An Adapted Deep Convolutional Neural Network for Automatic Measurement of Pancreatic Fat and Pancreatic Volume in Clinical Multi-Protocol Magnetic Resonance Images: A Retrospective Study with Multi-Ethnic External Validation
Abstract
1. Introduction
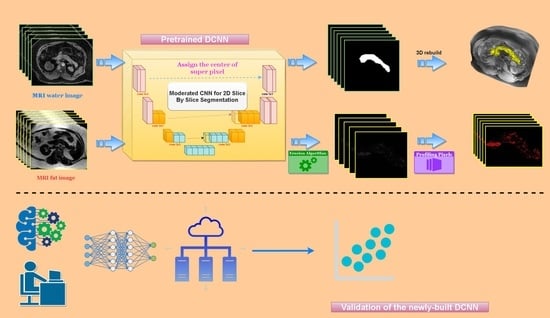
2. Methods
2.1. Study Participants and Datasets
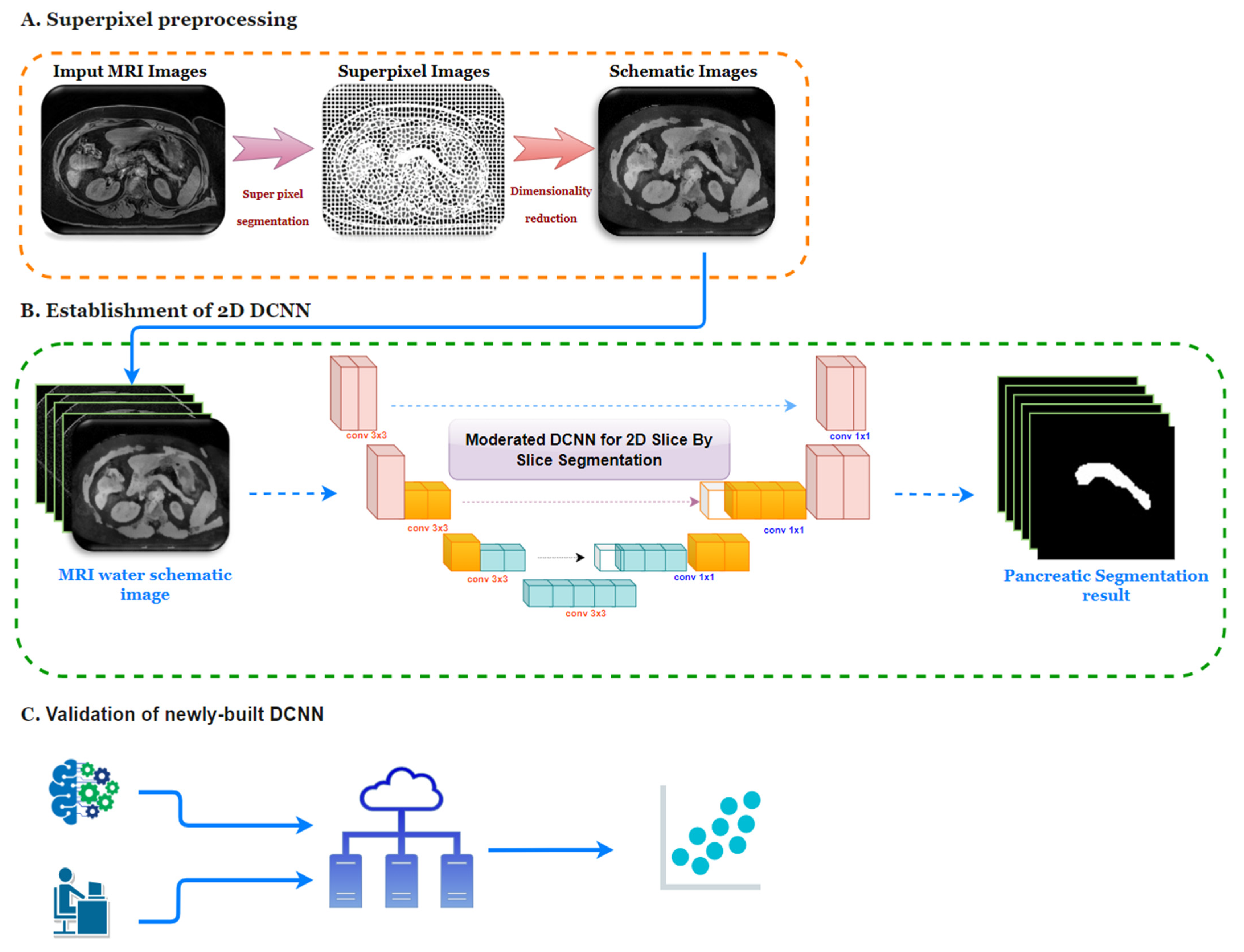
2.2. Image Preprocessing
2.2.1. Superpixel Segmentation
2.2.2. Dimensionality Reduction of Images
2.3. DCNN Establishment and Performance Evaluation
2.4. Principles of Measuring Pancreas Volume and Pancreatic Fat Deposition
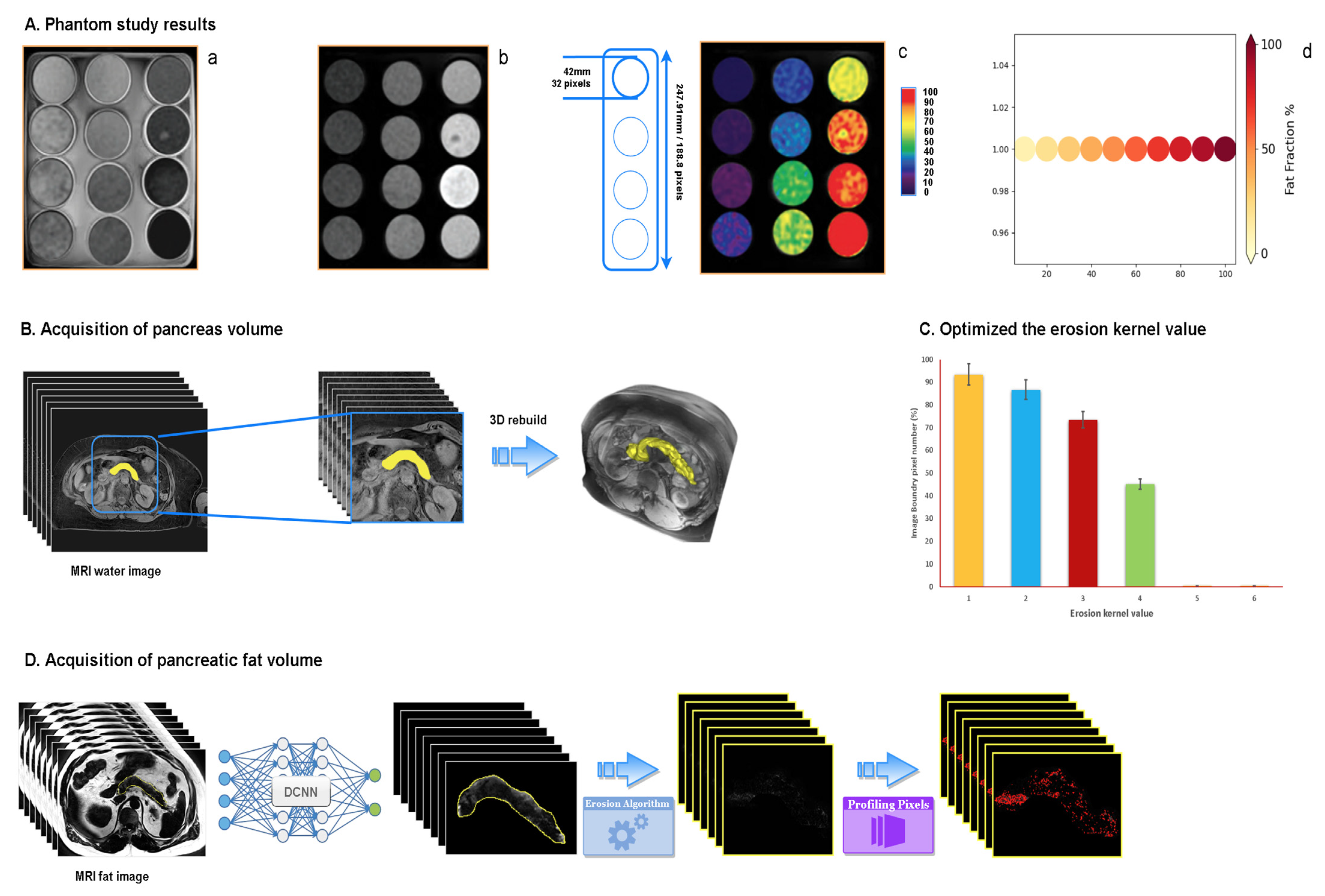
2.4.1. Phantom Study
2.4.2. Acquisition of Pancreas Volume
2.4.3. Acquisition of Pancreatic Fat Volume
2.5. Statistics
3. Results
3.1. Evaluation of the Correlation between the Pancreatic Fat Fraction and Type 2 Diabetes in Various Ethnic Groups
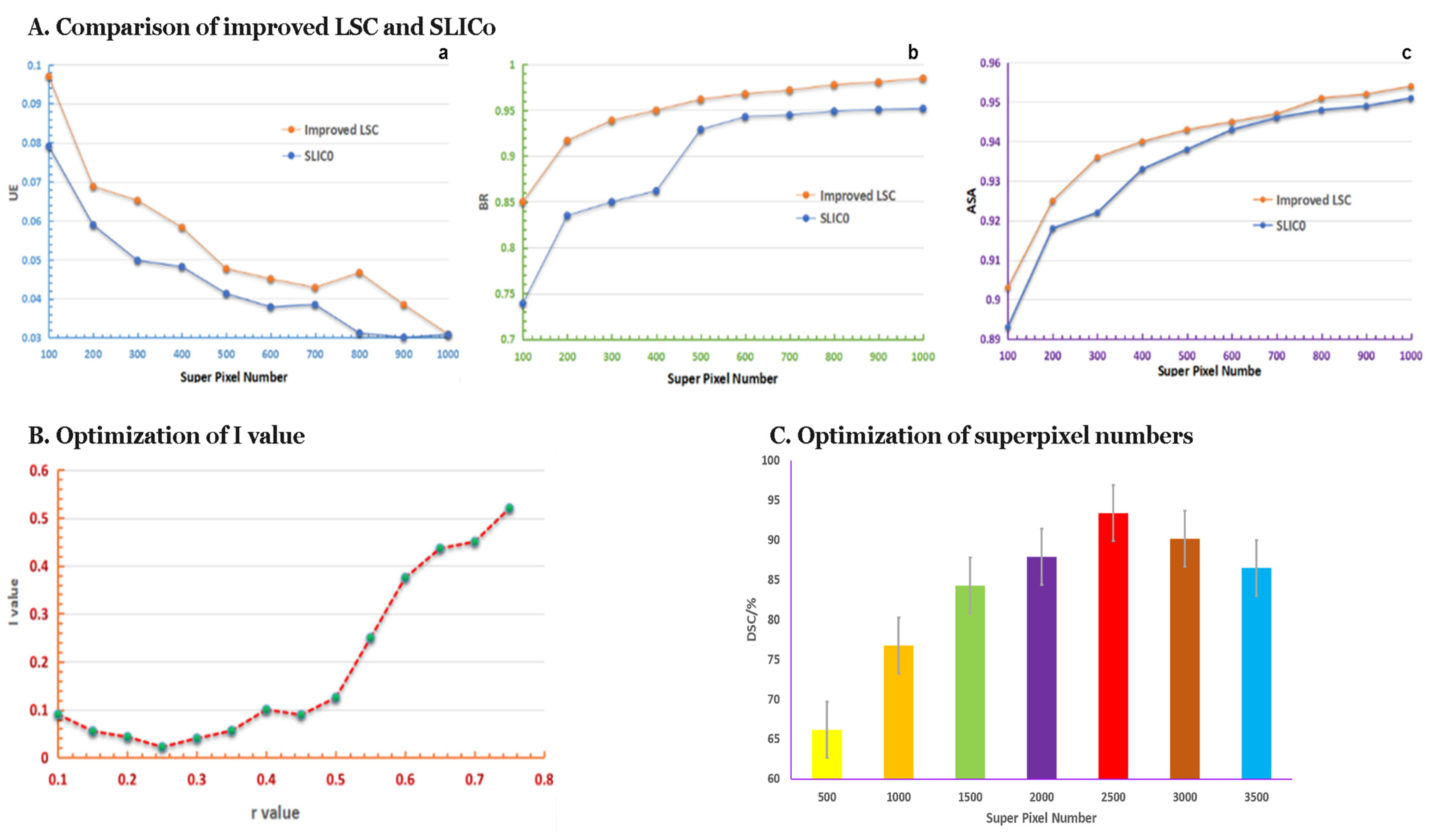
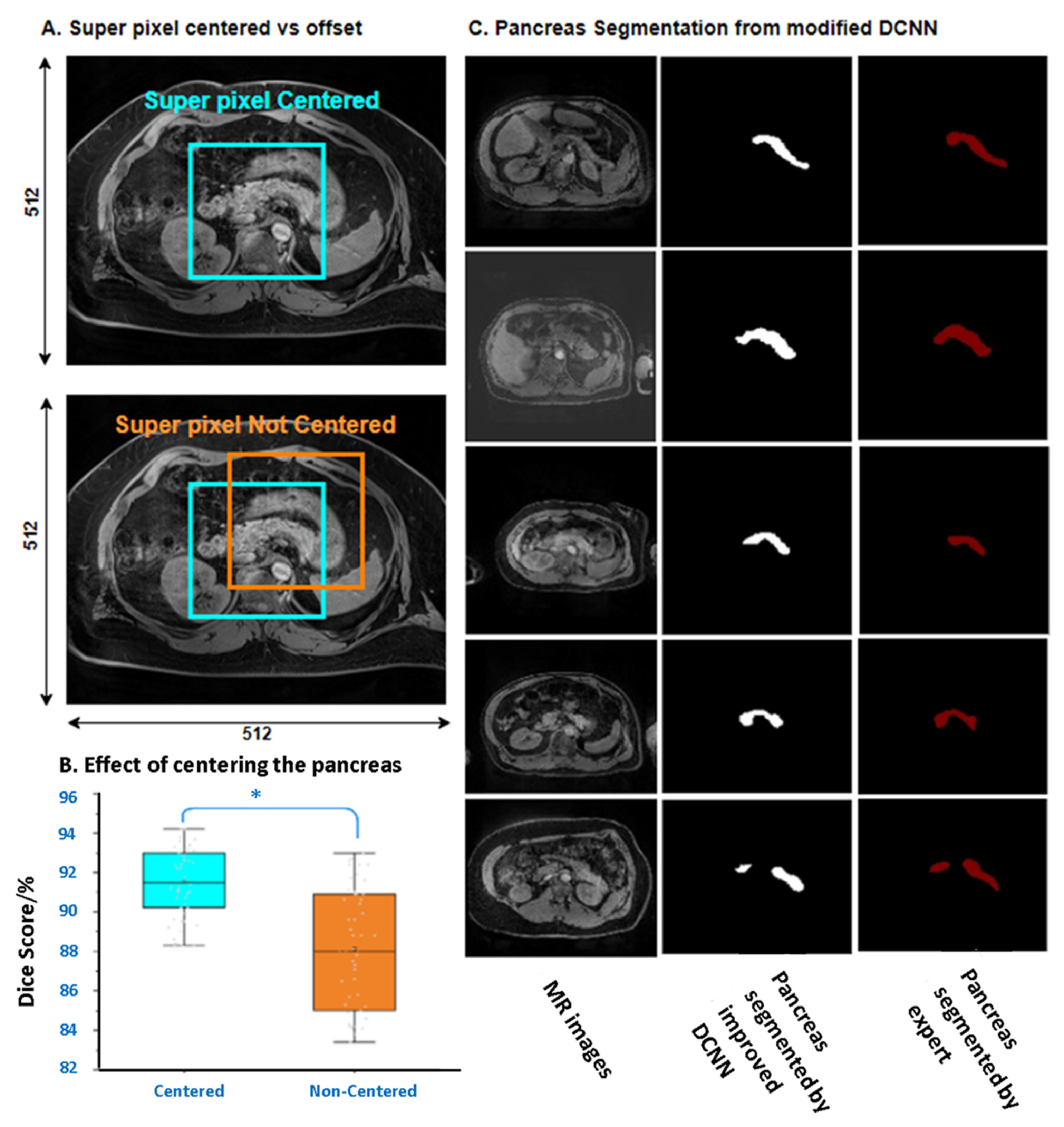
3.2. The Performance of Superpixel Segmentation
3.2.1. Results of the Comparison of Improved LSC and SLIC0
3.2.2. Request r-Value by Improved LSC Method
3.2.3. The Optimum Number of Superpixels
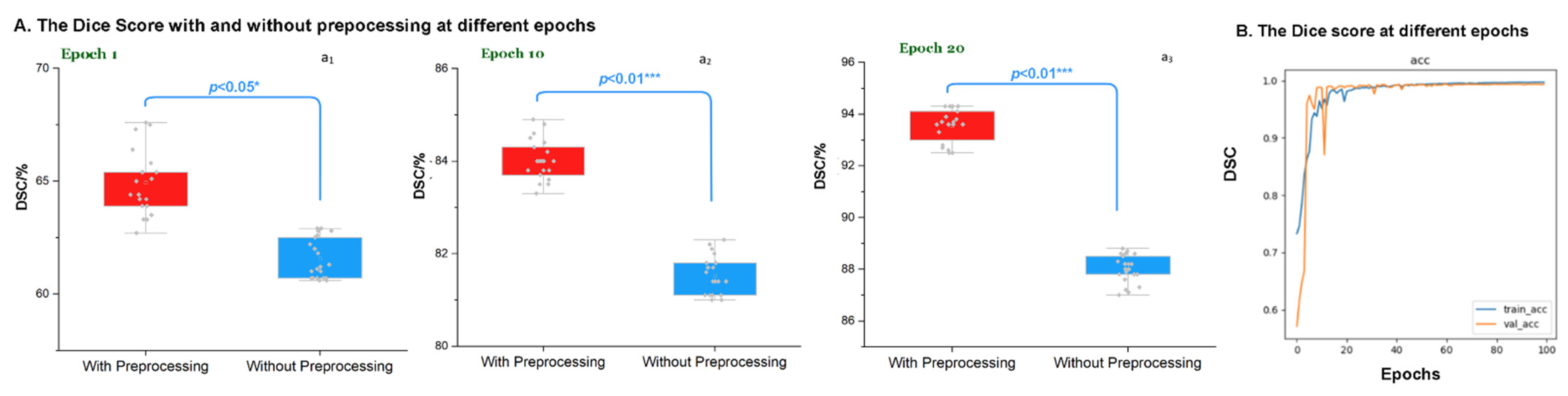
3.3. Evaluation of the Preprocessing at Different Epochs
3.4. Segmentation Integrity of Novel DCNN
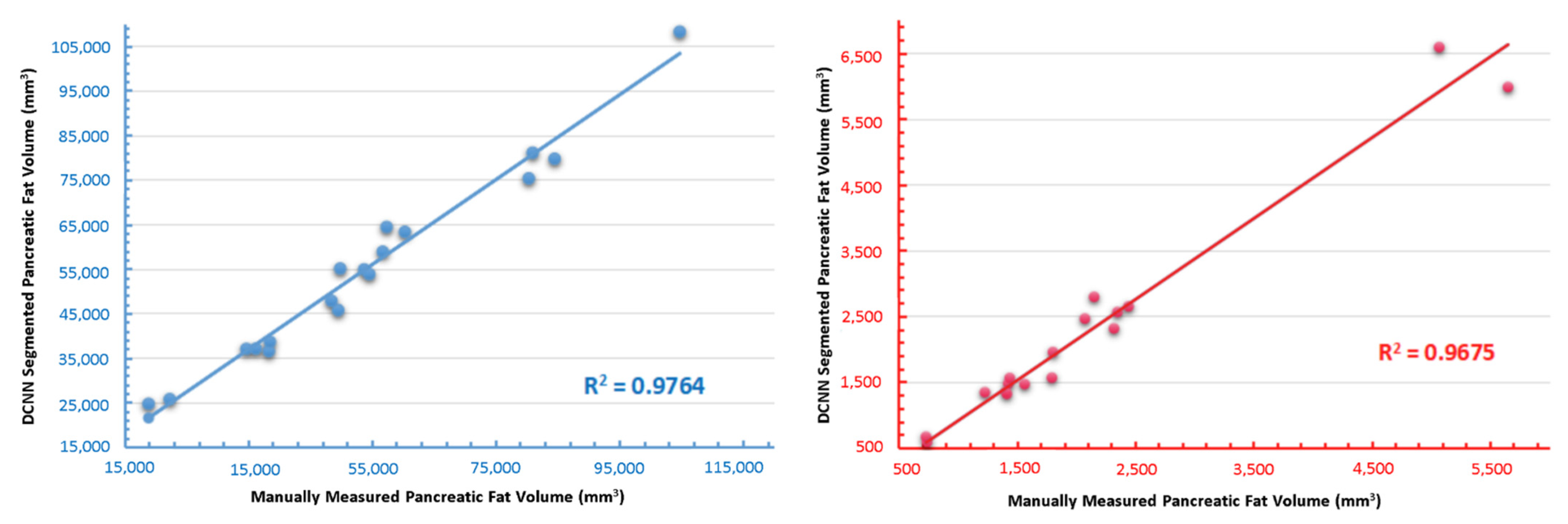
3.5. Independent Validation of the Novel DCNN
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nazir, M.A.; AlGhamdi, L.; AlKadi, M.; AlBeajan, N.; AlRashoudi, L.; AlHussan, M. The burden of Diabetes, Its Oral Complications and Their Prevention and Management. Open Access Maced. J. Med Sci. 2018, 6, 1545–1553. [Google Scholar] [CrossRef]
- DeSouza, S.V.; Singh, R.; Yoon, H.D.; Murphy, R.; Plank, L.D.; Petrov, M.S. Pancreas volume in health and disease: A systematic review and meta-analysis. Expert Rev. Gastroenterol. Hepatol. 2018, 12, 757–766. [Google Scholar] [CrossRef]
