Abstract
Renal clear cell carcinoma (ccRCC) comprises over 75% of all renal tumors and arises in the epithelial cells of the proximal convoluted tubule. Molecularly ccRCC is characterized by copy number alterations (CNAs) such as the loss of chromosome 3p and VHL inactivation. Additional driver mutations (SETD2, PBRM1, BAP1, and others) promote genomic instability and tumor cell metastasis through the dysregulation of various metabolic and immune-response pathways. Many researchers identified mutation, gene expression, and proteomic signatures for early diagnosis and prognostics for ccRCC. Despite a tremendous influx of data regarding DNA alterations, gene expression, and protein expression, the incorporation of these analyses for diagnosis and prognosis of RCC into the clinical application has not been implemented yet. In this review, we focused on the molecular changes associated with ccRCC development, along with gene expression and protein signatures, to emphasize the utilization of these molecular profiles in clinical practice. These findings, in the context of machine learning and precision medicine, may help to overcome some of the barriers encountered for implementing molecular profiles of tumors into the diagnosis and treatment of ccRCC.
1. Introduction
Renal cell carcinoma (RCC) originates in the renal cortex and comprises 80–85% of all primary renal neoplasms [1]. RCC accounts for 2% of global cancer diagnoses and is one of the ten most common types of cancer diagnosed in the United States [2]. In recent years, RCC has become one of the fastest-growing cancers in North America, with the incidence doubling from 1975 to 2016 [2]. According to recent Surveillance, Epidemiology, and End Results Program (SEER) statistics, mortality rates remained relatively stable from 1975 to 2016, which may be associated with improved diagnostic and prognostic measures [2,3]. Despite the tremendous advancements, particularly in targeted therapeutics, RCC remains the most lethal urogenital cancer with a 5-year survival rate of roughly 76% [2,3]. However, the survival statistics depend highly on the initial stage at diagnosis, with localized patients having 93% 5-year survival, while distant cases have only 15.3% [3]. The major subtypes of RCC include clear cell carcinoma (ccRCC, ~75% cases), papillary cell (pRCC, ~10–15% cases), and chromophobe (chRCC, ~5% cases), and other rare types [4]. Each of these types arises from histologically distinct cells [4]. Each subtype arises from a series of complex genetic driver events and molecular aberrations [4]. Over the years, our knowledge has broadened on genetic heterogeneity, including mutational burden and targetable markers by high throughput assays and sequencing technologies [5,6]. Until the recent development of proteomic signature data, all of the research in RCC biomarker identification has focused on genomic alterations and gene expression signatures, which have various limitations preventing their integration into the clinical practice [7].
Current genomic profiling approaches have limitations, such as small numbers of individual mutations, which are both difficult to target therapeutically and fail to capture phenotypic consequences of aberrant gene expression [7,8]. Transcriptomic analyses suffer from a high degree of variability among expression signatures within individual tumors with the absence of validation of the gene signatures in independent population [9]. With the recent integration of protein signature data, a more robust molecular “landscape” for ccRCC may be revealed as the number of protein signature profiles begins to approach the level of genomic and gene expression data currently available [7]. In this literature review, we provide developments over the past 15 years on proteogenomic characterizations of ccRCC and their implication for targeted therapy development by incorporating DNA mutations, gene expression, and proteomic signature data. Additionally, we provide our comments on the role of machine learning and deep learning algorithms that can improve diagnostic and prognostic measures using big data in RCC.
2. RCC Subtypes
Major subtypes of RCC include clear-cell (ccRCC), papillary (pRCC), and chromophobe (chRCC), as mentioned earlier [10] (Table 1). The vast majority of RCC cases are of clear-cell morphology (75%), while pRCC (10%), chRCC (5%), and other unclassified and rare subtypes make up the remainder of renal cancer [11]. Clear cell RCC tumors arise from epithelial cells of the proximal convoluted tubule in the nephron and are histologically confirmed by their abundant lipid and glycogen-rich cytoplasmic droplets [12]. Roughly 2–3% of ccRCC are hereditary, originating from VHL disease-induced renal neoplastic cysts [13,14]. Hereditary and sporadic tumors alike may degenerate into malignant tumors as the result of a combination of early driver and somatic mutations, DNA methylation, and copy number alterations (CNAs) [14]. These molecular changes promote oncogenesis through the proliferation of a multitude of growth factors and dysregulated pathways, i.e., VEGF, PDGF, and HIF pathways [15]. PRCC tumors are histologically classified as type 1 or type 2, which have distinct molecular and survival differences [16] (Table 1). Most pRCC cases are sporadic; however, type 1 tumors have a hereditary component arising from germline mutations of MET [17]. In comparison, type 2 tumors are linked to a greater number of chromosomal aberrations and are associated with higher grade, stage, and an overall worse prognosis [18]. ChRCC tumors are histologically subdivided into a classical type, consisting of pale and eosinophilic cells, and an eosinophilic variant, which contains predominantly eosinophilic cells [19] (Table 1). These tumors are generally viewed as less aggressive compared to the more frequent RCC subtypes [20]. Molecular features unique to chRCC include copy number variations involving complete loss of chromosomes 1, 2, 6, 10, 13, and 17 [19]. Despite these distinctions, much of the current multi-omics analyses have been directed towards ccRCC, as it is the most frequently diagnosed and most lethal subtype [21] (Table 1).
Over 50% of cases of RCC in the clinic are discovered incidentally, showing no common clinical symptoms of flank pain, hematuria, and/or palpable abdominal mass(es), usually associated with RCC [14]. Surgical removal of tumors is the preferred treatment for RCC when patients are in stages I-III; however, up to 1/3 of these patients will experience disease recurrence [22]. For advanced-stage disease, intratumor heterogeneity and tumor clonality are important factors for predicting prognostic outcome [5].

Table 1.
Renal Cell Carcinoma Types.
Table 1.
Renal Cell Carcinoma Types.
| RCC Type | Tumor Type | Histology | 5-Year Survival (%) | Ref |
|---|---|---|---|---|
| Clear-Cell | Malignant | Large, lipid-rich cells with abundant cytoplasm | 85.5 * | [12,23] |
| Papillary | Malignant | Type 1: thin, single-cell layered papillae, basophilic, scant cytoplasm; Type 2: thick, large-celled papillae, abundant eosinophilic cytoplasm | 90.9 | [16,23] |
| Chromophobe | Malignant | Large cells, distinct membrane, abundant pale cytoplasm | 95.2 | [19,23] |
* 5-year survival for metastatic ccRCC is 12%.
Risk stratification and targeted therapeutic development for these patients have generally relied on certain physiological and biochemical markers expressed within the genome, transcriptome, and proteome [24]. Recent progress in whole genome sequencing techniques has led to the identification of a number of genes with clinical and prognostic relevance for the ccRCC [21]. Additional analysis techniques, such as functional impact mutation ranking, phylogenetic analysis, and ploidy profiling, have revealed distinct driver mutations in the early development of ccRCC tumors [6,25]. The identification of common mutation patterns that initiate tumor progression can improve early detection and prognostication methods, which are two important factors for RCC survival outcomes [11,26].
We queried PubMed and Google Scholar to investigate studies revealing novel gene and protein expression signatures in RCC. PubMed and Google Scholar searches were performed on 21 September 2022, using the keywords “gene expression signature”, “protein expression signature”, and “ccRCC”. The search was restricted to the years 2007–2022. During the literature review, the inclusion criteria consisted of studies reporting sensitivity and specificity (AUROC) values greater than 75% and hazard ratios falling between 0.0–0.5 and >2.0 to capture both protective and detrimental signatures. The exclusion criteria consisted of publications prior to 2007 and studies with discovery sample sizes of less than 40.
