1. Introduction
Cutaneous melanoma is a highly aggressive malignancy with growing incidence and limited therapy strategies [
1,
2]. Cutaneous melanomas are classified into four subtypes: (i) mutation of the B-Raf proto-oncogene serine/threonine kinase (BRAFmut), (ii) mutation of the NRAS proto-oncogene GTPase (NRASmut), (iii) mutation of neurofibromin 1 (NF1mut), and (iv) none of the three, triple BRAF/NRAS/NF1 wild-type (WT) [
3,
4,
5]. The development of targeted therapies and immunotherapies has led to significant improvement in overall survival, particularly in non-NRAS-mutated melanoma (BRAF). Unfortunately, effective targeted therapies are lacking in patients with NRASmut melanomas due to emerging resistance and high aggressiveness, generally resulting in a poorer prognosis [
6]. As a result, to date, there is an unmet clinical need for sustained effective therapeutic strategies for NRASmut melanoma patients. Considerable efforts have been made to inhibit the MAPK pathway alone or in combination with other drugs. New classes of inhibitors such as the proteolysis targeting chimera (PROTAC), which involves the use of novel mono-, dual-, or even triple-targeting therapies, or mRNA vaccines that can stimulate a targeted immune response against personalized tumor antigens are being adequately investigated [
6,
7]. Nevertheless, intrinsic and acquired resistance is likely to occur, and sometimes toxicities and side effects are massively burdensome for the patient. For the majority of advanced melanoma patients, new classes of pharmacological agents need to be considered to achieve a long and well-tolerated PFS (progression-free survival). Combined with the highly pigmented melanogenic cell line MUG-Mel2 derived from a cutaneous metastasis of an aggressive NRAS p Q61R-mutated melanoma [
8] and innovative therapeutic options from the field of peptides, we demonstrate a new potential therapy [
9,
10,
11,
12,
13,
14].
Cationic antitumor peptides derived from the human host defense peptide Lactoferricin [
15] have been shown to selectively kill cancer cells of different types including malignant melanoma and their metastases [
9,
11,
12,
13,
14] in 2D and 3D in vitro and in vivo [
16] by targeting the negatively charged lipid phosphatidylserine (PS), specifically exposed by the outer leaflet of the plasma membrane of cancer cells [
17,
18]. PS exposure was not only shown for different cancer types such as melanoma, glioblastoma, prostate and renal cancer compared to absence in non-neoplastic control cells, but it was further demonstrated that PS exposure was not attributed to ongoing apoptosis [
17]. One of these peptides, RDP22 (R-DIM-P-LF11-322, PCT/EP2014/050330; US 14/760,445; EP 14700349.5), exhibits selective activity against the cancer cell marker PS in model systems and cytotoxicity against melanoma without significant effect on non-neoplastic cells as melanocytes or fibroblasts at antitumor-active concentrations [
9,
13,
14].
For enhanced output of in vitro experiments, before in vivo studies, a number of 3D studies were applied such as tumor spheroids and second organotypic melanoma models. Accrued advantages of the latter such as the recently published mOS-REp [
19] are traced back to the fact that these 3D models bridge the gap between the simplicity of 2D cell culture or spheroids lacking a microenvironment and the complexity of full-thickness skin equivalents and animal models [
20,
21]. The models in which the tumors mature within a physiologically differentiated epidermis enable the investigation of melanomagenesis, melanoma treatment and cellular or molecular crosstalk between the tumor and its surrounding skin cells.
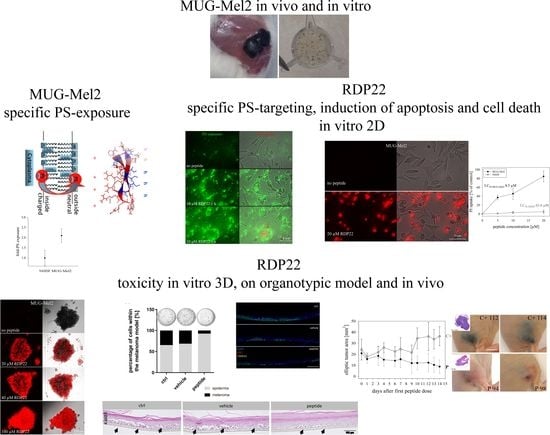
The study investigates the sensitivity of the aggressive NRAS mutated melanoma MUG-Mel2 for treatment with antitumor peptide RDP22. Application of elevated model systems up to 3D tumor spheroids and organotypic melanoma models can further help to significantly reduce the number of animal studies within the preclinical phases of the development of anti-melanoma therapies.
2. Materials and Methods
2.1. Materials
The peptide R-DIM-P-LF11-322 (RDP22) (PFWRIRIRRPRRIRIRWFP-NH2, M = 2677.26 g/mol) and the fluorescently labeled peptide ((5–6)-FAM-) RDP22 ((5–6)-FAM-) PFWRIRIRR-P-RRIRIRWFP-NH2, M = 3035.7 g/mol) were purchased from PolyPeptide (San Diego, CA, USA). The purity was >96% as determined by RP-HPLC. The peptides were dissolved in acetic acid (0.1%, v/v) at concentrations of 3 mg/mL for model and in vitro experiments and in Dulbecco’s Phosphate Buffered Saline, DPBS (no calcium, no magnesium; Gibco®, Thermo Fisher Scientific, Waltham, MA, USA) buffer at concentrations of 2 mg/mL before in vivo experiments. The peptide concentration was determined by measurement of UV-absorbance of tryptophan at a wavelength of 280 nm using a NanoDrop photometer (ND 1000, Peqlab, VWR International, Inc. Erlangen, Germany). All peptide stocks were stored at 4 °C till use. The lipid 1,2-dipalmitoyl-sn-glycero-3-phospho-L-serine (sodium salt) (DPPS) was purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA).
2.2. Model Studies
2.2.1. Liposomal Preparations
Lipid films of 5 mg DPPS (microscopy) were prepared by drying respective amounts of lipid under a stream of nitrogen and storage in a vacuum overnight. The dry lipid film was then dispersed in respective volumes of phosphate-buffered saline (PBS, 20 mM NaPi, 130 mM NaCl, pH 7.4) and hydrated at temperatures well above the gel-to-fluid phase transition of the respective phospholipid under intermittent vigorous vortex-mixing, as described previously [
14]. The lipid concentration was 0.25–0.50 mM for microscopic inspection. DPPS hydration was carried out at 65 °C for two hours by vortexing every 15 min. The fully hydrated samples were stored at room temperature until measurement.