- Martin, S.; Sorokin, E.P.; Thomas, E.L.; Sattar, N.; Cule, M.; Bell, J.D.; Yaghoostkar, H. Estimating the Effect of Liver and Pancreas Volume and Fat Content on Risk of Diabetes: A Mendelian Randomization Study. Diabetes Care 2021, 45, 460–468. [Google Scholar] [CrossRef]
- Goda, K.; Sasaki, E.; Nagata, K.; Fukai, M.; Ohsawa, N.; Hahafusa, T. Pancreatic volume in type 1 und type 2 diabetes mellitus. Geol. Rundsch. 2001, 38, 145–149. [Google Scholar] [CrossRef]
- Burute, N.; Nisenbaum, R.; Jenkins, D.J.; Mirrahimi, A.; Anthwal, S.; Colak, E.; Kirpalani, A. Pancreas volume measurement in patients with Type 2 diabetes using magnetic resonance imaging-based planimetry. Pancreatology 2014, 14, 268–274. [Google Scholar] [CrossRef]
- DeSouza, S.V.; Yoon, H.D.; Singh, R.G.; Petrov, M.S. Quantitative determination of pancreas size using anatomical landmarks and its clinical relevance: A systematic literature review. Clin. Anat. 2018, 31, 913–926. [Google Scholar] [CrossRef]
- Macauley, M.; Percival, K.; Thelwall, P.E.; Hollingsworth, K.G.; Taylor, R. Altered Volume, Morphology and Composition of the Pancreas in Type 2 Diabetes. PLoS ONE 2015, 10, e0126825. [Google Scholar] [CrossRef]
- Garcia, T.S.; Rech, T.H.; Leitao, C. Pancreatic size and fat content in diabetes: A systematic review and meta-analysis of imaging studies. PLoS ONE 2017, 12, e0180911. [Google Scholar] [CrossRef]
- Garcia, B.C.; Wu, Y.; Nordbeck, E.B.; Musovic, S.; Olofsson, C.S.; Rorsman, P.; Asterholm, I.W. Pancreatic Adipose Tissue in Diet-Induced Type 2 Diabetes. Diabetes 2018, 67, 2431-PUB. [Google Scholar] [CrossRef]
- Singh, R.; Cervantes, A.; Kim, J.U.; Nguyen, N.N.; DeSouza, S.V.; Dokpuang, D.; Lu, J.; Petrov, M.S. Intrapancreatic fat deposition and visceral fat volume are associated with the presence of diabetes after acute pancreatitis. Am. J. Physiol. Liver Physiol. 2019, 316, G806–G815. [Google Scholar] [CrossRef]
- Singh, R.G.; Pendharkar, S.A.; Gillies, N.A.; Miranda-Soberanis, V.; Plank, L.D.; Petrov, M.S. Associations between circulating levels of adipocytokines and abdominal adiposity in patients after acute pancreatitis. Clin. Exp. Med. 2017, 17, 477–487. [Google Scholar] [CrossRef]