3. Molecular Changes in RCC
The identification of genomic and transcriptomic biomarkers has added tremendous biological value for ccRCC characterization [27]. The development of ccRCC has been described by a series of molecular changes associated with tumor initiation, driver gene mutations, lethal events, and, ultimately, tumor metastasis [28]. Various DNA alterations are involved in tumor development and progression, including copy number alterations (CNAs), methylation, and mutations that drive genomic instability [28]. The resulting biological state of these alterations is often reflected in the gene expression profiles of tumor cells, from which expression signatures may be identified and associated with clinical metrics, such as diagnosis and prognosis [29]. When analyzed together, the correlation between mRNA transcripts and protein expression for RCC tumors has been shown to be quite variable [27]. As such, protein expression signatures may adequately summarize the consequences of genomic and transcriptomic alterations, while also providing new targetable agents for precision medicine [27].
3.1. DNA Alterations
DNA mutations are the most common form of alteration found in all cancers including ccRCC. Genomic alterations in ccRCC are summarized by copy number variations involving whole chromosome alterations (7 and 9), arm-level deletions (3p and 14q) and gains (5q), and additional somatic mutations [30,31]. Many of the mutations associated with ccRCC development follow the two-hit hypothesis of tumorigenesis; the loss of heterozygosity (LOH) occurs via the loss of 3p and inactivation of the remaining allele by somatic mutation [31] (Table 2). The loss of 3p leads to the loss of one copy of VHL, PBRM1, SETD2, and BAP1, the most commonly mutated genes in ccRCC. In addition to chromosomal aberrations, promoter CpG hypermethylation, missense, and truncating mutations account for a large percentage of the observed DNA dysregulation [6,31]. The most commonly dysregulated pathways include the well-known VHL/HIF pathway, chromatin remodeling/histone methylation activity, and the PI3K/AKT/mTOR pathway [31] (Table 2). The models for ccRCC development and progression consistently depict the importance of early driver mutations, which leave surroundings cells vulnerable to additional subclonal mutations [26]. Somatic mutations of genes with chromatin remodeling and histone modification capabilities (PBRM1, BAP1, SETD2, KDM5C) contribute to increased chromosome instability and alterations in gene expression control, which has been associated with higher-grade tumors and poorer survival [32,33].

Table 2.
Driver Mutations for ccRCC (Clark et al. [30]).
Driver mutations are defined as a specific group of mutations that arise in the early stages of cancer and are highly influential in the malignant transformation of tumor cells [34] (Table 2). In certain analyses of the evolution of ccRCC tumor mutations, driver mutations are differentiated by the time at which they occur along phylogenetic trees [5,6]. “Truncal” mutations represent the earliest mutational events in tumor progression, while “branched” mutations occur later and characterize distinct trajectories of the tumor development [5,6]. Despite largely ubiquitous VHL inactivation and 3p loss in ccRCC tumors, there is a wide variation in clinical outcomes, which brings into question the role of subclonal and passenger mutations in tumor progression and drug resistance [5,6]. Branched mutations and epigenetic changes often involve gene products associated with chromatin remodeling complexes and hypermethylation, which present unique challenges to the therapeutic targeting [32,35]. Somatic mutations of genes with chromatin remodeling and histone modification capabilities (PBRM1, BAP1, SETD2, KDM5C) contribute to increased chromosome instability and alterations in gene expression control, which has been associated with higher grade tumors [32,33] (Table 2). The role of DNA hypermethylation has also been investigated extensively in ccRCC, as silencing of tumor suppressor genes, such as FBN2, PCDH8, BNC1, and SFRP1, plays an integral role in the tumor progression [36].
3.2. Gene Expression Signatures
The application of gene expression signatures for clinical use has remained a long-standing question since the advent of expression analysis over 20 years ago [37]. Gene expression signatures are defined as a single gene, or group of genes, with an expression pattern that associates with some clinically relevant metric such as diagnosis, prognosis, or predictive treatment response [37]. As a potential biomarker, RNA expression provides a readily and easily available resource for detecting cellular changes reflected in mRNA and other types of extracellular RNA (exRNA) [38]. Extracellular RNA transcripts are also stable in a number of bio-fluids, including urine, serum, and plasma, providing a potentially promising resource for non-invasive collection methods [38]. Changes in gene expression patterns are directly correlated with biologically diseased states and ultimately may represent a surrogate phenotype for the cancer [29]. With recent developments in next-generation sequencing (NGS) transcriptomic signatures can be easily identified. It can also predict splice variants, gene fusions, and epigenetic changes, which are missed in the DNA analysis [39]. Low sample numbers and lack of validation are major obstacles to the clinical transition [40]. The gene expression signatures provided in Table 3 incorporate diagnostic, prognostic, and predictive response outcomes, representing the current state of expression signature analyses.
Classification of RCC, based on gene expression and survival outcomes, was proposed in 2010 as a molecular stratification tool to investigate metastasis and tumor aggressiveness [41,42]. This approach suggested using clear cell type A (ccA) and clear cell type B (ccB) for the classification of RCC to include metastasis and aggressive nature of the RCC tumors [41]. Built off of the ccA/ccB classifiers, ClearCode34 is a prognostic signature that can reliably predict the ccRCC recurrence risk [43]. This gene signature has been shown to identify patients who would benefit from surveillance versus adjuvant therapy following surgery [43]. Gene expression signatures can also be used to differentiate tumor and normal tissue [40]. Therefore, targeting driver mutation-specific expression profiles is a logical strategy to detect early oncogenic changes in pre-neoplastic cells as well as to supplement diagnosis by tumor biopsy and guide treatment decisions [44,45]. Ujfaludi et al. (2022) analyzed the transcriptomic signature of key ccRCC driver genes, VHL, SETD2, PBRM1, and BAP1, to find that the median transcription of these genes distinguished ccRCC from normal tissue with a moderate level of sensitivity and specificity (87% and 77%, respectively) [44]. There have been a number of recent clinical trials [46] which have revolutionized the treatment landscape of ccRCC, from which a wealth of biomarker data can be extracted. Biomarker analysis from the recent phase III CheckMate 214 clinical trial compared survival outcomes, progression-free survival (PFS), and overall survival (OS), of combination immune checkpoint inhibitor (ICI) treatment versus sunitinib, with established gene expression signatures [47]. Their findings suggest that combined signatures, such as tumor inflammation with angiogenesis or myeloid changes, may predict better response to immunotherapy versus tyrosine kinase inhibitor (TKI) alone [47]. However, the accumulated wealth of signature data has not been successfully implemented in the clinical setting [48].
One of the greatest barriers to signature implementation in the clinic is reliable data reproducibility, from which further analyses can build upon [49]. In an effort to overcome this, The Cancer Genome Atlas (TCGA) compiled pan-cancer data sets, which have been used as both discovery and validation sets for novel expression signatures [49,50,51]. Other issues in expression signature development include opposing views in the method of analysis, such as “top-down” and “bottom-up” supervised approaches. Supervised, “top-down”, approaches attempt to associate some clinical outcome (survival or metastasis) with an expression profile. Conversely, supervised, “bottom-up”, approaches utilize a biological basis for gene expression, which can be connected to some factors associated with tumor progression [52]. Predictive treatment responses encompass a much smaller range of the available expression signatures, as the individual signatures are tied to specific therapeutic agents [52]. Additionally, expression profiles can represent downstream alterations in proteins which may eventually become therapeutic targets [52].

Table 3.
List of gene expression studies for diagnostic (ccRCC vs. normal tissue), prognostic (overall survival, recurrence, disease-free survival, and cancer-specific survival), and therapeutic outcomes.
Table 3.