2.2.2. Microscopy Studies on Liposomes
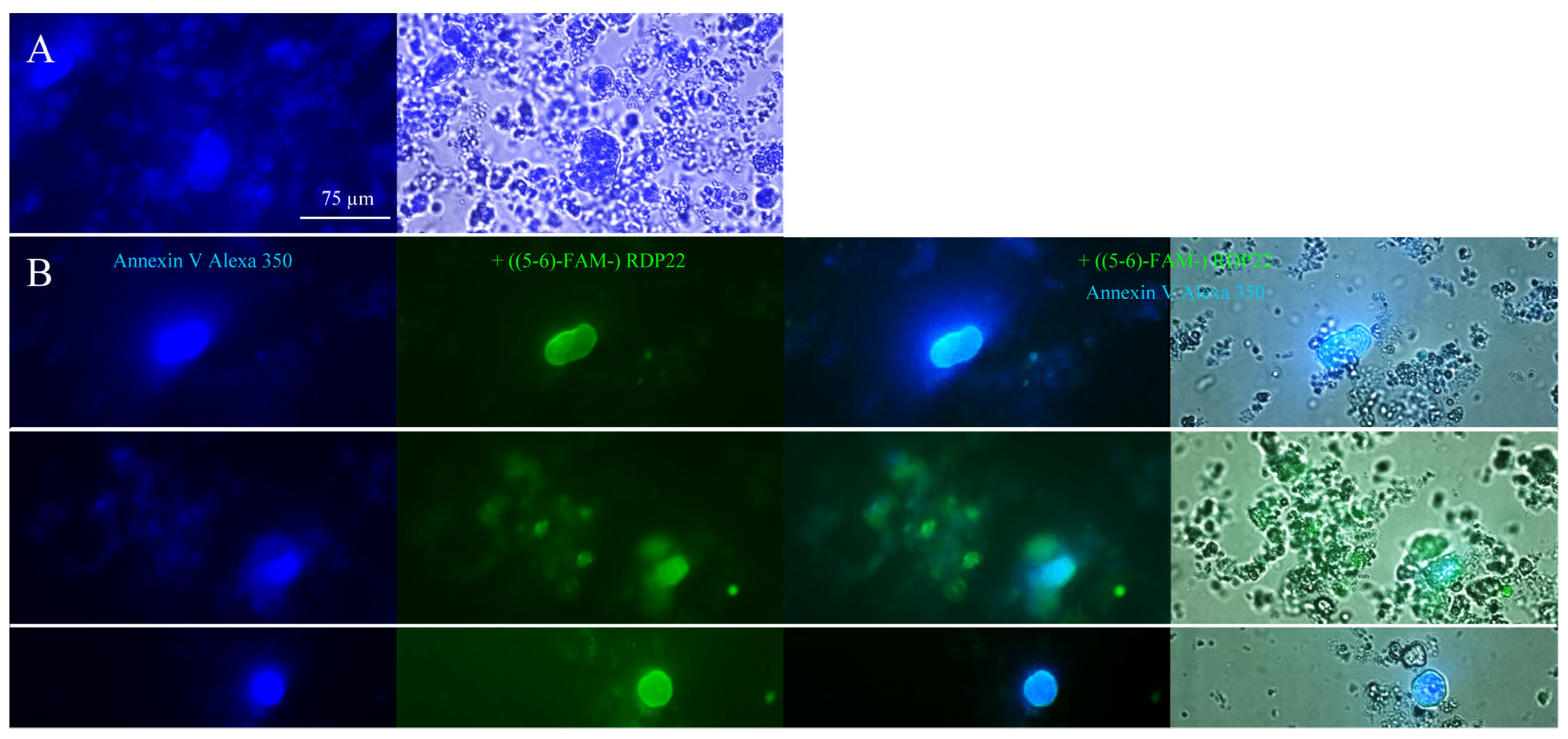
For microscopic studies, 250–500 µg liposomes of DPPS were centrifuged at 10,000× g for 10 min at room temperature. The liposomal pellet was resuspended in Annexin binding buffer (ABB) and stained with Annexin V Alexa Fluor 350 conjugate according to the manufacturer’s protocol (Molecular Probes Inc., Eugene, OR, USA). PS exposure by Annexin V binding was visualized with a Leica DMI6000 B with IMC using a Leica DFC360 FX camera and AF 6000 software (Leica Microsystems, Vienna, Austria) (excitation wavelength 346 nm; emission wavelength 442 nm). For investigation of potential PS-peptide co-localization prior to PS-staining, incubation of 0.25–0.50 mM DPPS liposomes with 2 μM fluorescently labeled RDP22, ((5-6)-FAM-) RDP22 (excitation wavelength λex = 495 nm; emission wavelength λem = 519 nm) for 10 min was performed.
2.3. Cell Culture
Melanoma cells MUG-Mel2 (p33) were cultured in RPMI 1640 medium with GlutaMAX™ (Gibco®, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (fetal bovine serum; Gibco®, Thermo Fisher Scientific, Waltham, MA, USA). Normal human dermal fibroblasts NHDF (PromoCell Inc., Heidelberg, Germany) were used as non-neoplastic controls. NHDF were cultured in Fibroblast Growth Medium 2 (PromoCell Inc., Heidelberg, Germany). At 90% confluence cells were passaged with accutase (Gibco®, Thermo Fisher Scientific, Waltham, MA, USA). All cells and spheroids were kept in 5% CO2 at 37 °C and periodically checked for mycoplasma. Cell line authentication was performed by using the Power Plex® 16 HS System (Promega, Madison, WI, USA).
2.4. PS Exposure
PS exposure of MUG-Mel2 was determined using the Annexin V Alexa 488 apoptosis detection kit (Molecular Probes Inc., Eugene, OR, USA). Briefly, 105 cells were seeded into black 96-well plates and incubated O/N at 5% CO2 and 37 °C. Subsequently, cells were washed once with Annexin V binding buffer and stained with 5 µL Annexin V-Alexa 488 and 2 µL PI (50 µg/µL) for 5 min at room temperature in the dark. Cells were washed again and fluorescence (Annexin V Alexa Fluor 488, λex = 488 nm and λem = 530 nm; PI, λex = 536 nm and λem = 617 nm) was recorded immediately using the GloMax® Discover Microplate Reader (Promega, Madison, WI, USA).
2.5. Two-Dimensional In Vitro Experiments
2.5.1. Analysis of Cell Death—Fluorescence Spectroscopy—PI Uptake
Cells were collected, resuspended in respective media (see cell culture) and diluted to a concentration of 10
6 cells/mL. Aliquots of 10
5 cells in suspension were incubated in presence of respective concentrations of peptide (0 µM up to 20 µM) for up to 8 h in the presence of propidium iodide PI (2 µL/10
5 cells of 50 µg/mL, Molecular Probes Inc., Eugene, OR, USA) at room temperature in black 96-well plates. PI-uptake was measured after 0, 1, 2, 4 and 8 h using the GloMax
® Discover Microplate Reader (Promega, Madison, WI, USA). Cytotoxicity was calculated from the percentage of PI-positive cells in media alone (P
0) and in the presence of peptide (P
X) (Equation (1)). Triton-X-100 (10%) was used to determine 100% of PI-positive cells (P
100).
Excitation and emission wavelengths were 536 nm and 617 nm, respectively. Each experiment was repeated at least three times.
2.5.2. Visualization of Cell Death 2D—Fluorescence Microscopy—PI Uptake
Cells (1–5 × 104) were seeded on Ibidi µ-Slide 8 wells (ibidi GmbH, Martinsried, Germany) and grown in 300 µL media for 2–3 days to a confluent layer. The peptide was added at different concentrations and incubated for 8 h at 37 °C and 5% CO2. Propidium iodide PI (2 µL/well of 50 µg/mL, Molecular Probes Inc., Eugene, OR, USA) was added to the wells and incubated for another 5 min in the dark. Experiments were performed with a Leica DMI6000 B with IMC using a Leica DFC360 FX camera and AF 6000 software (Leica Microsystems, Vienna, Austria). Excitation and emission wavelengths were 536 nm and 617 nm, respectively. PI can only enter cells and intercalate with the DNA upon potential peptide-induced membrane damage. For the visualization of the interaction of the peptide with PS and induced cell death, MUG-Mel2 cells seeded and grown for 2–3 days on Ibidi µ-Slides were incubated with 10 μM ((5-6)-FAM-) RDP22 for up to 6 h. Cells were then stained and examined with Annexin V Alexa Fluor 350 conjugate and PI as described in 2.2.2. For the visualization of the interaction of RDP22 with non-malignant control cells, NHDF were analyzed on Ibidi µ-Slides upon incubation with 10 μM ((5-6)-FAM-) RDP22 for up to 4 h and stained with Annexin V-Alexa 488 and PI as described in 2.4.