- Singh, R.G.; Yoon, H.D.; Wu, L.M.; Lu, J.; Plank, L.D.; Petrov, M.S. Ectopic fat accumulation in the pancreas and its clinical relevance: A systematic review, meta-analysis, and meta-regression. Metabolism 2017, 69, 1–13. [Google Scholar] [CrossRef]
- Singh, R.G.; Yoon, H.D.; Poppitt, S.D.; Plank, L.D.; Petrov, M.S. Ectopic fat accumulation in the pancreas and its biomarkers: A systematic review and meta-analysis. Diabetes/Metabolism Res. Rev. 2017, 33, e2918. [Google Scholar] [CrossRef]
- Al-Mrabeh, A.; Hollingsworth, K.G.; Steven, S.; Taylor, R. Morphology of the pancreas in type 2 diabetes: Effect of weight loss with or without normalisation of insulin secretory capacity. Diabetologia 2016, 59, 1753–1759. [Google Scholar] [CrossRef]
- Patel, N.S.; Peterson, M.R.; Brenner, D.; Heba, E.; Sirlin, C.; Loomba, R. Association between novel MRI-estimated pancreatic fat and liver histology-determined steatosis and fibrosis in non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2013, 37, 630–639. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, H.; Cho, J.Y.; Lim, S.; Cha, K.; Lee, K.H.; Kim, Y.H.; Kim, J.H.; Yoon, Y.-S.; Han, H.-S.; et al. Quantitative Assessment of Pancreatic Fat by Using Unenhanced CT: Pathologic Correlation and Clinical Implications. Radiology 2014, 271, 104–112. [Google Scholar] [CrossRef]
- Stine, J.G.; Munaganuru, N.; Barnard, A.; Wang, J.L.; Kaulback, K.; Argo, C.K.; Singh, S.; Fowler, K.J.; Sirlin, C.B.; Loomba, R. Change in MRI-PDFF and Histologic Response in Patients With Nonalcoholic Steatohepatitis: A Systematic Review and Meta-Analysis. Clin. Gastroenterol. Hepatol. 2021, 19, 2274–2283.e2275. [Google Scholar] [CrossRef]
- Bs, A.M.; Railkar, R.; Yokoo, T.; Levin, Y.; Clark, L.; Fox-Bosetti, S.; Middleton, M.S.; Riek, J.; Kauh, E.; Dardzinski, B.J.; et al. Reproducibility of hepatic fat fraction measurement by magnetic resonance imaging. J. Magn. Reson. Imaging 2013, 37, 1359–1370. [Google Scholar]