List of gene expression studies for diagnostic (ccRCC vs. normal tissue), prognostic (overall survival, recurrence, disease-free survival, and cancer-specific survival), and therapeutic outcomes.
| Serial | No. of Genes | Metric | No. of Samples | Measure | Outcome | Ref |
|---|---|---|---|---|---|---|
| 1. | 3 | AUC = 0.912 | 413 | 5-year survival post-nephrectomy | Prognostic | [53] |
| 2. | 34 | RFS: HR = 2.3 (1.6–3.3); CSS: HR = 2.9 (1.6–5.6); OS: HR = 2.4, (1.6–3.7) | 530 | RFS, CSS, and OS (ccA vs. ccB) | Prognostic | [43] |
| 3. | 5 | AUC = 0.783 | 523 | Overall Survival (OS) | Prognostic | [54] |
| 4. | 16 | HR = 3.37 | 615 | RFS, CSS, and OS | Prognostic | [55] |
| 5. | 10 | HR = 2.85 | 468 | Overall Survival (OS) | Prognostic | [56] |
| 6. | 8 | AUC = 0.821 | 888 | Fuhrman grade (high grade) | Prognostic | [57] |
| 7. | 9 | OR = 3.08 | 443 | Recurrence post-nephrectomy, immune signature | Prognostic | [51] |
| 8. | 1 | AUC = 0.9451 | 605 | Overall Survival (OS) and DFS | Prognostic | [58] |
| 9. | 3 | AUC = 0.9235–0.9451 | 605 | Normal vs. ccRCC tissue | Diagnostic | [58] |
| 10. | 17 | HR = 51.37 | 46 | Overall Survival | Prognostic | [50] |
| 11. | 4 | SN = 87%/SP = 77% | 60 | Normal vs. ccRCC tissue | Diagnostic | [44] |
SN/SP: Sensitivity/Specificity. The names of all genes in the signatures are presented in Supplemental Table S1.
3.3. Protein Signatures
Next-generation sequencing (NGS) and other high-throughput analyses provide a wealth of information regarding the abundance of genomic and transcriptomic alterations but often fail to fully characterize the biological state of the tumor as protein modifications are missed at these levels of analysis [59]. Post-translational modifications (PTMs) are enzyme-catalyzed additions of specific functional groups to proteins that promote a variety of biological functions [60]. Certain PTMs, such as phosphorylation and glycosylation, are highly relevant for tumor progression as they regulate cellular processes such as adhesion, migration, signaling, and growth [61,62]. Additionally, PTMs are key events in the dysregulation of metabolic events associated with ccRCC, such as the upregulation of glycolysis and downregulation of the Krebs cycle and electron transport chain, permitting an oncogenic metabolic shift or Warburg effect [11,31]. Proteomic analyses have also been influential in the categorizing of ccRCC, as molecular subgrouping is a useful tool in stratifying patients for precision therapeutics, such as for the use of VEGF inhibitors versus immune-based therapies [30,63]. Despite a multitude of studies over the past 15 years that have uncovered a generalized proteomic profile for ccRCC, none have achieved an adequately comprehensive characterization depicting the necessary linkage between genomic and transcriptomic aberrations, phenotypic presentation, and independent validation to promote clinical utility [59]. Matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) was the most commonly utilized technique to analyze peptide fractions for diagnostic studies, with validation by western- and immune-blotting (serum/plasma and urine) and immunohistochemistry (IHC) (tissue) (Table 4). The results of these experiments were used in statistical and machine learning models to identify correlation and association with disease vs. control populations. Output metrics are AUC (Area under the receiver operative curve), Sensitivity (SN), Specificity (SP), and hazard ratio (HR). Enrichment analyses were also used to identify upregulation and downregulated pathways in tumor vs. normal tissue. The use of alternate biomarker sources, such as serum/plasma and urine, will likely have important implications for the progression of signatures into clinical practice.

Table 4.
Studies identifying proteomic signature for clinical management and treatment of renal cell carcinoma patients.
Table 4.
Studies identifying proteomic signature for clinical management and treatment of renal cell carcinoma patients.
| Serial | No. of Proteins | Metric | SN/SP | No. of Samples | Measure | Outcome | Ref |
|---|---|---|---|---|---|---|---|
| 1. | 3 † | AUC = 0.90–0.94 | - | 70 | Diagnostic | ccRCC confirmed (NAT vs. tumor) | [64] |
| 2. | 3 * | - | 88/92% | 162 | Diagnostic | ccRCC confirmed | [65] |
| 3. | 5 * | - | 88.80/91.00% | 189 | Predictive | ccRCC confirmed (recurrence or initial) | [66] |
| 4. | 1 * | AUC = 0.86 | 87/80% | 80 | Diagnostic | ccRCC confirmed | [67] |
| 5. | 1 † | - | 96.61/71.43% | 881 | Prognostic | reduced OS and DFS | [68] |
| 6. | 1 ** | ACC = 0.930 | - | 93 | Diagnostic | ccRCC vs. Healthy Subjects (HS) | [69] |
| 7. | 10 † | AUC = 0.771 | - | 611 | Prognostic | Increased overall survival * | [70] |
| 8. | 9 † | AUC = 0.689 | - | 512 | Prognostic | Decreased OS | [71] |
| 9. | 7 * | AUC = 0.769 | - | 445 | Prognostic | High/Low-Risk Grouping | [63] |
| 10. | 4 †,** | HR = 2.76 | - | 552 | Prognostic | Higher risk of death (decreased OS/RFS) | [72] |
| 11. | 20 * | AUC = 0.870 | - | 232 | Prognostic | decreased PFS | [73] |
* Serum/Plasma, † Tissue, ** Urine as an analyte. SN/SP: Sensitivity/Specificity. The list of proteins in the signature(s) are presented in Supplemental Table S2.
Reviews of recent ccRCC proteomic studies revealed a wide variation of dysregulated pathways ranging from metabolic alterations to disruptions of the cell division [64,65]. Clark et al. (2019) characterized the proteogenomic profile of ccRCC by evaluating the role of genomic alterations in promoting the phenotypic presentation of ccRCC tumors. Their efforts, through the Clinical Proteomics Tumor Analysis Consortium (CPTAC), identified a lack of correlation between gene and protein expression, particularly related to oxidative phosphorylation, ribosome, spliceosome, and metabolic pathways [30]. Diagnostic studies found protein signatures associated with protein folding and binding mediation, cell-signaling regulation, tubulin formation, and heat shock protein response in addition to similarities with CPTAC data [64,66,67,68,69] (Table 4). Prognostic protein signatures exhibited dysregulation in the mTORC1 signaling pathway, lipid metabolism, intracellular/vesicle-mediated transport systems, cytokine response and receptor interaction, ribosomal binding proteins (RBPs) functions, as well as a multitude of metabolic and biosynthetic pathways [63,70,71,72,73,74] (Table 4).
Tumor tissue generally contains higher protein concentration than normal tissue, however, the investigation of blood serum/plasma as well as the urine secretome, offer alternative methods for noninvasive biofluid analysis [75]. Tissue-based sampling represents the most direct route of protein extraction, where fresh frozen (FF) tissues, obtained via fine needle aspiration (FNA) biopsy or surgical resection, and formalin-fixed paraffin-embedded (FFPE) tissues are most commonly used for analysis [75,76]. Limited access to biopsy samples is invasive and challenging, while urine and blood collection require less technical expertise and are minimally invasive [77]. Serum samples provide an advantageous route of analysis not only for their less-invasive collection methods but also for potentially early predictive and diagnostic detection of the ccRCC [64,67,68,78]. Urine sampling is the least-invasive peripheral fluid analysis technique, and it contains a less-dynamic complement of proteins compared to the highly abundant proteome of plasma and serum, which make identification of lower molecular weight proteins difficult [59,79]. While the direct relationship of urine analysis to kidney dysfunction may offer promising diagnostic capabilities for the slow-to-moderate onset of tumors, orthogonal models of study incorporating multiple sample types will likely be required to develop consistency in the proteomic characterization of ccRCC [80].