2.5.3. Apoptosis/Necrosis Assay
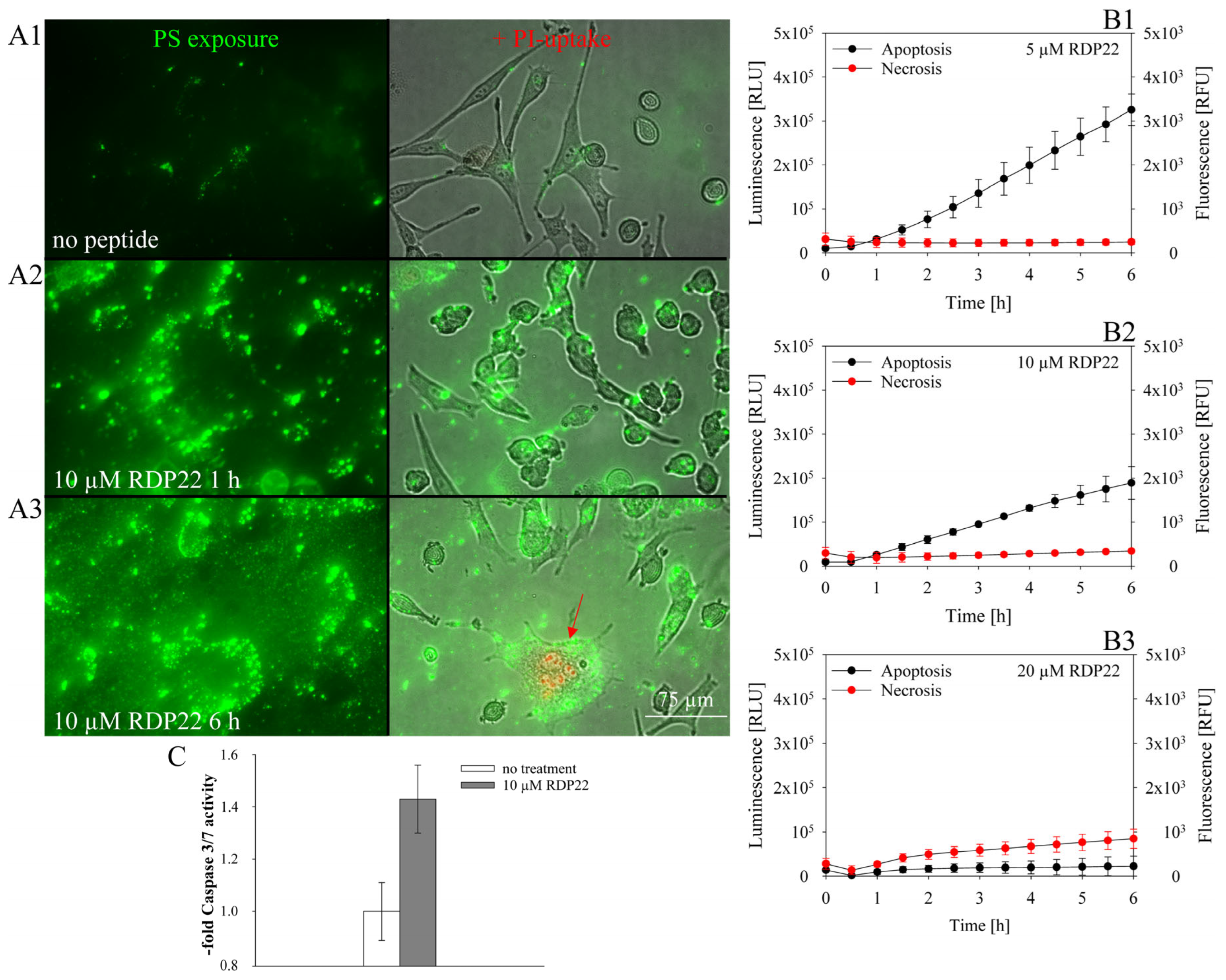
To assess whether the peptide induces apoptosis or necrosis, the RealTime-Glo™ Annexin V Apoptosis (luminescence) and Necrosis (fluorescence) Assay from Promega (Promega, Madison, WI, USA) were used. Briefly, cells were seeded at 105 cells/100 µL in a white 96-multiwell plate with clear bottom and incubated overnight at 5% CO2 and 37 °C. The reagent stock solution (1000×) was diluted in medium according to the manufacturer’s protocol. Before the measurement, 100 µL of the reagent solution was added to the cell suspension. For the fluorophore, excitation and emission wavelengths were 490 nm and 525 nm, respectively. Before peptide addition, the signals at time zero were measured with the GloMax® Discover Microplate Reader (Promega, Madison, WI, USA). Thereupon, the cells were treated with peptides at different concentrations (5–20 µM). Apoptosis and necrosis signals were measured every 30 min for 6 h.
2.5.4. Caspase-3/7 Assay
To further assess whether cells undergo apoptosis the Caspase-Glo® 3/7 Assay (Promega, Madison, WI, USA) was used; 2 × 105 cells/mL were seeded in a white 96-multiwell plate with clear bottom and grown for 48 h at 5% CO2 and 37 °C. Peptide at 0 and 10 µM was added for incubation over 4 h. Caspase solution was added with a 1:1 ratio upon 30 min at RT. Luminescence was measured with a GloMax® Discover Microplate Reader (Promega, Madison, WI, USA). Mean values of caspase 3/7 activity were analyzed as a multiple of non-peptide treated cells. Three repeats were performed.
2.6. Three-Dimensional In Vitro Experiments
2.6.1. Preparation of Tumor Spheroids—Hanging Drop Method
For fluorescence microscopy experiments spheroids were generated using the hanging drop method. Briefly, small volumes (25 µL) of cell suspension (2 × 105 cells/mL) were placed onto the bottom of a µ-Dish (50 mm, low, uncoated) from ibidi GmbH (Martinsried, Germany) and the dish was inverted afterward forming “hanging drops”; 500 µL of Dulbecco’s Phosphate Buffered Saline, DPBS (no calcium, no magnesium; Gibco®, Thermo Fisher Scientific, Waltham, MA, USA) were placed into the lid of the µ-Dish to create a hydrated atmosphere.
2.6.2. Visualization of Cell Death 3D—Fluorescence Microscopy—PI Uptake
Cell death induced by antitumor peptides on tumor cell spheroids was also analyzed by detection of PI-uptake by microscopy. After the generation of hanging drops for 72 h as described above, the peptide was carefully added at different concentrations (0–100 µM) to the hanging drops. Spheroids were then incubated in presence of peptide at 37 °C and 5% CO2 for 24 h. After the addition of 2.5 µL PI (10 µg/mL), spheroids were incubated for another 10 min before microscopic images were taken. Experiments were performed with a Leica DMI6000 B with IMC using a Leica DFC360 FX camera and AF 6000 software (Leica Microsystems, Vienna, Austria). Excitation and emission wavelengths were 536 nm and 617 nm, respectively.
2.6.3. Generation of Tumor Spheroids—Ultra-Low Attachment Plates
The MUG-Mel2 cells were seeded at 104 cells per 100 µL in the respective medium into black Corning® 96-well Spheroid Microplates with a round clear bottom (Ultra-Low Attachment surface) from Sigma Aldrich (Deisenhofen, Germany) and grown for 5 days at 37 °C and 5% CO2. About 60% of the medium was partially exchanged every second day.