- Reeder, S.B.; Hu, H.H.; Sirlin, C. Proton density fat-fraction: A standardized mr-based biomarker of tissue fat concentration. J. Magn. Reson. Imaging 2012, 36, 1011. [Google Scholar] [CrossRef]
- Szczepaniak, E.W.; Malliaras, K.; Nelson, M.; Szczepaniak, L.S. Measurement of Pancreatic Volume by Abdominal MRI: A Validation Study. PLOS ONE 2013, 8, e55991. [Google Scholar] [CrossRef]
- Lingvay, I.; Esser, V.; Legendre, J.L.; Price, A.L.; Wertz, K.M.; Adams-Huet, B.; Szczepaniak, L.S. Non-invasive quantification of pancreatic fat in humans. J. Clin. Endocrinol. Metab. 2009, 94, 4070–4076. [Google Scholar] [CrossRef]
- Hu, H.H.; Kim, H.-W.; Nayak, K.; Goran, M.I. Comparison of Fat-Water MRI and Single-voxel MRS in the Assessment of Hepatic and Pancreatic Fat Fractions in Humans. Obesity 2010, 18, 841–847. [Google Scholar] [CrossRef]
- Pinnick, K.E.; Collins, S.C.; Londos, C.; Gauguier, D.; Clark, A.; Fielding, B.A. Pancreatic Ectopic Fat Is Characterized by Adipocyte Infiltration and Altered Lipid Composition. Obesity 2008, 16, 522–530. [Google Scholar] [CrossRef]
- Al-Mrabeh, A.; Hollingsworth, K.G.; Steven, S.; Tiniakos, D.; Taylor, R. Quantification of intrapancreatic fat in type 2 diabetes by MRI. PLoS ONE 2017, 12, e0174660. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Krähenbühl, P.; Koltun, V. Efficient inference in fully connected crfs with gaussian edge potentials. Adv. Neural Inf. Process. Syst. 2011, 24, 109–117. [Google Scholar]
- Roth, H.R.; Lu, L.; Lay, N.; Harrison, A.P.; Farag, A.; Sohn, A.; Summers, R.M. Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med Image Anal. 2018, 45, 94–107. [Google Scholar] [CrossRef]
- Zhou, Y.; Xie, L.; Shen, W.; Wang, Y.; Fishman, E.K.; Yuille, A.L. A fixed-point model for pancreas segmentation in abdominal CT scans. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017. [Google Scholar]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv 2018, arXiv:180206955. [Google Scholar]
- Cai, J.; Lu, L.; Zhang, Z.; Xing, F.; Yang, L.; Yin, Q. Pancreas segmentation in MRI using graph-based decision fusion on convolutional neural networks. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, MA, USA, 2016; pp. 442–450). [Google Scholar]
