicki, J.E.; Gan, L. SIRT1 deacetylates tau and reduces pathogenic tau spread in a mouse model of tauopathy. J. Neurosci. 2018, 38, 3680–3688. [Google Scholar] [CrossRef]
- Cockram, P.E.; Kist, M.; Prakash, S.; Chen, S.-H.; Wertz, I.E.; Vucic, D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021, 28, 591–605. [Google Scholar] [CrossRef]
- Harris, L.D.; Jasem, S.; Licchesi, J.D.F. The ubiquitin system in AlzheimerTs. Proteostasis Dis. Basic Mech. Clin. 2020, 1233, 195. [Google Scholar]
- Thomas, S.N.; Funk, K.E.; Wan, Y.; Liao, Z.; Davies, P.; Kuret, J.; Yang, A.J. Dual modification of Alzheimer’s disease PHF-tau protein by lysine methylation and ubiquitylation: A mass spectrometry approach. Acta Neuropathol. 2011, 123, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, J.P.; Corbett, N.J.; Kellett, K.A.B.; Hooper, N.M. Tau Proteolysis in the Pathogenesis of Tauopathies: Neurotoxic Fragments and Novel Biomarkers. J. Alzheimer’s Dis. 2018, 63, 13–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novak, P.; Cehlar, O.; Skrabana, R.; Novak, M. Tau Conformation as a Target for Disease-Modifying Therapy: The Role of Truncation. J. Alzheimer’s Dis. 2018, 64, S535–S546. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kotilinek, L.A.; Smith, B.; Hlynialuk, C.; Zahs, K.; Ramsden, M.; Cleary, J.; Ashe, K.H. Caspase-2 cleavage of tau reversibly impairs memory. Nat. Med. 2016, 22, 1268–1276. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xu, W.; Jin, N.; Li, L.; Zhou, Y.; Chu, D.; Gong, C.-X.; Iqbal, K.; Liu, F. Truncation of Tau selectively facilitates its pathological activities. J. Biol. Chem. 2020, 295, 13812–13828. [Google Scholar] [CrossRef] [PubMed]
- Kampers, T.; Friedhoff, P.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 1996, 399, 344–349. [Google Scholar] [CrossRef]
- Dinkel, P.D.; Holden, M.R.; Matin, N.; Margittai, M. RNA Binds to Tau Fibrils and Sustains Template-Assisted Growth. Biochemistry 2015, 54, 4731–4740. [Google Scholar] [CrossRef] [Green Version]
- Kapeli, K.; Martinez, F.J.; Yeo, G.W. Genetic mutations in RNA-binding proteins and their roles in ALS. Qual. Life Res. 2017, 136, 1193–1214. [Google Scholar] [CrossRef] [Green Version]
- Tomé, S.O.; Gomes, L.A.; Li, X.; Vandenberghe, R.; Tousseyn, T.; Thal, D.R. TDP-43 interacts with pathological τ protein in Alzheimer’s disease. Acta Neuropathol. 2021, 141, 795–799. [Google Scholar] [CrossRef]
- Montalbano, M.; McAllen, S.; Cascio, F.L.; Sengupta, U.; Garcia, S.; Bhatt, N.; Ellsworth, A.; Heidelman, E.A.; Johnson, O.D.; Doskocil, S.; et al. TDP-43 and Tau Oligomers in Alzheimer’s Disease, Amyotrophic Lateral Sclerosis, and Frontotemporal Dementia. Neurobiol. Dis. 2020, 146, 105130. [Google Scholar] [CrossRef]
- Maziuk, B.F.; Apicco, D.; Cruz, A.L.; Jiang, L.; Ash, P.E.A.; da Rocha, E.L.; Zhang, C.; Yu, W.H.; Leszyk, J.; Abisambra, J.F.; et al. RNA binding proteins co-localize with small tau inclusions in tauopathy. Acta Neuropathol. Commun. 2018, 6, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apicco, D.; Ash, P.E.A.; Maziuk, B.; LeBlang, C.; Medalla, M.; Al Abdullatif, A.; Ferragud, A.; Botelho, E.; Balance, H.I.; Dhawan, U.; et al. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat. Neurosci. 2018, 21, 72–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauderlier, A.; Gilles, M.; Spolcova, A.; Caillierez, R.; Chwastyniak, M.; Kress, M.; Drobecq, H.; Bonnefoy, E.; Pinet, F.; Weil, D.; et al. Tau/DDX6 interaction increases microRNA activity. Biochim. Biophys. Acta (BBA)-Bioenerg. 2018, 1861, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Kedersha, N.; Low, W.-K.; Romo, D.; Gorospe, M.; Kaufman, R.; Anderson, P.; Liu, J.O. Eukaryotic Initiation Factor 2α-independent Pathway of Stress Granule Induction by the Natural Product Pateamine A. J. Biol. Chem. 2006, 281, 32870–32878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilks, N.; Kedersha, N.; Ayodele, M.; Shen, L.; Stoecklin, G.; Dember, L.M.; Anderson, P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 2004, 15, 5383–5398. [Google Scholar] [CrossRef] [Green Version]