2.6.4. Three-Dimensional Viability Assay
The CellTiter-Glo
® 3D-Cell Viability Assay from Promega (Promega, Madison, WI, USA) was used to determine the cell viability in 3D MUG-Mel2 spheroids prepared with Ultra-Low Attachment plates. The 3D assay reagent measures ATP as an indicator of viability and generates a detectable luminescent readout. For the assay, the spheroids were cultivated and seeded as described above and treated with peptide for 48 h. In addition, a positive control was treated with 2 µL of Triton-X-100 (10%) representing 0% of cell viability (V
0). The luminescence signal was measured using the GloMax
® Discover Microplate Reader (Promega, Madison, WI, USA) at room temperature for peptide concentrations from 0 to 100 µM. The plate was shaken vigorously for 5 min and afterward, incubated for 25 min. The viability was calculated from the percentage of viable cells without peptide (V
100) and in the presence of peptide (V
X) (Equation (2)).
2.7. Human 3D In Vitro MUG-Mel2 Melanoma Organotypic Models (mOS-REpMUG-Mel2)
2.7.1. General Cell Culture
According to a previously published protocol [
22], primary human epidermal keratinocytes were isolated from foreskin biopsies from juvenile donors between 3 and 6 years.
Keratinocytes were cultured in E1 medium (EpiLife® medium supplemented with 1% human keratinocytes growth supplements (HKGS) and 1% penicillin/streptomycin (all Thermo Fisher Scientific, Waltham, MA, USA)). MUG-Mel2 was cultured in RPMI medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal calf serum (FCS; Bio & Sell, Nürnberg, Germany) and 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA, USA).
2.7.2. Generation of Melanoma Open Source Reconstructed Epidermis Models (mOS-REpMUG-Mel2)
Melanoma organotypic models were generated according to a previously published protocol [
19]. Briefly, 5 × 105 keratinocytes at passage 3 were mixed with a pre-defined ratio of melanoma cells in E2 medium (E1 medium supplemented with 1.44 mM CaCl2) and seeded on a polycarbonate membrane (pore size 0.4 µm) with 12-well inserts (Greiner Bio-One, Frickenhausen, Germany). After 24 h, the models were set to the air-liquid-interphase and the medium was changed to E3 medium (E2 medium containing 10 ng/mL Keratinocyte Growth Factor (Thermo Fisher Scientific, Waltham, MA, USA) and 73 μg/mL ascorbin-2-phosphat). For a culture period of 21 days, the medium was changed three times per week.
2.7.3. Treatment of Melanoma Models with Peptide RDP22
Fifty grams of peptide RDP22 were applied topically on day 1 and day 2, respectively.
2.7.4. Non-Destructive Fully Automated Detection of Tumor Progression and Regression
Tumor progression and regression were non-destructively detected using a fully automated, in-house developed MediTOM device. Images were taken with the transmitted light microscope and analyzed with ImageJ (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, MD, USA).
2.7.5. Determination of Cellular Integrity
Cellular integrity was analyzed photometrically in cell culture supernatants using the Cedex Bio Analyzer (Roche, Königswinter, Germany). Values were determined by measurement of the concentration of the intracellular protein Lactate dehydrogenase (LDH) indicative of the rupture of the cell membrane.
2.7.6. Assessment of Metabolic Activity
To analyze the metabolic activity of the models, a 3-(4,5-dimethyldiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed on day 21. Tissues were incubated in 1 mg/mL MTT (Merck, Darmstadt, Germany) at 37 °C for 3 h. After extraction of formazan salt from the models using 2 mL 2-propanol, a quantitative analysis via absorbance measurement at 570 nm using a spectrophotometer (Infinite 200 M; Tecan, Männedorf, Switzerland) was conducted. Values were normalized to the untreated replicates of each group.
2.7.7. Histological Assessment
For histological assessment, 4 µm histological cross-sections were prepared from tissue samples which were fixed in 4% paraformaldehyde and embedded in paraffin. Hydrated cross-sections were stained with hematoxylin and eosin (HE) for a morphological overview of the models. For immunofluorescence analysis, hydrated cross-sections were blocked using 5% BSA in phosphate-buffered saline (Merck, Darmstadt, Germany) containing 0.2% TritonTM X-100 (Merck, Darmstadt, Germany) for 20 min at room temperature. Subsequently, slides were incubated with primary antibodies against Ki67 (recombinant monoclonal rabbit anti-Ki67 (Cat# ab16667, 1:100, Abcam, Cambridge, United Kingdom)) and HMB45 (monoclonal mouse anti-melanosome/HMB-45 (Cat# M0634, 1:50, Agilent Dako, Santa Clara, CA, USA)) overnight at 4 °C. Slides were washed and incubated with fluorophore-conjugated secondary antibody (polyclonal donkey anti-mouse Alexa Fluor 647 (Cat# A31571) or polyclonal donkey anti-rabbit Alexa Fluor 555 (Cat# A31572), respectively, both 1:400, Thermo Fisher Scientific, Waltham, MA, USA) for 1 h at room temperature. After repeated washing, slides were covered with Fluoromount-DAPI (Thermo Fisher Scientific, Waltham, MA, USA).