- Wolz, R.; Chu, C.; Misawa, K.; Fujiwara, M.; Mori, K.; Rueckert, D. Automated Abdominal Multi-Organ Segmentation With Subject-Specific Atlas Generation. IEEE Trans. Med. Imaging 2013, 32, 1723–1730. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Z.; Zhou, Y.; Xia, Y.; Shen, W.; Fishman, E.K.; Yuille, A.L. Volumetric Medical Image Segmentation: A 3D Deep Coarse-to-Fine Framework and Its Adversarial Examples. In Deep Learning and Convolutional Neural Networks for Medical Imaging and Clinical Informatics; Springer: Berlin/Heidelberg, Germany, 2019; pp. 69–91. [Google Scholar]
- Zeng, G.; Zheng, G. Holistic decomposition convolution for effective semantic segmentation of medical volume images. Med. Image Anal. 2019, 57, 149–164. [Google Scholar] [CrossRef]
- Yu, Q.; Xie, L.; Wang, Y.; Zhou, Y.; Fishman, E.K.; Yuille, A.L. Recurrent Saliency Transformation Network: Incorporating Multi-stage Visual Cues for Small Organ Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Zhu, Z.; Xia, Y.; Shen, W.; Fishman, E.; Yuille, A. A 3D coarse-to-fine framework for volumetric medical image segmentation. In Proceedings of the 2018 International Conference on 3D Vision (3DV), Verona, Italy, 5–8 September 2018. [Google Scholar]
- Yang, J.Z.; Dokpuang, D.; Nemati, R.; He, K.H.; Zheng, A.B.; Petrov, M.S.; Lu, J. Evaluation of Ethnic Variations in Visceral, Subcutaneous, Intra-Pancreatic, and Intra-Hepatic Fat Depositions by Magnetic Resonance Imaging among New Zealanders. Biomedicines 2020, 8, 174. [Google Scholar] [CrossRef]
- Russell, B.C.; Torralba, A.; Murphy, K.P.; Freeman, W.T. LabelMe: A Database and Web-Based Tool for Image Annotation. Int. J. Comput. Vis. 2008, 77, 157–173. [Google Scholar] [CrossRef]
- Yoon, A.P.; Lee, Y.L.; Kane, R.L.; Kuo, C.F.; Lin, C.; Chung, K.C. Development and validation of a deep learning model using convolutional neural networks to identify scaphoid fractures in radiographs. JAMA Netw. Open 2021, 4(5), e216096. [Google Scholar] [CrossRef]