- Kedersha, N.; Cho, M.R.; Li, W.; Yacono, P.W.; Chen, S.; Gilks, N.; Golan, D.E.; Anderson, P. Dynamic Shuttling of Tia-Accompanies the Recruitment of mRNA to Mammalian Stress Granules. J. Cell Biol. 2000, 151, 1257–1268. [Google Scholar] [CrossRef]
- Kedersha, N.; Chen, S.; Gilks, N.; Li, W.; Miller, I.J.; Stahl, J.; Anderson, P. Evidence that ternary complex (eIF2-GTP-tRNAiMet)-Deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 2002, 13, 195–210. [Google Scholar] [CrossRef] [Green Version]
- Mazroui, R.; Sukarieh, R.; Bordeleau, M.-E.; Kaufman, R.J.; Northcote, P.; Tanaka, J.; Gallouzi, I.; Pelletier, J. Inhibition of Ribosome Recruitment Induces Stress Granule Formation Independently of Eukaryotic Initiation Factor 2α Phosphorylation. Mol. Biol. Cell 2006, 17, 4212–4219. [Google Scholar] [CrossRef]
- McEwen, E.; Kedersha, N.; Song, B.; Scheuner, D.; Gilks, N.; Han, A.; Chen, J.-J.; Anderson, P.; Kaufman, R.J. Heme-regulated Inhibitor Kinase-mediated Phosphorylation of Eukaryotic Translation Initiation Factor Inhibits Translation, Induces Stress Granule Formation, and Mediates Survival upon Arsenite Exposure. J. Biol. Chem. 2005, 280, 16925–16933. [Google Scholar] [CrossRef] [Green Version]
- Mokas, S.; Mills, J.R.; Garreau, C.; Fournier, M.J.; Robert, F.; Arya, P.; Kaufman, R.J.; Pelletier, J.; Mazroui, R. Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell 2009, 20, 2673–2683. [Google Scholar] [CrossRef] [Green Version]
- Buchan, J.R.; Parker, R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asadi, M.R.; Moslehian, M.S.; Sabaie, H.; Jalaiei, A.; Ghafouri-Fard, S.; Taheri, M.; Rezazadeh, M. Stress Granules and Neurodegenerative Disorders: A Scoping Review. Front. Aging Neurosci. 2021, 13, 650740. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Ash, P.E.A.; Maziuk, B.F.; Balance, H.I.; Boudeau, S.; Al Abdullatif, A.; Orlando, M.; Petrucelli, L.; Ikezu, T.; Wolozin, B. TIA1 regulates the generation and response to toxic tau oligomers. Acta Neuropathol. 2019, 137, 259–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, J.; Rodrigues, S.; Sampaio-Marques, B.; Gomes, P.; Carvalho, A.; Dioli, C.; Soares-Cunha, C.; Mazuik, B.F.; Takashima, A.; Ludovico, P.; et al. Dysregulation of autophagy and stress granule-related proteins in stress-driven Tau pathology. Cell Death Differ. 2019, 26, 1411–1427. [Google Scholar] [CrossRef] [PubMed]
- Vanderweyde, T.; Yu, W.H.; Varnum, M.; Liu-Yesucevitz, L.; Citro, A.; Ikezu, T.; Duff, K.; Wolozin, B. Contrasting Pathology of the Stress Granule Proteins TIA-1 and G3BP in Tauopathies. J. Neurosci. 2012, 32, 8270–8283. [Google Scholar] [CrossRef] [Green Version]
- Vanderweyde, T.; Apicco, D.J.; Youmans-Kidder, K.; Ash, P.E.A.; Cook, C.; da Rocha, E.L.; Jansen-West, K.; Frame, A.A.; Citro, A.; Leszyk, J.D.; et al. Interaction of tau with the RNA-Binding Protein TIARegulates tau Pathophysiology and Toxicity. Cell Rep. 2016, 15, 1455–1466. [Google Scholar] [CrossRef] [Green Version]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.-C.; Krach, F.; Yang, D.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604.e13. [Google Scholar] [CrossRef] [Green Version]
- Apicco, D.; Zhang, C.; Maziuk, B.; Jiang, L.; Balance, H.I.; Boudeau, S.; Ung, C.; Li, H.; Wolozin, B. Dysregulation of RNA Splicing in Tauopathies. Cell Rep. 2019, 29, 4377–4388.e4. [Google Scholar] [CrossRef] [Green Version]