2.8. In Vivo Experiments
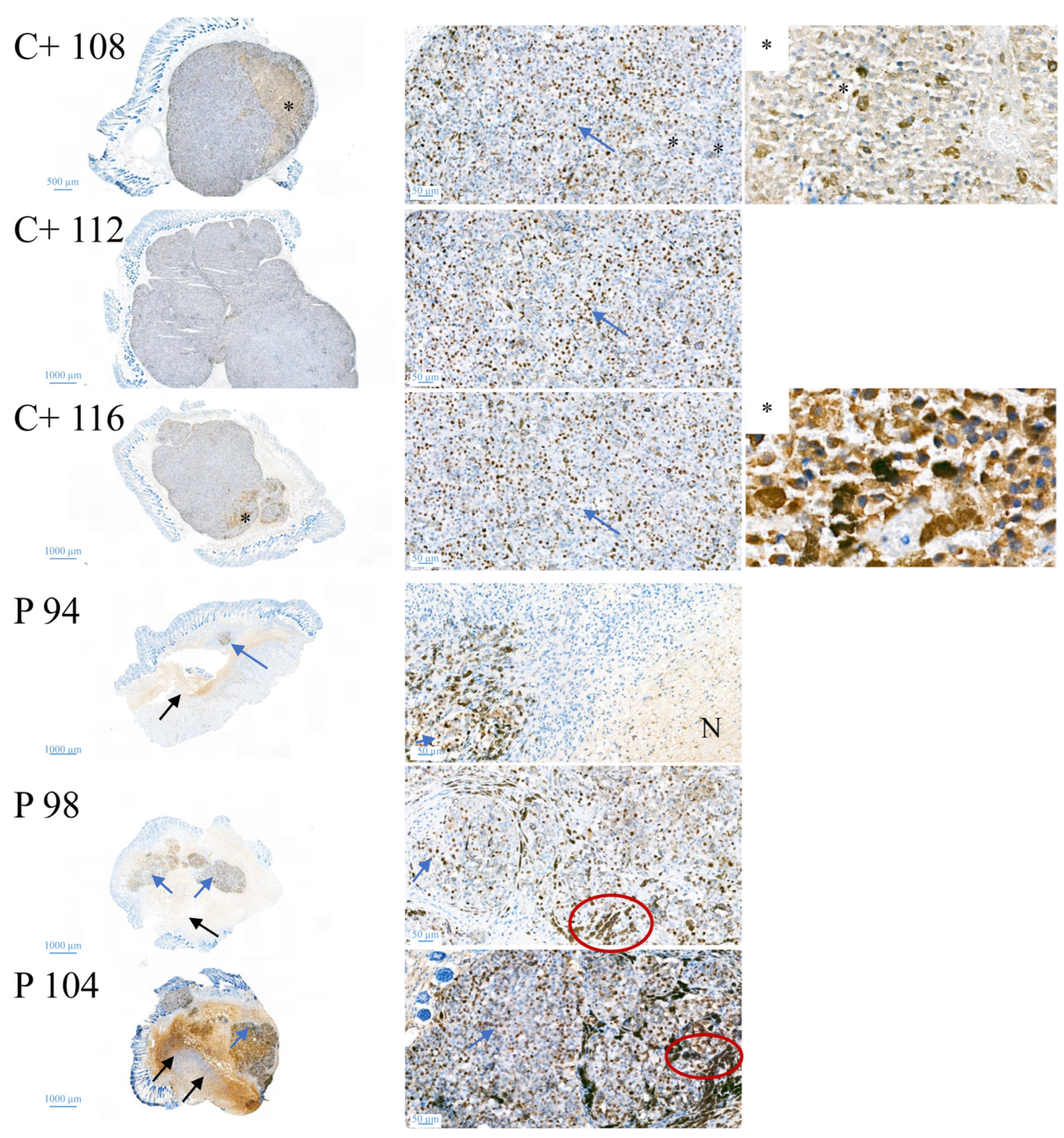
CR ATH HO mice (Crl:NU(NCr)-Foxn1nu, Charles River Laboratories, Kent, UK) were maintained in-house (4–5 weeks of age, weight between 15 and 20 g, 5 in each group). In accordance with a protocol approved by the committee for institutional animal care and use at the Austrian Federal Ministry of Science and Research (BMWFW) (vote 66.010/0046-WF/V/3b/2016), animal work was carefully carried out. All mice were maintained under specific pathogen free (SPF) conditions in individually ventilated cages with ad libitum access to food and water. For cell injection and tumor volume measurement, mice were anesthetized with constant administration of 2% isoflurane in a constant airflow of 2.5 L/min (2.5 × 106 cells in 100 µL PBS). Inoculation of the tumor was monitored daily by using a caliper, starting on day eight post-injection. After sacrifice, a histopathological examination was performed. The mice were dissected and the tumors were extracted. All other organs were checked visually for structural changes. The tissue was then fixed in a 4% paraformaldehyde solution for 24 h and further embedded in paraffin. After embedment in paraffin, four-micron sections were stained with hematoxylin and eosin. Immunohistochemical studies were performed in three cases of control and three peptide treated cases to evaluate the proliferative activity. Therefore, Ki67 staining was performed by using the anti-human Ki67-antigen clone MIB1, with EnVision FLEX (Agilent Dako Omnis, Santa Clara, CA, USA).
2.9. Statistical Analysis
Values are presented as the mean ± SEM (standard error of the mean). Cell culture and microscopy studies were repeated at least three times. For microscopy studies, a set of data being representative of the respective results is shown. Where applicable, analyses of organotypic and in vivo studies were performed by applying the unpaired student’s t-test, where differences with p-values < 0.001 were considered statistically significant.
4. Discussion
Within this study, the effect of a so far well-characterized antitumor peptide, RDP22 (R-DIM-P-LF11-322) on the highly aggressive melanoma metastasis cell line MUG-Mel2, carrying an NRAS mutation, was investigated. RDP22 was originally derived by elongation of the short peptide moiety LF11-322, which has shown strong antibacterial activity, but was lacking antitumor effects [
14]. Since it has been reported by Yang et al. [
23] that a requirement for antitumor peptides is a minimum net charge near +7 and a sequence length of at least 14 amino acids, the retro sequence of the short peptide stretch was added and separated by a proline to induce a turn for enabling the formation of a structurally stabilized peptide. Indeed, whereas LF11-322 (net charge +5; 9 amino acids) exhibited considerable antibacterial activity with a minimal inhibitory concentration for
E. coli of 8–16 µg/mL [
24] but only negligible antitumor activity (LC
50 Sbcl-2 >100 µM), R-DIM-P-LF11-322 (RDP22) (net charge +9; 19 amino acids) showed highly increased (LC
50 Sbcl-2 2.5 µM) and specific antitumor activity [
14]. The idea of the addition of the retro sequence was to design a ‘‘twin’’ peptide with increased amphipathicity and a higher propensity to form a stable secondary structure. The elongated di-retro-peptide RDP22 exhibited highly increased antitumor activity with a cancer-specificity of more than 1000-fold for cell lines of primary melanoma (Sbcl-2, NRAS mutated) and more than 500-fold for melanoma metastases (WM164, BRAF mutated) at a peptide concentration of 20 µM as compared to normal melanocytes [
14]. Additionally, high toxicity in glioblastoma and rhabdomyosarcoma cells was shown [
13]. Propidium iodide-uptake of melanoma cells upon incubation with peptide RDP22 further nicely demonstrated that the peptide operates via a membrane-mediated mechanism since propidium iodide can only be taken up by cells that suffer membrane disintegration [
14]. It was, moreover, demonstrated that the peptide-induced apoptosis in SBcl-2 or A375 melanoma cells occurred due to the specific interaction with the PS exposed by their membranes and the formation of a bioactive structure to enter the cell and reach the Golgi upon hours of incubation, followed by mitochondrial swelling, cytochrome C release, lipid trafficking, apoptotic blebbing and finally apoptotic cell death [
12,
13]. The necessity for PS exposure was further confirmed by the conversion of PS to phosphatidylethanolamine and the consequent significant decrease in peptide activity toward melanoma [
12].