4. Utility of Molecular Information in Clinical Management and Treatment of RCC
Diagnosis and staging of RCC are currently performed by anatomical evaluation through imaging techniques (MRI, CT) followed by histopathological confirmation [10]. Diagnosis in the early stage is the most important factor for survival, and at presentation, 25% of the patients already have distant metastasis [81]. The symptoms of the classical triad (hematuria, flank pain, and abdominal mass) are seen in only 10% of the patients’ [2]. Therefore, an early diagnosis of tumor can play a significant role in the survival of the patients. The germline and driver gene mutations can be captured using gene signature assays for diagnostic purposes in both tumor and pre-neoplastic cells [34] (Table 3). Moreover, gene and protein signatures can be used in small renal mass biopsies to identify benign, malignant, and normal tissue to direct therapy [82]. Changes in genetic and epigenetic profiles can also be detected in the patient’s serum and urine analysis for methylation, miRNA, and lncRNA. Genetic markers such as VHL [69,71], APC [83,84,85], and P16 [71,83,84,85] are most commonly mentioned in the literature seen in both serum and urine samples in the form of cell-free DNA/ RNA/ methylomes. These tests are non-invasive and can be an excellent screening tool for RCC detection as well as are commonly used in biomarker panels to increase the accuracy of tumor detection. For example, Nuzzo et al. recently presented 300 differentially expressed methylomes in a study with 148 patients for the detection of ccRCC and pRCC with AUROC of 0.99 in serum and 0.86 in urine (patient vs. control). Similarly, miRNA [86] and lncRNA panels [87] are also available for the detection of RCC in urine and serum, respectively. Screening with such tools in high-risk patients can help to identify the tumors early in Stages I-III, where surgical intervention is curative. For imaging, machine learning and deep learning models with the neural network can be used in image classification to identify smaller tumors in suspecting cases [88].
CcRCC treatment depends on the stage at the time of diagnosis. Surgical interventions, such as partial or radical nephrectomy, can be curative in Stages I-III, but about 33% of patients eventually recur. Initial systemic therapy, for locally advanced or metastatic disease, is immune checkpoint inhibitors (ICI) in various combinations of PD-1 (nivolumab & pembrolizumab), PD-L1 (avelumab and atezolizumab), anti-CTLA-4 (ipilimumab) with or without the combination of VEGF inhibitors (axitinib, sunitinib, pazopanib, bevacizumab, etc.), and mTOR inhibitors (everolimus) [89]. The selection of therapy depends on risk stratification using IMDC risk stratification criteria. Response to ICI depends on the expression of many different markers, including the following: PD-L1 [70], tumor-infiltrating lymphocytes [90], tumor mutation burden [91,92], mismatched repair [93], PTEN inactivation [94], POLE mutation [95], co-mutation of KRAS and STK11 [96], and EGFR mutation [97]. There are other markers in peripheral blood, such as neutrophil to lymphocyte ratio, LDH, peripheral immune cells, circulating tumor DNA, soluble PD-L1, peripheral blood T-cell receptor, and peripheral cytokine [98]. Immune-related factors, such as Beta 2 microglobulin, B7-H4, TOX, and gut microbiota, also play a role in the prediction of ICI response [98].
Prognostic signatures using specific biomarkers (genes and proteins, Table 3 and Table 4) can stratify the patients into groups. These groups can then be used for targeting specific genomic aberrations for predicting drug responses [32]. The best example of this approach is the Prosigna breast cancer signature [99] which quantifies gene expression for 50 genes and uses that information to predict the recurrence [100]. Another example is the use of cytogenetics for diagnosis/stratification of leukemia and lymphomas patients with high and low risk and to identify minimal residual disease [101], i.e., the mutation in FLT3 gene causes an aggressive form of acute myeloblastic leukemia which is likely to relapse [102]. Similar methods could also be used in ccRCC using mutational, transcriptomic, and proteomic profiling. With metabolic dysfunction being central to tumor progression and aggressiveness in nearly all cancers, including ccRCC, can provide a potential prognostic tool based on metabolic signatures [35]. TCGA analysis found survival outcomes are associated with alternations of mRNA and miRNA expression of multiple metabolic pathways, including glycolysis, Krebs cycle, pentose phosphate pathway (PPP), fatty acid synthesis, PI3K, and AMPK pathway [26]. Dynamic changes to the tumor microenvironment occur in ccRCC development as well as in response to systemic therapies, and clinical trial data have been evaluated to better stratify patients to specific treatments based on gene expression data [9,36,37,38]. Integration of machine learning can identify complex relationships between the gene-gene and gene-protein interactions in regard to survival, recurrence, and treatment responses. The drawback is that overfitting or underfitting the data can lead to false discovery. To overcome that, larger validation studies are needed in a clinical trial controlling all the variables. The eventual application of such signatures in the clinic can guide the physicians for early diagnosis and prognosis to stratify the patients to be treated aggressively or conservatively.
5. Conclusions
A lot of progress has been made in the fields of genomics and proteomics for the high-throughput discovery of novel biomarkers for ccRCC, connecting genomic and molecular data to biological significance. These biomarkers are measured in the tumor tissue itself, serum, or urine and have generated large-scale data. This data can extensively be used for molecular characterization for diagnostic and prognostic signatures. This profiling can identify the tumors in high-risk cases for early diagnosis and predict the prognosis of the patients (Figure 1). The drawback is the lack of validation in a larger multi-institutional cohort in randomized clinical trials. With the validation and implementation of these signatures, it is possible to screen the general population for renal tumors, provide curative therapy in early stages, and predict response and recurrence rates. This would inadvertently lower the physical and emotional burden of the patients as well as the economic burden for society.
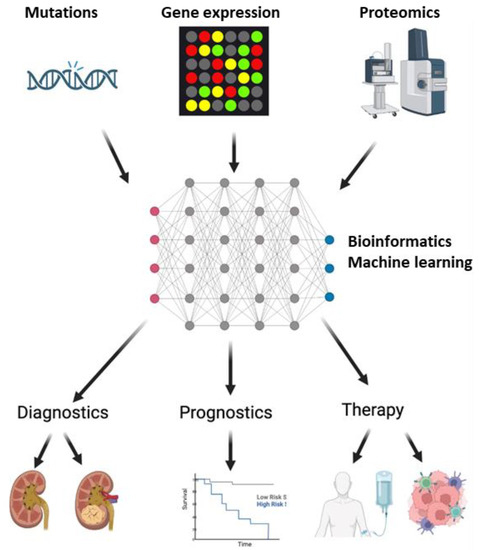
Figure 1.
Implementation of biomarkers in Clear Cell Renal Carcinoma (ccRCC). DNA sequencing, microarray, and mass spectrometry with liquid chromatography identify the mutation, gene, and protein expression profiles. These profiles create big data, that, when analyzed by machine learning algorithms, can identify markers for diagnosis, prognosis, and therapeutic decisions for ccRCC. In terms of diagnosis, these biomarkers can help in distinguishing ccRCC patients from healthy individuals, as well as from other patients with benign or malignant renal masses. Disease progression and survival, as an outcome of therapy, can be monitored by prognostic biomarkers. Finally, these biomarkers can provide information, which can offer precision medicine for patients.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10112953/s1, Table S1: Supplementary Table S1: Gene symbols with correlating serial number as presented in Table 3; Table S2: Supplementary Table S2: Protein names and symbols with correlating serial number as presented in Table 4.
Author Contributions
Conceptualization, C.W., K.B.S. and S.P.; methodology, C.W., K.B.S., K.P.R. and S.P.; investigation, C.W., K.B.S. and K.P.R.; resources, S.P.; writing—original draft preparation, C.W.; writing—review and editing: C.W., K.B.S., K.P.R., P.M.H.T., L.K.H.T. and K.P.R.; supervision, S.P.; project administration, S.P.; funding acquisition, S.P.; visualization, K.B.S. and S.P. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was covered by the Center for Biotechnology and Genomic Medicine and BAT Cancer study.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Atkins, M.; Choueiri, T. Epidemiology, Pathology, and Pathogenesis of Renal Cell Carcinoma-Up to Date. Available online: https://www.uptodate.com/contents/epidemiology-pathology-and-pathogenesis-of-renal-cell-carcinoma (accessed on 2 March 2021).