- Santacruz, K.; Lewis, J.; Spires, T.; Paulson, J.; Kotilinek, L.; Ingelsson, M.; Guimaraes, A.; DeTure, M.; Ramsden, M.; McCowan, E.; et al. Medicine: Tau suppression in a neurodegenerative mouse model improves memory function. Science 2005, 309, 476–481. [Google Scholar] [CrossRef] [Green Version]
- Zempel, H.; Mandelkow, E. Lost after translation: Missorting of Tau protein and consequences for Alzheimer disease. Trends Neurosci. 2014, 37, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Buratti, E.; Brindisi, A.; Giombi, M.; Tisminetzky, S.; Ayala, Y.M.; Baralle, F.E. TDP-43 binds heterogeneous nuclear ri-bonucleoprotein A/B through its C-terminal tail: An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005, 280, 37572–37584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercado, P.A.; Ayala, Y.M.; Romano, M.; Buratti, E.; Baralle, F.E. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005, 33, 6000–6010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butti, Z.; Patten, S.A. RNA dysregulation in amyotrophic lateral sclerosis. Front. Genet. 2019, 10, 712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ederle, H.; Dormann, D. TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett. 2017, 591, 1489–1507. [Google Scholar] [CrossRef] [Green Version]
- Prasad, A.; Bharathi, V.; Sivalingam, V.; Girdhar, A.; Patel, B.K. Molecular mechanisms of TDP-43 misfolding and pa-thology in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 2019, 12, 25. [Google Scholar] [CrossRef]
- Monahan, Z.; Shewmaker, F.; Pandey, U.B. Stress granules at the intersection of autophagy and ALS. Brain Res. 2016, 1649, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Ratti, A.; Buratti, E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 2016, 138, 95–111. [Google Scholar] [CrossRef]
- Liu-Yesucevitz, L.; Bilgutay, A.; Zhang, Y.-J.; Vanderweyde, T.; Citro, A.; Mehta, T.; Zaarur, N.; McKee, A.; Bowser, R.; Sherman, M.; et al. Tar DNA Binding Protein-43 (TDP-43) Associates with Stress Granules: Analysis of Cultured Cells and Pathological Brain Tissue. PLoS ONE 2010, 5, e13250. [Google Scholar] [CrossRef] [Green Version]
- Gasset-Rosa, F.; Lu, S.; Yu, H.; Chen, C.; Melamed, Z.; Guo, L.; Shorter, J.; Da Cruz, S.; Cleveland, D.W. Cytoplasmic TDP-De-mixing Independent of Stress Granules Drives Inhibition of Nuclear Import, Loss of Nuclear TDP-43, and Cell Death. Neuron 2019, 102, 339–357.e7. [Google Scholar] [CrossRef] [Green Version]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreedharan, J.; Blair, I.P.; Tripathi, V.B.; Hu, X.; Vance, C.; Rogelj, B.; Ackerley, S.; Durnall, J.C.; Williams, K.L.; Buratti, E.; et al. TDP-Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Science 2008, 319, 1668–1672. [Google Scholar] [CrossRef] [PubMed]
- Tomé, S.O.; Vandenberghe, R.; Ospitalieri, S.; Van Schoor, E.; Tousseyn, T.; Otto, M.; Von Arnim, C.A.F.; Thal, D.R. Distinct molecular patterns of TDP-43 pathology in Alzheimer’s disease: Relationship with clinical phenotypes. Acta Neuropathol. Commun. 2020, 8, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Dickson, D.W.; Trojanowski, J.Q.; Jack, C.R.; Boyle, P.A.; Arfanakis, K.; Rademakers, R.; Alafuzoff, I.; Attems, J.; Brayne, C.; et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): Consensus working group report. Brain 2019, 142, 1503–1527. [Google Scholar] [CrossRef] [Green Version]
- Hirose, T.; Virnicchi, G.; Tanigawa, A.; Naganuma, T.; Li, R.; Kimura, H.; Yokoi, T.; Nakagawa, S.; Bénard, M.; Fox, A.H.; et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 2014, 25, 169–183. [Google Scholar] [CrossRef]
- Lee, M.; Sadowska, A.; Bekere, I.; Ho, D.; Gully, B.; Lu, Y.; Iyer, K.S.; Trewhella, J.; Fox, A.H.; Bond, C.S. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 2015, 43, 3826–3840. [Google Scholar] [CrossRef]