Within the current study, the peptide RDP22 was now studied for its toxicity on MUG-Mel2 [
8]. The cell line showed extremely aggressive and fast growth behavior, even in 3D cultures and animal models with persisting phenotypes [
8]. The cell line was derived from a highly aggressive cutaneous metastasis that did not respond to any treatment, neither targeted, immune or cytotoxic therapy, and had the characteristic of persisting pigmentation [
8] (
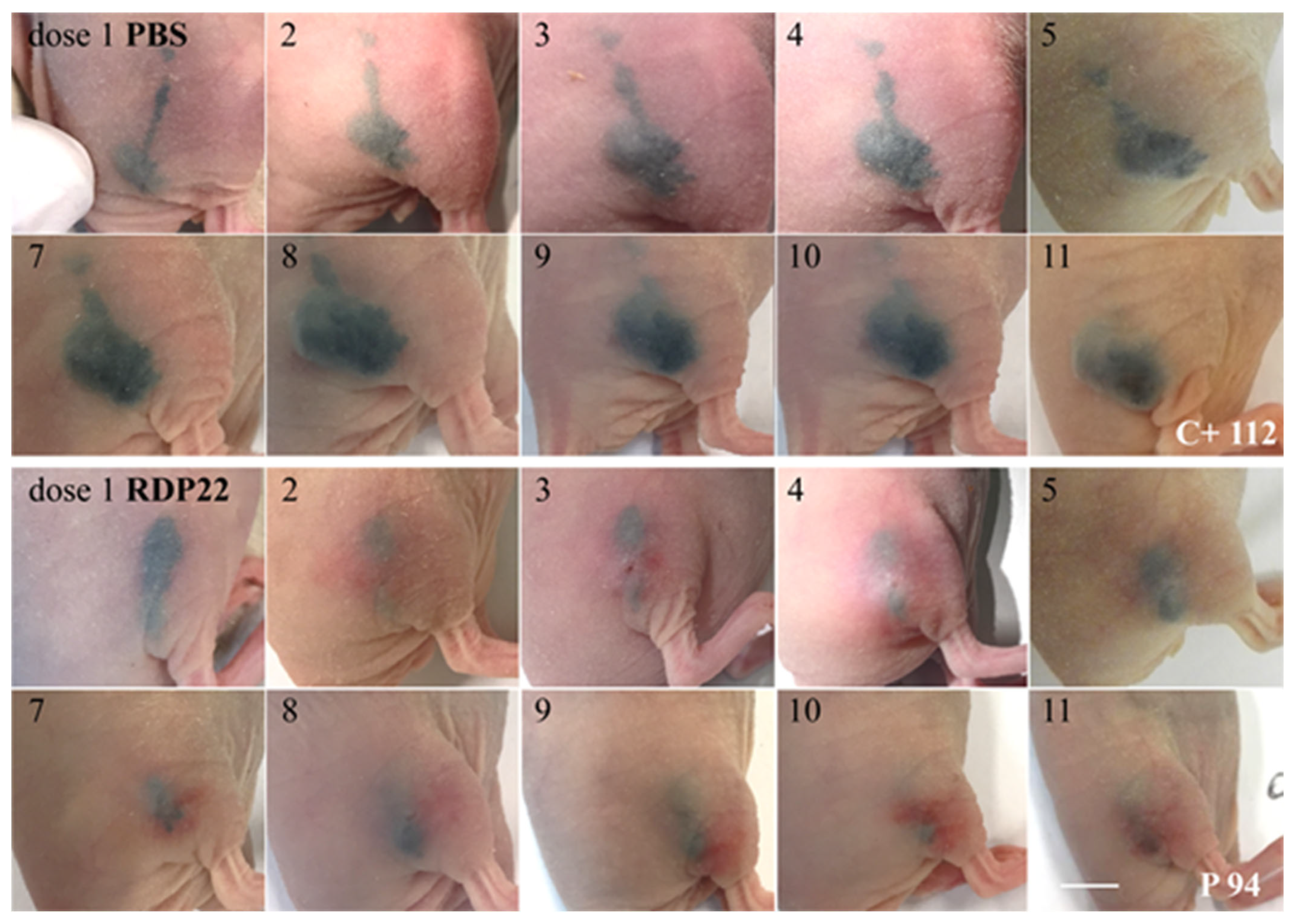
Figure S1); MUG-Mel2 was chosen to be a perfect model for studies on new therapies in the form of antitumor peptides, such as RDP22. The natural pigmentation of the cell line allows for morphologic observations in in vivo models without using any staining additives.
To reduce the number of animals, a thorough sequence of model studies was now lined up in front of the in vivo experiment. Starting with studies on PS exposure of MUG-Mel2, liposomal and cellular PS-peptide interaction studies, and toxicity in vitro studies with 2D and 3D cultures and a melanoma organotypic model, the in vivo studies with a MUG-Mel2 xenograft followed.
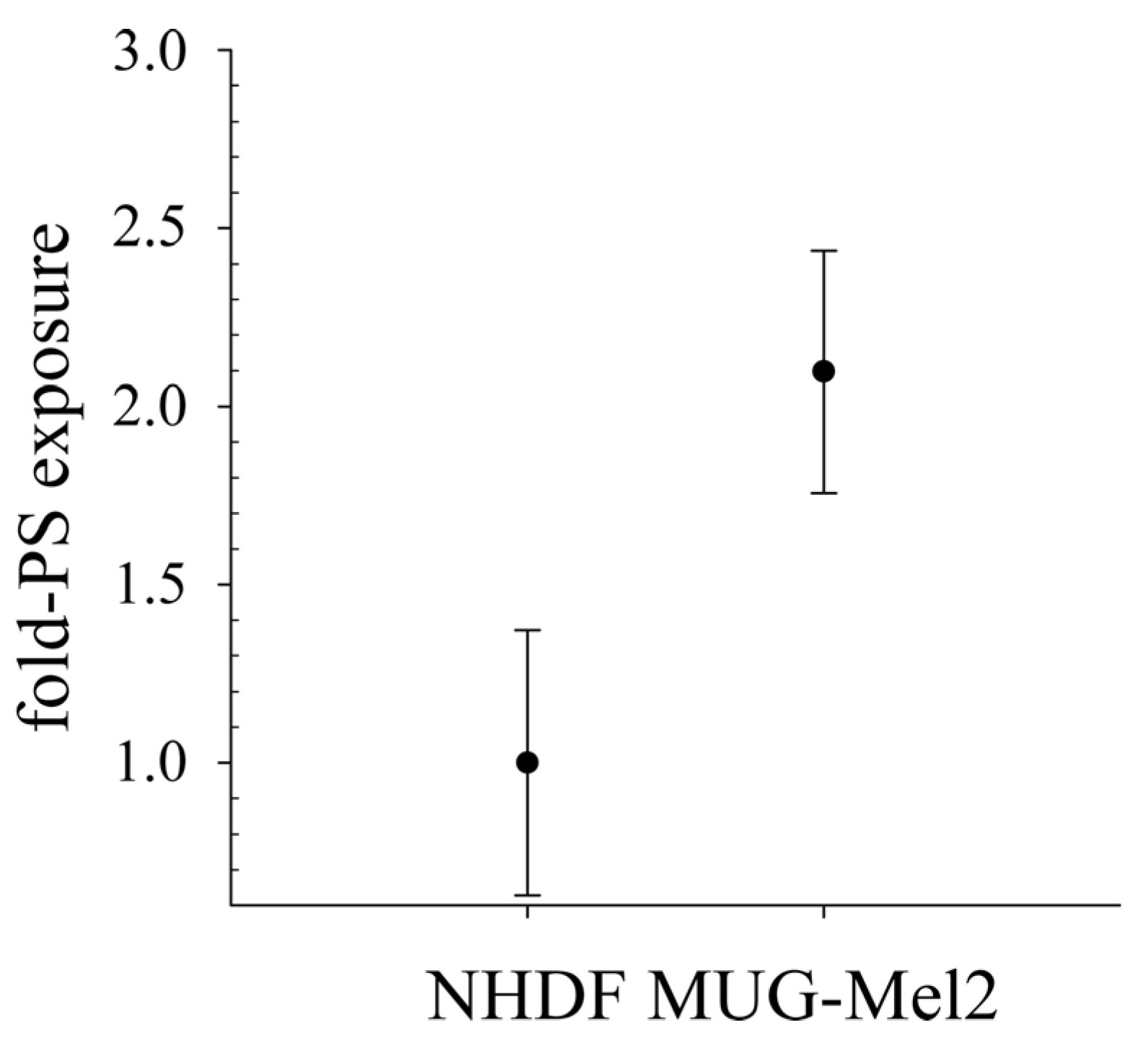
As already shown for different cancer cell types as glioblastoma, rhabdomyosarcoma [
17], breast cancer [
25,
26], leukemia [
27] and melanoma and their metastasis [
17,
28], MUG-Mel2 exposes significantly increased levels of PS, two-fold compared to non-malignant cells, such as normal human dermal fibroblasts (NHDF). This negative charge on cancer cells can be discriminated by cationic amphipathic peptides, such as RDP22, from neutral non-PS-exposing plasma membranes of non-malignant cells and thereby be selectively targeted [
18]. Indeed, a colocalization of PS to liposomes as well as MUG-Mel2 with the peptide RDP22 could be shown within the present study. Such a colocalization of antitumor peptide and PS was also reported for peptide D-K
6L
9 with PS exposed by prostate cancer cells [
29].