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol. 2020, 11, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. (Eds.) SEER Cancer Statistics Review, 1975–2017. Available online: https://seer.cancer.gov/csr/1975_2017/index.html (accessed on 10 September 2022).
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 313–326.e5. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Horswell, S.; Larkin, J.; Rowan, A.J.; Salm, M.P.; Varela, I.; Fisher, R.; McGranahan, N.; Matthews, N.; Santos, C.R.; et al. Genomic Architecture and Evolution of Clear Cell Renal Cell Carcinomas Defined by Multiregion Sequencing. Nat. Genet. 2014, 46, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Mani, D.R.; Krug, K.; Zhang, B.; Satpathy, S.; Clauser, K.R.; Ding, L.; Ellis, M.; Gillette, M.A.; Carr, S.A. Cancer Proteogenomics: Current Impact and Future Prospects. Nat. Rev. Cancer 2022, 22, 298–313. [Google Scholar] [CrossRef]
- Hofstatter, E.W.; Bale, A.E. The Promise and Pitfalls of Genomics-Driven Cancer Medicine. AMA J. Ethics 2013, 15, 681–686. [Google Scholar] [CrossRef]
- Motzer, R.J.; Banchereau, R.; Hamidi, H.; Powles, T.; McDermott, D.; Atkins, M.B.; Escudier, B.; Liu, L.-F.; Leng, N.; Abbas, A.R.; et al. Molecular Subsets in Renal Cancer Determine Outcome to Checkpoint and Angiogenesis Blockade. Cancer Cell 2020, 38, 803–817.e4. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal Cell Carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17009. [Google Scholar] [CrossRef]
- Li, Q.K.; Pavlovich, C.P.; Zhang, H.; Kinsinger, C.R.; Chan, D.W. Challenges and Opportunities in the Proteomic Characterization of Clear Cell Renal Cell Carcinoma (CcRCC): A Critical Step towards the Personalized Care of Renal Cancers. Semin. Cancer Biol. 2019, 55, 8–15. [Google Scholar] [CrossRef]
- Muglia, V.F.; Prando, A. Renal Cell Carcinoma: Histological Classification and Correlation with Imaging Findings. Radiol. Bras. 2015, 48, 166–174. [Google Scholar] [CrossRef]
- Kaelin, W.G. Molecular Basis of the VHL Hereditary Cancer Syndrome. Nat. Rev. Cancer 2002, 2, 673–682. [Google Scholar] [CrossRef]
- Escudier, B.; Porta, C.; Schmidinger, M.; Rioux-Leclercq, N.; Bex, A.; Khoo, V.; Grünwald, V.; Gillessen, S.; Horwich, A. Renal Cell Carcinoma: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up†. Ann. Oncol. 2019, 30, 706–720. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshizato, T.; Shiraishi, Y.; Maekawa, S.; Okuno, Y.; Kamura, T.; Shimamura, T.; Sato-Otsubo, A.; Nagae, G.; Suzuki, H.; et al. Integrated Molecular Analysis of Clear-Cell Renal Cell Carcinoma. Nat. Genet. 2013, 45, 860–867. [Google Scholar] [CrossRef]
- Akhtar, M.; Al-Bozom, I.A.; Al Hussain, T. Papillary Renal Cell Carcinoma (PRCC): An Update. Adv. Anat. Pathol. 2019, 26, 124–132. [Google Scholar] [CrossRef]
- Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N. Engl. J. Med. 2016, 374, 135–145. [CrossRef]
- Klatte, T.; Pantuck, A.J.; Said, J.W.; Seligson, D.B.; Rao, N.P.; LaRochelle, J.C.; Shuch, B.; Zisman, A.; Kabbinavar, F.F.; Belldegrun, A.S. Cytogenetic and Molecular Tumor Profiling for Type 1 and Type 2 Papillary Renal Cell Carcinoma. Clin. Cancer Res. 2009, 15, 1162–1169. [Google Scholar] [CrossRef]
- Garje, R.; Elhag, D.; Yasin, H.A.; Acharya, L.; Vaena, D.; Dahmoush, L. Comprehensive Review of Chromophobe Renal Cell Carcinoma. Crit. Rev. Oncol. Hematol. 2021, 160, 103287. [Google Scholar] [CrossRef]
- Lopez-Beltran, A.; Montironi, R.; Cimadamore, A.; Cheng, L. Grading of Chromophobe Renal Cell Carcinoma: Do We Need It? Eur. Urol. 2021, 79, 232–233. [Google Scholar] [CrossRef]
- Dizman, N.; Philip, E.J.; Pal, S.K. Genomic Profiling in Renal Cell Carcinoma. Nat. Rev. Nephrol. 2020, 16, 435–451. [Google Scholar] [CrossRef]
- Motzer, R.J.; Jonasch, E.; Agarwal, N.; Alva, A.; Baine, M.; Beckermann, K.; Carlo, M.I.; Choueiri, T.K.; Costello, B.A.; Derweesh, I.H.; et al. Kidney Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 71–90. [Google Scholar] [CrossRef]
- Palumbo, C.; Pecoraro, A.; Knipper, S.; Rosiello, G.; Luzzago, S.; Deuker, M.; Tian, Z.; Shariat, S.F.; Simeone, C.; Briganti, A.; et al. Contemporary Age-Adjusted Incidence and Mortality Rates of Renal Cell Carcinoma: Analysis According to Gender, Race, Stage, Grade, and Histology. Eur. Urol. Focus 2021, 7, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Linehan, W.M.; Ricketts, C.J. The Cancer Genome Atlas of Renal Cell Carcinoma: Findings and Clinical Implications. Nat. Rev. Urol. 2019, 16, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, A.; Perez-Llamas, C.; Deu-Pons, J.; Tamborero, D.; Schroeder, M.P.; Jene-Sanz, A.; Santos, A.; Lopez-Bigas, N. IntOGen-Mutations Identifies Cancer Drivers across Tumor Types. Nat. Methods 2013, 10, 1081–1082. [Google Scholar] [CrossRef] [PubMed]
- Brugarolas, J. Molecular Genetics of Clear-Cell Renal Cell Carcinoma. JCO 2014, 32, 1968–1976. [Google Scholar] [CrossRef] [PubMed]
- Creighton, C.J. Proteomic Signatures of Clear Cell Renal Cell Carcinoma. Nat. Rev. Nephrol. 2020, 16, 133–134. [Google Scholar] [CrossRef]