- Chen, J.; Li, Z.; Huang, B. Linear Spectral Clustering Superpixel. IEEE Trans. Image Process. 2017, 26, 3317–3330. [Google Scholar] [CrossRef]
- Qiao, N.; Sun, C.; Sun, J.; Song, C. Superpixel Combining Region Merging for Pancreas Segmentation. In Proceedings of the 2021 36th Youth Academic Annual Conference of Chinese Association of Automation (YAC), Nanchang, China, 28–30 May 2021. [Google Scholar]
- Schick, A.; Fischer, M.; Stiefelhagen, R. An evaluation of the compactness of superpixels. Pattern Recognit. Lett. 2014, 43, 71–80. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:14091556. [Google Scholar]
- Bernard, C.P.; Liney, G.P.; Manton, D.J.; Turnbull, L.W.; Langton, C.M. Comparison of fat quantification methods: A phantom study at 3.0 T. J. Magn. Reson. Imaging Off. J. Int. Soc. Magn. Reson. Med. 2008, 27, 192–197. [Google Scholar] [CrossRef]
- Sahin, B.; Emirzeoglu, M.; Uzun, A.; Incesu, L.; Bek, Y.; Bilgic, S.; Kaplan, S. Unbiased estimation of the liver volume by the Cavalieri principle using magnetic resonance images. Eur. J. Radiol. 2003, 47, 164–170. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Rueckert, D. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:180403999. [Google Scholar]
- Zhang, D.; Zhang, J.; Zhang, Q.; Han, J.; Zhang, S.; Han, J. Automatic pancreas segmentation based on lightweight DCNN modules and spatial prior propagation. Pattern Recognit. 2021, 114, 107762. [Google Scholar] [CrossRef]
- Ma, J.; Lin, F.; Wesarg, S.; Erdt, M. A Novel Bayesian Model Incorporating Deep Neural Network and Statistical Shape Model for Pancreas Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018. [Google Scholar]
- Szczepaniak, L.S.; Nurenberg, P.; Leonard, D.; Browning, J.D.; Reingold, J.S.; Grundy, S.; Hobbs, H.H.; Dobbins, R.L. Magnetic resonance spectroscopy to measure hepatic triglyceride content: Prevalence of hepatic steatosis in the general population. Am. J. Physiol. Metab. 2005, 288, E462–E468. [Google Scholar] [CrossRef]