Recently, we were able to prove that the PS exposed by melanoma cells of A375 is like a door-lock for specific entrance for peptides, such as RDP22, into the cancer cell for subsequent effective induction of cell death via apoptosis upon interaction with inner targets such as the Golgi [
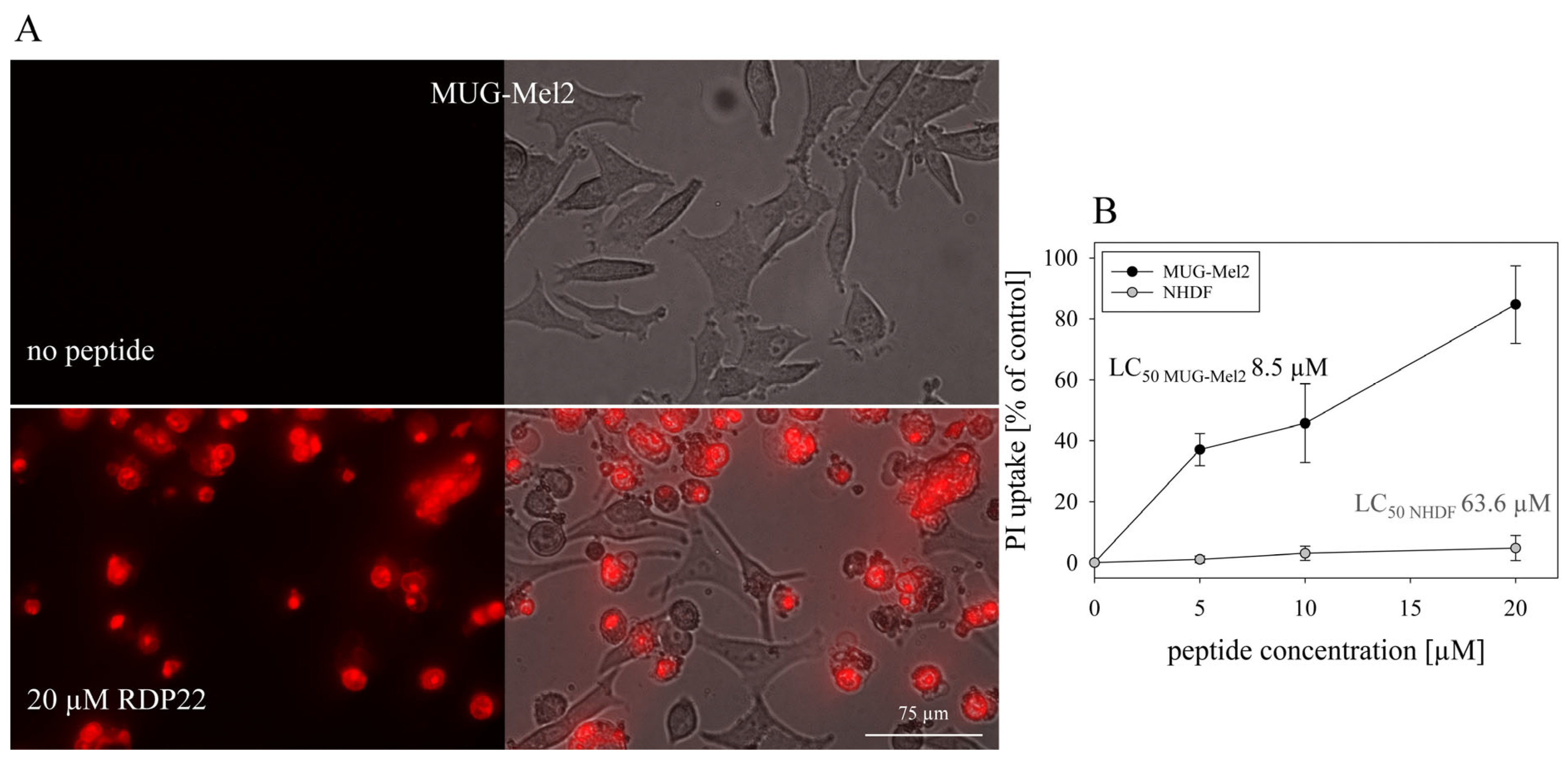
12]. In vitro studies with MUG-Mel2 also show high specificity of about 7.5-fold for MUG-Mel2 (LC
50 MUG-Mel2 8.5 µM) compared to normal dermal fibroblasts NHDF (LC
50 NHDF 63.6 µM) [
9]. Likewise, mainly apoptosis was shown to be induced by RDP22 in the MUG-Mel2 cells. As previously reported for RDP22 apoptosis-induced cell death is correlated with cancer-specific toxicity compared to non-specific peptides such as R-DIM-LF11-318, inducing necrosis, which harms malignant and non-malignant cells [
12].
To more closely mimic in vivo conditions within the study, in vitro experiments were expanded to 3D cultures. Before, we were already able to show that RDP22 induces cell death in tumor spheroids of melanoma cells of A375 and even mixtures of melanoma and normal dermal fibroblasts, mimicking the tumor with tumor stroma, whereas spheroids of normal fibroblasts were not significantly harmed at the antitumor active concentrations [
9]. Capability for entrance into the core and efficient killing of MUG-Mel2 spheroids was similarly observed within the present study, which is a necessary skill of a potent antitumor agent.
Further, MUG-Mel2 [
8] was used to successfully generate in vitro melanoma organotypic models, which were treated topically with the peptide or the vehicle. As shown before melanoma matured within a physiologically stratified epidermis. The persisting pigmentation of MUG-Mel2 within the in vitro models (
Figure S1) enabled the non-destructive detection of tumor progression and regression throughout the whole culture duration via the in-house developed MediTOM device. Transmission light microscopy revealed a massive regression in tumor growth upon treatment with RDP22 and allowed the assessment of the efficacy of the potential antitumor therapeutic. Additionally, the studied metabolic activity and cellular integrity as well as proliferation rates after treatment revealed a selectively preventive killing of MUG-Mel2 within the 3D skin models. Although the organotypic models already adequately mimic the complex animal models and human tissue, they still disadvantageously lack important components of the tumor microenvironment, e.g., immune system components. Thus, animal studies cannot be completely replaced yet but reduced significantly and help to decide independently of species-specific differences whether animal experiments are indicated or not.
Thus, a reasonable number of mice xenografts was consequently treated with peptide RDP22, due to the successful confirmation of PS exposed by MUG-Mel2 as a potent biomarker and target for cationic, amphipathic and PS-specific peptide RDP22; the proof of interaction of the peptide with PS in the lipid model systems in present and previous studies [
13,
14]; the peptide-PS interaction in cellular (MUG-Mel2) systems; the effective and specific killing of so far non-treatable MUG-Mel2 in vitro in more highly developed tumor spheroids alone or integrated into organotypic models.