- Jonasch, E.; Walker, C.L.; Rathmell, W.K. Clear Cell Renal Cell Carcinoma Ontogeny and Mechanisms of Lethality. Nat. Rev. Nephrol. 2021, 17, 245–261. [Google Scholar] [CrossRef]
- Nevins, J.R.; Potti, A. Mining Gene Expression Profiles: Expression Signatures as Cancer Phenotypes. Nat. Rev. Genet. 2007, 8, 601–609. [Google Scholar] [CrossRef]
- Clark, D.J.; Dhanasekaran, S.M.; Petralia, F.; Pan, J.; Song, X.; Hu, Y.; da Veiga Leprevost, F.; Reva, B.; Lih, T.-S.M.; Chang, H.-Y.; et al. Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell 2019, 179, 964–983.e31. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network Comprehensive Molecular Characterization of Clear Cell Renal Cell Carcinoma. Nature 2013, 499, 43–49. [CrossRef]
- Bihr, S.; Ohashi, R.; Moore, A.L.; Rüschoff, J.H.; Beisel, C.; Hermanns, T.; Mischo, A.; Corrò, C.; Beyer, J.; Beerenwinkel, N.; et al. Expression and Mutation Patterns of PBRM1, BAP1 and SETD2 Mirror Specific Evolutionary Subtypes in Clear Cell Renal Cell Carcinoma. Neoplasia 2019, 21, 247–256. [Google Scholar] [CrossRef]
- Joosten, S.C.; Smits, K.M.; Aarts, M.J.; Melotte, V.; Koch, A.; Tjan-Heijnen, V.C.; van Engeland, M. Epigenetics in Renal Cell Cancer: Mechanisms and Clinical Applications. Nat. Rev. Urol. 2018, 15, 430–451. [Google Scholar] [CrossRef]
- Poulos, R.C.; Wong, J.W.H. Finding Cancer Driver Mutations in the Era of Big Data Research. Biophys. Rev. 2018, 11, 21–29. [Google Scholar] [CrossRef]
- Helming, K.C.; Wang, X.; Roberts, C.W.M. Vulnerabilities of Mutant SWI/SNF Complexes in Cancer. Cancer Cell 2014, 26, 309–317. [Google Scholar] [CrossRef]
- Ricketts, C.J.; Hill, V.K.; Linehan, W.M. Tumor-Specific Hypermethylation of Epigenetic Biomarkers, Including SFRP1, Predicts for Poorer Survival in Patients from the TCGA Kidney Renal Clear Cell Carcinoma (KIRC) Project. PLoS ONE 2014, 9, e85621. [Google Scholar] [CrossRef]
- Qian, Y.; Daza, J.; Itzel, T.; Betge, J.; Zhan, T.; Marmé, F.; Teufel, A. Prognostic Cancer Gene Expression Signatures: Current Status and Challenges. Cells 2021, 10, 648. [Google Scholar] [CrossRef]
- Xi, X.; Li, T.; Huang, Y.; Sun, J.; Zhu, Y.; Yang, Y.; Lu, Z.J. RNA Biomarkers: Frontier of Precision Medicine for Cancer. Non-Coding RNA 2017, 3, 9. [Google Scholar] [CrossRef]
- Kamps, R.; Brandão, R.D.; van den Bosch, B.J.; Paulussen, A.D.C.; Xanthoulea, S.; Blok, M.J.; Romano, A. Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. Int. J. Mol. Sci. 2017, 18, 308. [Google Scholar] [CrossRef]
- Kapoor, J.; Ischia, J. Clinical Utility of Biomarkers in Renal Cell Carcinoma. In Molecular Biomarkers in Urologic Oncology, 1st ed.; Lotan, Y., Lawrentschuk, N., Schalken, J., Eds.; World Urologic Oncology Federation: Toronto, ON, Canada, 2020; pp. 365–380. [Google Scholar]
- Brannon, A.R.; Reddy, A.; Seiler, M.; Arreola, A.; Moore, D.T.; Pruthi, R.S.; Wallen, E.M.; Nielsen, M.E.; Liu, H.; Nathanson, K.L.; et al. Molecular Stratification of Clear Cell Renal Cell Carcinoma by Consensus Clustering Reveals Distinct Subtypes and Survival Patterns. Genes Cancer 2010, 1, 152–163. [Google Scholar] [CrossRef]
- Serie, D.J.; Joseph, R.W.; Cheville, J.C.; Ho, T.H.; Parasramka, M.; Hilton, T.; Thompson, R.H.; Leibovich, B.C.; Parker, A.S.; Eckel-Passow, J.E. Clear Cell Type A and B Molecular Subtypes in Metastatic Clear Cell Renal Cell Carcinoma: Tumor Heterogeneity and Aggressiveness. Eur. Urol. 2017, 71, 979–985. [Google Scholar] [CrossRef]
- Brooks, S.A.; Brannon, A.R.; Parker, J.S.; Fisher, J.C.; Sen, O.; Kattan, M.W.; Hakimi, A.A.; Hsieh, J.J.; Choueiri, T.K.; Tamboli, P.; et al. ClearCode34: A Prognostic Risk Predictor for Localized Clear Cell Renal Cell Carcinoma. Eur. Urol. 2014, 66, 77–84. [Google Scholar] [CrossRef]
- Ujfaludi, Z.; Kuthi, L.; Pankotai-Bodó, G.; Bankó, S.; Sükösd, F.; Pankotai, T. Novel Diagnostic Value of Driver Gene Transcription Signatures to Characterise Clear Cell Renal Cell Carcinoma, CcRCC. Pathol. Oncol. Res. 2022, 28, 1610345. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive Analysis of Normal Adjacent to Tumor Transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef] [PubMed]
- George, D.J.; Lee, C.-H.; Heng, D. New Approaches to First-Line Treatment of Advanced Renal Cell Carcinoma. Ther. Adv. Med. Oncol. 2021, 13, 17588359211034708. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Choueiri, T.K.; McDermott, D.F.; Powles, T.; Vano, Y.-A.; Gupta, S.; Yao, J.; Han, C.; Ammar, R.; Papillon-Cavanagh, S.; et al. Biomarker Analysis from CheckMate 214: Nivolumab plus Ipilimumab versus Sunitinib in Renal Cell Carcinoma. J. Immunother. Cancer 2022, 10, e004316. [Google Scholar] [CrossRef]
- Why Most Gene Expression Signatures of Tumors Have Not Been Useful in the Clinic|Science Translational Medicine. Available online: https://www.science.org/doi/abs/10.1126/scitranslmed.3000313?casa_token=P8jvrda_vxAAAAAA:JKPbMf7JoE3ajbM4Xq1HNYCPT1Wik2iQ5Dh6DsJd52eEJGcaIlKF6Dfe0oEPXibBnxbi8T_AwRDC (accessed on 2 October 2022).