- Szczepaniak, L.S.; Victor, R.G.; Mathur, R.; Nelson, M.D.; Szczepaniak, E.W.; Tyer, N.; Lingvay, I. Pancreatic steatosis and its relationship to β-cell dysfunction in humans: Racial and ethnic variations. Diabetes Care 2012, 35, 2377–2383. [Google Scholar] [CrossRef] [PubMed]
- Lê, K.-A.; Ventura, E.E.; Fisher, J.Q.; Davis, J.N.; Weigensberg, M.J.; Punyanitya, M.; Hu, H.H.; Nayak, K.S.; Goran, M.I. Ethnic Differences in Pancreatic Fat Accumulation and Its Relationship With Other Fat Depots and Inflammatory Markers. Diabetes Care 2011, 34, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Virostko, J. Quantitative Magnetic Resonance Imaging of the Pancreas of Individuals With Diabetes. Front. Endocrinol. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.M.; Hilmes, M.A.; Archer, B.; Dulaney, A.; Du, L.; Kang, H.; Russell, W.E.; Powers, A.C.; Moore, D.J.; Virostko, J. Repeatability and Reproducibility of Pancreas Volume Measurements Using MRI. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Heller, N.; Isensee, F.; Maier-Hein, K.H.; Hou, X.; Xie, C.; Li, F.; Nan, Y.; Mu, G.; Lin, Z.; Han, M.; et al. The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 challenge. Med Image Anal. 2021, 67, 101821. [Google Scholar] [CrossRef]


| Characteristics | DCNN Training and Testing | Independent Validation | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | NZ. European | Māori/Pl | Asian/Others | Total | NZ. European | Māori/Pl | Asian/Others | |
| No. of participants | 394 | 167 | 106 | 121 | 10 | 4 | 4 | 2 |
| Age (years) | 47.8 ± 1.2 | 52 ± 2.4 | 55.3 ± 1.9 | 35.7 ± 2.3 | 38.8 ± 0.8 | 37.7 ± 1.1 | 32.9 ± 0.6 | 39 ± 1.2 |
| Sex | ||||||||
| Male | 249 | 98 | 71 | 81 | 4 | 2 | 1 | 1 |
| Female | 145 | 69 | 35 | 40 | 6 | 2 | 3 | 1 |
| BMI. | 28.8 ± 0.7 | 28.1 ± 0.5 | 33.7 ± 1.3 | 25.4 ± 1.0 | 35.6 ± 1.6 | 35.5 ± 1.2 | 38.8 ± 1.8 | 29.3 ± 0.6 |
| Pancreatic fat fraction% (MRS) | 31.2 ± 2.0 | 29.7 ± 1.5 | 38.4 ± 2.8 | 26.8 ± 2.2 | 46.2 ± 1.9 | 39.3 ± 1.4 | 46.8 ± 1.7 | 52.5 ± 2.6 |