The in vivo anti-melanoma activity of the related peptide R-DIM-P-LF11-334 (RDP22 -F2, -F18, A375 [
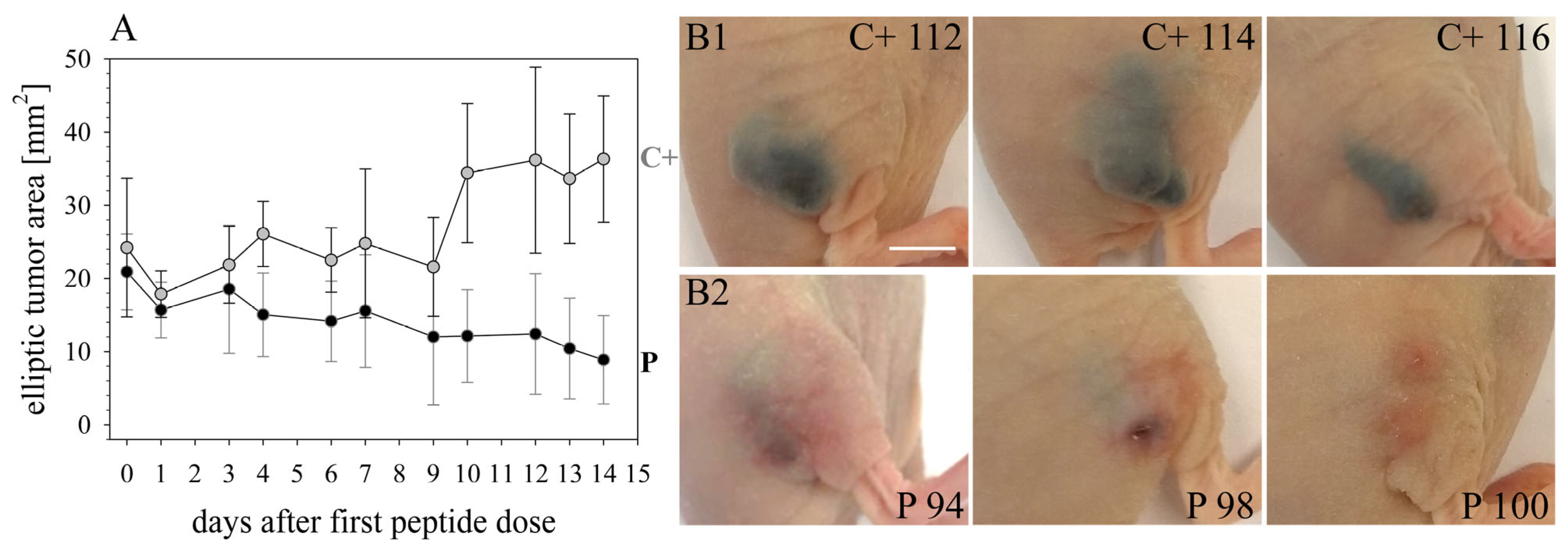
16]) was improved by RDP22 towards MUG-Mel2, which was shown by stronger regression of tumor growth and proliferation and massive reduction in tumor size in the mouse xenografts upon intratumoral peptide treatment. This was observable due to the advantageous fact that the tumor cells of MUG-Mel2 keep their pigmentation throughout their cultivation in vitro and even in vivo in the xenograft (
Figure S1), which makes the measurement of the external tumor area more reliable. To, however, more precisely calculate the real tumor area, histology of biopsies with HE was performed. As in the previous study [
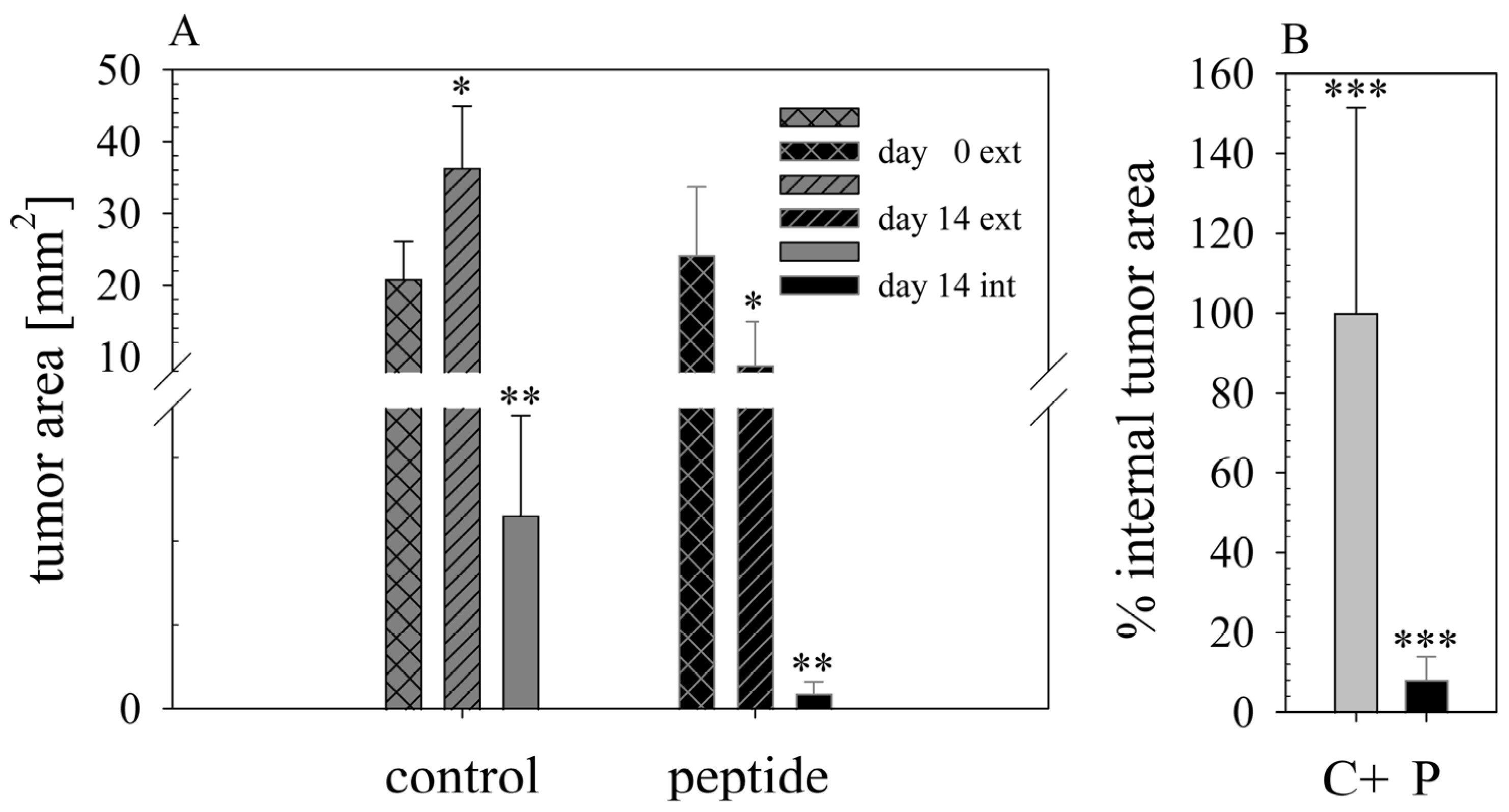
16] histologically studied biopsies of control (PBS) and peptide-treated tumors of MUG-Mel2 indicated a strong reduction not only of externally observable, but also of internal real tumor size. RDP22 thereby reduced the real tumor size/area by about 90%, compared to the control tumors with solely PBS injection. In comparison, R-DIM-P-LF11-334 reduced internal tumor size by about 70% within these groups in A375 xenografts [
16]. Similarly, within the present study, residual MUG-Mel-2 cells, if still present, were located in the inner core of the tumor surrounded by non-malignant freshly grown cells, whereas control biopsies not only were 75% more extensive but also completely comprised of viable tumor material. Proliferation marker Ki67 was further shown to be strongly reduced in peptide-treated tumors.
The fact that other antitumor peptides, e.g., LTX-315 [
30] besides killing melanoma cells, also induce a peptide-enhanced antitumor immune reaction opens the possibility for further RDP22 studies in more highly elevated melanoma organotypic/immune cell models and murine tumor studies to observe a potentially similar stimulation of the immune system by RDP22 besides the direct apoptotic effect in the tumor. For systemic application, the proteolytic stability of the peptide has to be improved eventually by the introduction of D-amino acids [
11]. A combination with immune therapies or cytostatic agents is also imaginable.
In conclusion, upon stepwise investigations of efficacy in increasingly sophisticated models, the antitumor peptide RDP22 was found to offer a new approach in the treatment of MUG-Mel2, an NRAS mutated melanoma with a high demand for curative treatment.