- Yuan, Y.; Van Allen, E.M.; Omberg, L.; Wagle, N.; Amin-Mansour, A.; Sokolov, A.; Byers, L.A.; Xu, Y.; Hess, K.R.; Diao, L.; et al. Assessing the Clinical Utility of Cancer Genomic and Proteomic Data across Tumor Types. Nat. Biotechnol. 2014, 32, 644–652. [Google Scholar] [CrossRef]
- Bassanelli, M.; Borro, M.; Roberto, M.; Giannarelli, D.; Giacinti, S.; Di Martino, S.; Ceribelli, A.; Russo, A.; Aschelter, A.; Scarpino, S.; et al. A 17-Gene Expression Signature for Early Identification of Poor Prognosis in Clear Cell Renal Cell Carcinoma. Cancers 2022, 14, 178. [Google Scholar] [CrossRef]
- Ghatalia, P.; Gordetsky, J.; Kuo, F.; Dulaimi, E.; Cai, K.Q.; Devarajan, K.; Bae, S.; Naik, G.; Chan, T.A.; Uzzo, R.; et al. Prognostic Impact of Immune Gene Expression Signature and Tumor Infiltrating Immune Cells in Localized Clear Cell Renal Cell Carcinoma. J. Immunother. Cancer 2019, 7, 139. [Google Scholar] [CrossRef]
- Chibon, F. Cancer Gene Expression Signatures–The Rise and Fall? Eur. J. Cancer 2013, 49, 2000–2009. [Google Scholar] [CrossRef]
- Yao, M.; Huang, Y.; Shioi, K.; Hattori, K.; Murakami, T.; Sano, F.; Baba, M.; Kondo, K.; Nakaigawa, N.; Kishida, T.; et al. A Three-Gene Expression Signature Model to Predict Clinical Outcome of Clear Cell Renal Carcinoma. Int. J. Cancer 2008, 123, 1126–1132. [Google Scholar] [CrossRef]
- Zhan, Y.; Guo, W.; Zhang, Y.; Wang, Q.; Xu, X.; Zhu, L. A Five-Gene Signature Predicts Prognosis in Patients with Kidney Renal Clear Cell Carcinoma. Comput. Math. Methods Med. 2015, 2015, e842784. [Google Scholar] [CrossRef]
- Rini, B.; Goddard, A.; Knezevic, D.; Maddala, T.; Zhou, M.; Aydin, H.; Campbell, S.; Elson, P.; Koscielny, S.; Lopatin, M.; et al. A 16-Gene Assay to Predict Recurrence after Surgery in Localised Renal Cell Carcinoma: Development and Validation Studies. Lancet Oncol. 2015, 16, 676–685. [Google Scholar] [CrossRef]
- Boguslawska, J.; Kedzierska, H.; Poplawski, P.; Rybicka, B.; Tanski, Z.; Piekielko-Witkowska, A. Expression of Genes Involved in Cellular Adhesion and Extracellular Matrix Remodeling Correlates with Poor Survival of Patients with Renal Cancer. J. Urol. 2016, 195, 1892–1902. [Google Scholar] [CrossRef]
- Wan, F.; Zhu, Y.; Han, C.; Xu, Q.; Wu, J.; Dai, B.; Zhang, H.; Shi, G.; Gu, W.; Ye, D. Identification and Validation of an Eight-Gene Expression Signature for Predicting High Fuhrman Grade Renal Cell Carcinoma. Int. J. Cancer 2017, 140, 1199–1208. [Google Scholar] [CrossRef]
- Bao, L.; Zhao, Y.; Liu, C.; Cao, Q.; Huang, Y.; Chen, K.; Song, Z. The Identification of Key Gene Expression Signature and Biological Pathways in Metastatic Renal Cell Carcinoma. J. Cancer 2020, 11, 1712–1726. [Google Scholar] [CrossRef]
- Clark, D.J.; Zhang, H. Proteomic Approaches for Characterizing Renal Cell Carcinoma. Clin. Proteom. 2020, 17, 28. [Google Scholar] [CrossRef]
- Chen, L.; Kashina, A. Post-Translational Modifications of the Protein Termini. Front. Cell Dev. Biol. 2021, 9, 719590. [Google Scholar] [CrossRef]
- Drake, P.M.; Cho, W.; Li, B.; Prakobphol, A.; Johansen, E.; Anderson, N.L.; Regnier, F.E.; Gibson, B.W.; Fisher, S.J. Sweetening the Pot: Adding Glycosylation to the Biomarker Discovery Equation. Clin. Chem. 2010, 56, 223–236. [Google Scholar] [CrossRef]
- Zhang, H.; Zha, X.; Tan, Y.; Hornbeck, P.V.; Mastrangelo, A.J.; Alessi, D.R.; Polakiewicz, R.D.; Comb, M.J. Phosphoprotein Analysis Using Antibodies Broadly Reactive against Phosphorylated Motifs*. J. Biol. Chem. 2002, 277, 39379–39387. [Google Scholar] [CrossRef]
- Chu, G.; Xu, T.; Zhu, G.; Liu, S.; Niu, H.; Zhang, M. Identification of a Novel Protein-Based Signature to Improve Prognosis Prediction in Renal Clear Cell Carcinoma. Front. Mol. Biosci. 2021, 8, 623120. [Google Scholar] [CrossRef]
- Yang, J.; Yang, J.; Gao, Y.; Zhao, L.; Liu, L.; Qin, Y.; Wang, X.; Song, T.; Huang, C. Identification of Potential Serum Proteomic Biomarkers for Clear Cell Renal Cell Carcinoma. PLoS ONE 2014, 9, e111364. [Google Scholar] [CrossRef]
- Neely, B.A.; Wilkins, C.E.; Marlow, L.A.; Malyarenko, D.; Kim, Y.; Ignatchenko, A.; Sasinowska, H.; Sasinowski, M.; Nyalwidhe, J.O.; Kislinger, T.; et al. Proteotranscriptomic Analysis Reveals Stage Specific Changes in the Molecular Landscape of Clear-Cell Renal Cell Carcinoma. PLoS ONE 2016, 11, e0154074. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T.M.; Seeley, E.H.; Fadare, O.; Caprioli, R.M.; Clark, P.E. Imaging the Clear Cell Renal Cell Carcinoma Proteome. J. Urol. 2013, 189, 1097–1103. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Y.; Cai, Y.; Yu, H.; Li, H. ITRAQ-Based Quantitative Proteomic Analysis Identified HSC71 as a Novel Serum Biomarker for Renal Cell Carcinoma. Biomed. Res. Int. 2015, 2015, 802153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, H.; Xu, G.; Chu, N.; Xu, N.; Wen, H.; Gu, B.; Liu, J.; Mao, S.; Na, R.; et al. ITRAQ-Based Quantitative Proteomic Analysis Reveals Potential Early Diagnostic Markers of Clear-Cell Renal Cell Carcinoma. BioSci. Trends 2016, 10, 210–219. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Papale, M.; Vocino, G.; Lucarelli, G.; Rutigliano, M.; Gigante, M.; Rocchetti, M.T.; Pesce, F.; Sanguedolce, F.; Bufo, P.; Battaglia, M.; et al. Urinary RKIP/p-RKIP Is a Potential Diagnostic and Prognostic Marker of Clear Cell Renal Cell Carcinoma. Oncotarget 2017, 8, 40412–40424. [Google Scholar] [CrossRef]
- Qu, Y.; Feng, J.; Wu, X.; Bai, L.; Xu, W.; Zhu, L.; Liu, Y.; Xu, F.; Zhang, X.; Yang, G.; et al. A Proteogenomic Analysis of Clear Cell Renal Cell Carcinoma in a Chinese Population. Nat. Commun. 2022, 13, 2052. [Google Scholar] [CrossRef]
- Senturk, A.; Sahin, A.T.; Armutlu, A.; Kiremit, M.C.; Acar, O.; Erdem, S.; Bagbudar, S.; Esen, T.; Tuncbag, N.; Ozlu, N. Quantitative Proteomics Identifies Secreted Diagnostic Biomarkers as Well as Tumor-Dependent Prognostic Targets for Clear Cell Renal Cell Carcinoma. Mol. Cancer Res. 2021, 19, 1322–1337. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, L.; Peng, Z.; Yang, Y.; Feng, D.; He, J. Low Level of PDZ Domain Containing 1 (PDZK1) Predicts Poor Clinical Outcome in Patients with Clear Cell Renal Cell Carcinoma. EBioMedicine 2017, 15, 62–72. [Google Scholar] [CrossRef]
- Zhong, W.; Huang, C.; Lin, J.; Zhu, M.; Zhong, H.; Chiang, M.-H.; Chiang, H.-S.; Hui, M.-S.; Lin, Y.; Huang, J. Development and Validation of Nine-RNA Binding Protein Signature Predicting Overall Survival for Kidney Renal Clear Cell Carcinoma. Front. Genet. 2020, 11, 568192. [Google Scholar] [CrossRef]
- Zhu, Z.; He, A.; Lin, L.; Xu, C.; Cai, T.; Lin, J. Biological Functions and Prognostic Value of RNA Binding Proteins in Clear Cell Renal Cell Carcinoma. J. Cancer 2020, 11, 6591–6600. [Google Scholar] [CrossRef]
- Chinello, C.; L’imperio, V.; Stella, M.; Smith, A.J.; Bovo, G.; Grasso, A.; Grasso, M.; Raimondo, F.; Pitto, M.; Pagni, F.; et al. The Proteomic Landscape of Renal Tumors. Expert Rev. Proteom. 2016, 13, 1103–1120. [Google Scholar] [CrossRef]
- Sio, G.D.; Smith, A.J.; Galli, M.; Garancini, M.; Chinello, C.; Bono, F.; Pagni, F.; Magni, F. A MALDI-Mass Spectrometry Imaging Method Applicable to Different Formalin-Fixed Paraffin-Embedded Human Tissues. Mol. BioSyst. 2015, 11, 1507–1514. [Google Scholar] [CrossRef]
- Rodríguez-Suárez, E.; Siwy, J.; Zürbig, P.; Mischak, H. Urine as a Source for Clinical Proteome Analysis: From Discovery to Clinical Application. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2014, 1844, 884–898. [Google Scholar] [CrossRef]
- Nuerrula, Y.; Rexiati, M.; Liu, Q.; Wang, Y.-J. Differential Expression and Clinical Significance of Serum Protein among Patients with Clear-Cell Renal Cell Carcinoma. Cancer Biomark. 2015, 15, 485–491. [Google Scholar] [CrossRef]
- Raimondo, F.; Corbetta, S.; Chinello, C.; Pitto, M.; Magni, F. The Urinary Proteome and Peptidome of Renal Cell Carcinoma Patients: A Comparison of Different Techniques. Expert Rev. Proteom. 2014, 11, 503–514. [Google Scholar] [CrossRef]
- Thomas, S.; Hao, L.; Ricke, W.A.; Li, L. Biomarker Discovery in Mass Spectrometry-Based Urinary Proteomics. Proteom. Clin. Appl. 2016, 10, 358–370. [Google Scholar] [CrossRef]
- Clinical Manifestations, Evaluation, and Staging of Renal Cell Carcinoma-Up To Date. Available online: https://www.uptodate.com/contents/clinical-manifestations-evaluation-and-staging-of-renal-cell-carcinoma?search=renal%20cell%20carcinoma&source=search_result&selectedTitle=2~150&usage_type=default&display_rank=2 (accessed on 23 October 2022).