| Pancreatic fat fraction% (Manual) | 8.4 ± 0.6 | 7.6 ± 0.4 | 9.9 ± 0.8 | 8.4 ± 0.6 | 8.5 ± 0.5 | 8.8 ± 0.3 | 8.6 ± 0.6 | 7.6 ± 0.5 |
| Method | Mean Dice Score/% |
|---|---|
| Attention U-net [46] | 81.5 |
| 3D FCN [28] | 76.8 |
| RSTN [35] | 84.5 |
| Graph-based decision fusion [31] | 76.1 |
| Lightweight DCNN modules [47] | 85.6 |
| Spatial aggregation [29] | 81.3 |
| Fixed-Point [40] | 83.2 |
| Bayesian Model [48] | 85.3 |
| Our DCNN | 91.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.Z.; Zhao, J.; Nemati, R.; Yin, X.; He, K.H.; Plank, L.; Murphy, R.; Lu, J. An Adapted Deep Convolutional Neural Network for Automatic Measurement of Pancreatic Fat and Pancreatic Volume in Clinical Multi-Protocol Magnetic Resonance Images: A Retrospective Study with Multi-Ethnic External Validation. Biomedicines 2022, 10, 2991. https://doi.org/10.3390/biomedicines10112991
Yang JZ, Zhao J, Nemati R, Yin X, He KH, Plank L, Murphy R, Lu J. An Adapted Deep Convolutional Neural Network for Automatic Measurement of Pancreatic Fat and Pancreatic Volume in Clinical Multi-Protocol Magnetic Resonance Images: A Retrospective Study with Multi-Ethnic External Validation. Biomedicines. 2022; 10(11):2991. https://doi.org/10.3390/biomedicines10112991
Chicago/Turabian StyleYang, John Zhiyong, Jichao Zhao, Reza Nemati, Xavier Yin, Kevin Haokun He, Lindsay Plank, Rinki Murphy, and Jun Lu. 2022. "An Adapted Deep Convolutional Neural Network for Automatic Measurement of Pancreatic Fat and Pancreatic Volume in Clinical Multi-Protocol Magnetic Resonance Images: A Retrospective Study with Multi-Ethnic External Validation" Biomedicines 10, no. 11: 2991. https://doi.org/10.3390/biomedicines10112991
APA StyleYang, J. Z., Zhao, J., Nemati, R., Yin, X., He, K. H., Plank, L., Murphy, R., & Lu, J. (2022). An Adapted Deep Convolutional Neural Network for Automatic Measurement of Pancreatic Fat and Pancreatic Volume in Clinical Multi-Protocol Magnetic Resonance Images: A Retrospective Study with Multi-Ethnic External Validation. Biomedicines, 10(11), 2991. https://doi.org/10.3390/biomedicines10112991