- Younas, N.; Zafar, S.; Shafiq, M.; Noor, A.; Siegert, A.; Arora, A.S.; Galkin, A.; Zafar, A.; Schmitz, M.; Stadelmann, C.; et al. SFPQ and Tau: Critical factors contributing to rapid progression of Alzheimer’s disease. Acta Neuropathol. 2020, 140, 317–339. [Google Scholar] [CrossRef]
- Jiang, L.; Lin, W.; Zhang, C.; Ash, P.E.; Verma, M.; Kwan, J.; van Vliet, E.; Yang, Z.; Cruz, A.L.; Boudeau, S.; et al. Interaction of tau with HNRNPA2B1 and N6-methyladenosine RNA mediates the progression of tauopathy. Mol. Cell 2021, 81, 4209–4227.e12. [Google Scholar] [CrossRef]
- Anders, M.; Chelysheva, I.; Goebel, I.; Trenkner, T.; Zhou, J.; Mao, Y.; Verzini, S.; Qian, S.-B.; Ignatova, Z. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 2018, 1, e201800113. [Google Scholar] [CrossRef] [Green Version]
- Arguello, A.E.; DeLiberto, A.N.; Kleiner, R.E. RNA Chemical Proteomics Reveals the N6-Methyladenosine (m6A)-Regulated Protein–RNA Interactome. J. Am. Chem. Soc. 2017, 139, 17249–17252. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Zhang, J.; Zhu, J.-S. The role of m6A RNA methylation in human cancer. Mol. Cancer 2019, 18, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koren, S.; Hamm, M.J.; Meier, S.E.; Weiss, B.E.; Nation, G.K.; Chishti, E.A.; Arango, J.P.; Chen, J.; Zhu, H.; Blalock, E.M.; et al. Tau drives translational selectivity by interacting with ribosomal proteins. Acta Neuropathol. 2019, 137, 571–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koren, S.A.; Galvis-Escobar, S.; Abisambra, J.F. Tau-mediated dysregulation of RNA: Evidence for a common molecular mechanism of toxicity in frontotemporal dementia and other tauopathies. Neurobiol. Dis. 2020, 141, 104939. [Google Scholar] [CrossRef]
- Montalbano, M.; McAllen, S.; Puangmalai, N.; Sengupta, U.; Bhatt, N.; Johnson, O.D.; Kharas, M.G.; Kayed, R. RNA-binding proteins Musashi and tau soluble aggregates initiate nuclear dysfunction. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Okano, H.; Blendy, J.A.; Montell, C. Musashi, a neural RNA-binding protein required for drosophila adult external sensory organ development. Neuron 1994, 13, 67–81. [Google Scholar] [CrossRef]
- Kaneko, J.; Chiba, C. Immunohistochemical analysis of Musashi-1 expression during retinal regeneration of adult newt. Neurosci. Lett. 2009, 450, 252–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunawardana, C.G.; Mehrabian, M.; Wang, X.; Mueller, I.; Lubambo, I.B.; Jonkman, J.; Wang, H.; Schmitt-Ulms, G. The Human Tau Interactome: Binding to the Ribonucleoproteome, and Impaired Binding of the Proline-to-Leucine Mutant at Position 301 (P301L) to Chaperones and the Proteasome. Mol. Cell. Proteom. 2015, 14, 3000–3014. [Google Scholar] [CrossRef] [Green Version]
- Mathys, H.; Basquin, J.Ô.; Ozgur, S.; Czarnocki-Cieciura, M.; Bonneau, F.; Aartse, A.; Dziembowski, A.; Nowotny, M.; Conti, E.; Filipowicz, W. Structural and Biochemical Insights to the Role of the CCR4-NOT Complex and DDXATPase in Mi-croRNA Repression. Mol. Cell 2014, 54, 751–765. [Google Scholar] [CrossRef] [Green Version]
- Maina, M.B.; Al-Hilaly, Y.K.; Serpell, L.C. Nuclear Tau and Its Potential Role in Alzheimer’s Disease. Biomolecules 2016, 6, 9. [Google Scholar] [CrossRef] [Green Version]
- Maina, M.B.; Bailey, L.J.; Doherty, A.J.; Serpell, L.C. The Involvement of Aβ42 and Tau in Nucleolar and Protein Synthesis Machinery Dysfunction. Front. Cell. Neurosci. 2018, 12, 220. [Google Scholar] [CrossRef]
- Cruz, A.; Verma, M.; Wolozin, B. The Pathophysiology of Tau and Stress Granules in Disease. In Tau Biology; Springer: Singapore, 2019; Volume 1184, pp. 359–372. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).