- Bin Satter, K.; Ramsey, Z.; Tran, P.M.H.; Hopkins, D.; Bearden, G.; Richardson, K.P.; Terris, M.K.; Savage, N.M.; Kavuri, S.K.; Purohit, S. Development of a Single Molecule Counting Assay to Differentiate Chromophobe Renal Cancer and Oncocytoma in Clinics. Cancers 2022, 14, 3242. [Google Scholar] [CrossRef]
- Battagli, C.; Uzzo, R.G.; Dulaimi, E.; Ibanez de Caceres, I.; Krassenstein, R.; Al-Saleem, T.; Greenberg, R.E.; Cairns, P. Promoter Hypermethylation of Tumor Suppressor Genes in Urine from Kidney Cancer Patients. Cancer Res. 2003, 63, 8695–8699. [Google Scholar]
- Hauser, S.; Zahalka, T.; Fechner, G.; Müller, S.C.; Ellinger, J. Serum DNA Hypermethylation in Patients with Kidney Cancer: Results of a Prospective Study. Anticancer Res. 2013, 33, 4651–4656. [Google Scholar]
- Hoque, M.O.; Begum, S.; Topaloglu, O.; Jeronimo, C.; Mambo, E.; Westra, W.H.; Califano, J.A.; Sidransky, D. Quantitative Detection of Promoter Hypermethylation of Multiple Genes in the Tumor, Urine, and Serum DNA of Patients with Renal Cancer. Cancer Res. 2004, 64, 5511–5517. [Google Scholar] [CrossRef]
- Cochetti, G.; Cari, L.; Nocentini, G.; Maulà, V.; Suvieri, C.; Cagnani, R.; Rossi De Vermandois, J.A.; Mearini, E. Detection of Urinary MiRNAs for Diagnosis of Clear Cell Renal Cell Carcinoma. Sci. Rep. 2020, 10, 21290. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Y.-Q.; Weng, W.-W.; Zhang, Q.-Y.; Yang, X.-Q.; Gan, H.-L.; Yang, Y.-S.; Zhang, P.-P.; Sun, M.-H.; Xu, M.-D.; et al. A Serum-Circulating Long Noncoding RNA Signature Can Discriminate between Patients with Clear Cell Renal Cell Carcinoma and Healthy Controls. Oncogenesis 2016, 5, e192. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.A.; Kondrashova, O.; Bradley, A.; Williams, E.D.; Pearson, J.V.; Waddell, N. Deep Learning in Cancer Diagnosis, Prognosis and Treatment Selection. Genome Med. 2021, 13, 152. [Google Scholar] [CrossRef] [PubMed]
- George, D.; Jonasch, E. Systemic Therapy of Advanced Clear Cell Renal Carcinoma-UpToDate. Available online: https://www.uptodate.com/contents/systemic-therapy-of-advanced-clear-cell-renal-carcinoma (accessed on 9 November 2022).
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.M.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 Blockade Induces Responses by Inhibiting Adaptive Immune Resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Samstein, R.M.; Lee, C.-H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor Mutational Load Predicts Survival after Immunotherapy across Multiple Cancer Types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch Repair Deficiency Predicts Response of Solid Tumors to PD-1 Blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, A.X.; Gartrell, R.D.; Silverman, A.M.; Aparicio, L.; Chu, T.; Bordbar, D.; Shan, D.; Samanamud, J.; Mahajan, A.; et al. Immune and Genomic Correlates of Response to Anti-PD-1 Immunotherapy in Glioblastoma. Nat. Med. 2019, 25, 462–469. [Google Scholar] [CrossRef]
- Mehnert, J.M.; Panda, A.; Zhong, H.; Hirshfield, K.; Damare, S.; Lane, K.; Sokol, L.; Stein, M.N.; Rodriguez-Rodriquez, L.; Kaufman, H.L.; et al. Immune Activation and Response to Pembrolizumab in POLE-Mutant Endometrial Cancer. J. Clin. Investig. 2016, 126, 2334–2340. [Google Scholar] [CrossRef]
- Skoulidis, F.; Goldberg, M.E.; Greenawalt, D.M.; Hellmann, M.D.; Awad, M.M.; Gainor, J.F.; Schrock, A.B.; Hartmaier, R.J.; Trabucco, S.E.; Gay, L.; et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018, 8, 822–835. [Google Scholar] [CrossRef]
- Li, X.; Lian, Z.; Wang, S.; Xing, L.; Yu, J. Interactions between EGFR and PD-1/PD-L1 Pathway: Implications for Treatment of NSCLC. Cancer Lett. 2018, 418, 1–9. [Google Scholar] [CrossRef]
- Wang, C.; Wang, H.; Wang, L. Biomarkers for Predicting the Efficacy of Immune Checkpoint Inhibitors. J. Cancer 2022, 13, 481–495. [Google Scholar] [CrossRef]
- Dowsett, M.; Sestak, I.; Lopez-Knowles, E.; Sidhu, K.; Dunbier, A.K.; Cowens, J.W.; Ferree, S.; Storhoff, J.; Schaper, C.; Cuzick, J. Comparison of PAM50 Risk of Recurrence Score with Oncotype DX and IHC4 for Predicting Risk of Distant Recurrence after Endocrine Therapy. J. Clin. Oncol. 2013, 31, 2783–2790. [Google Scholar] [CrossRef]
- Wallden, B.; Storhoff, J.; Nielsen, T.; Dowidar, N.; Schaper, C.; Ferree, S.; Liu, S.; Leung, S.; Geiss, G.; Snider, J.; et al. Development and Verification of the PAM50-Based Prosigna Breast Cancer Gene Signature Assay. BMC Med. Genom. 2015, 8, 54. [Google Scholar] [CrossRef]
- ESMO Interactive Guidelines. Available online: http://interactiveguidelines.esmo.org/esmo-web-app/gl_toc/index.php?GL_id=54 (accessed on 23 October 2022).
- Kiyoi, H.; Kawashima, N.; Ishikawa, Y. FLT3 Mutations in Acute Myeloid Leukemia: Therapeutic Paradigm beyond Inhibitor Development. Cancer Sci. 2020, 111, 312–322. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).