Increased Extrasynaptic Glutamate Escape in Stochastically Shaped Probabilistic Synaptic Environment
Abstract
1. Introduction
1.1. High-Affinity Astroglial Transporters Control Extrasynaptic Actions of Glutamate
1.2. Theoretical Models to Estimate Extrasynaptic Glutamate Escape
1.3. Glutamate Binding by Transporter-Enriched Astroglial Surfaces
2. Methods
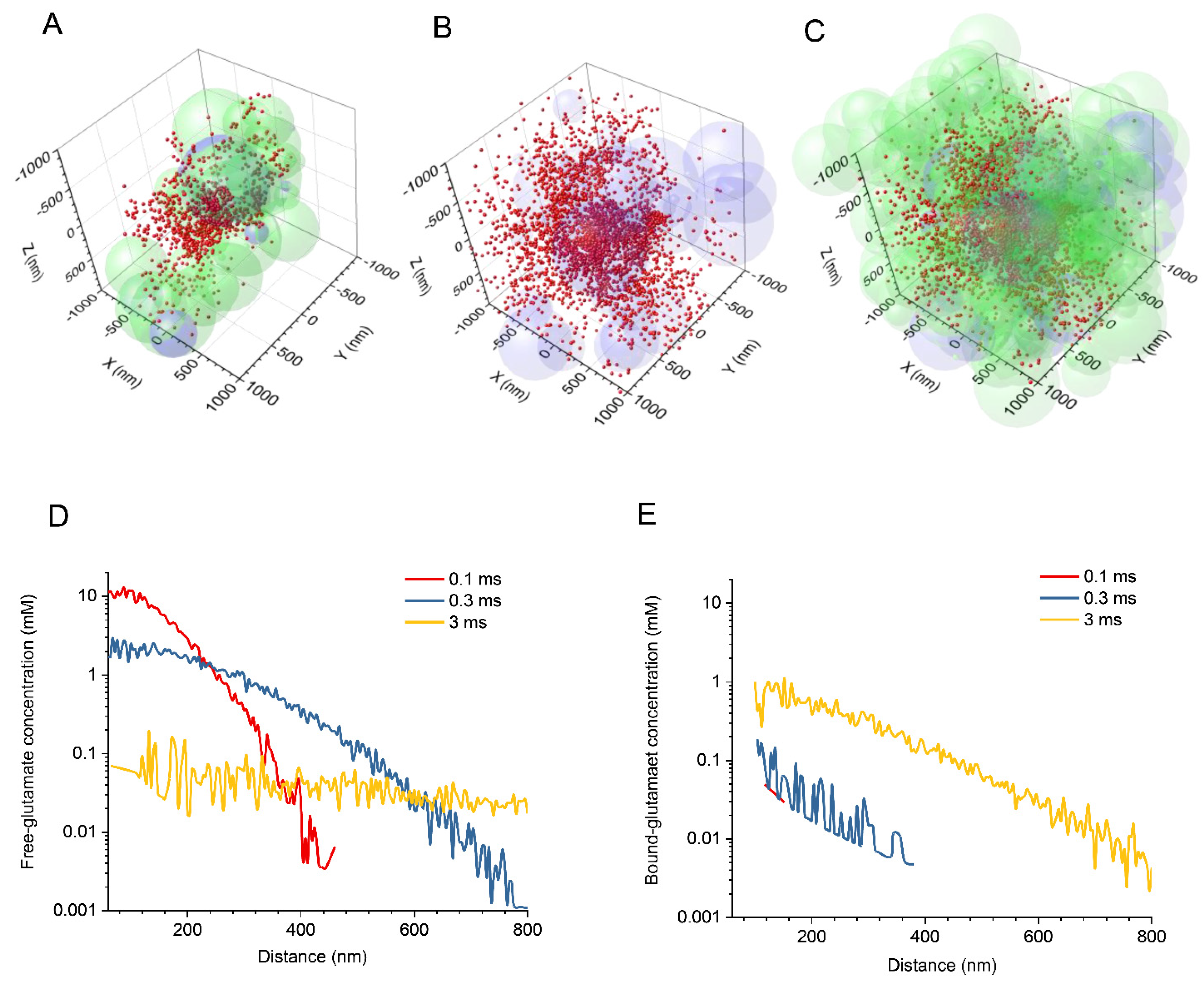
2.1. Stochastic Model of Neuropil Geometry
2.2. Interaction between Diffusing Molecules and Simulated Cell Surfaces
3. Results
3.1. Glutamate Release, Diffusion, and Transporter Binding
3.2. Exploring Free Parameters
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Motta, A.; Berning, M.; Boergens, K.M.; Staffler, B.; Beining, M.; Loomba, S.; Hennig, P.; Wissler, H.; Helmstaedter, M. Dense connectomic reconstruction in layer 4 of the somatosensory cortex. Science 2019, 366, eaay3134. [Google Scholar] [CrossRef]
- Bopp, R.; Holler-Rickauer, S.; Martin, K.A.; Schuhknecht, G.F. An Ultrastructural Study of the Thalamic Input to Layer 4 of Primary Motor and Primary Somatosensory Cortex in the Mouse. J. Neurosci. 2017, 37, 2435–2448. [Google Scholar] [CrossRef]
- Rusakov, D.A.; Kullmann, D.M. Extrasynaptic glutamate diffusion in the hippocampus: Ultrastructural constraints, uptake, and receptor activation. J. Neurosci. 1998, 18, 3158–3170. [Google Scholar] [CrossRef]
- Spacek, J. 3-Dimensional analysis of dendritic spines 3 Glial sheath. Anat. Embryol. 1985, 171, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Ventura, R.; Harris, K.M. Three-dimensional relationships between hippocampal synapses and astrocytes. J. Neurosci. 1999, 19, 6897–6906. [Google Scholar] [CrossRef]
- Witcher, M.R.; Park, Y.D.; Lee, M.R.; Sharma, S.; Harris, K.M.; Kirov, S.A. Three-dimensional relationships between perisynaptic astroglia and human hippocampal synapses. Glia 2010, 58, 572–587. [Google Scholar] [CrossRef]
- Patrushev, I.; Gavrilov, N.; Turlapov, V.; Semyanov, A. Subcellular location of astrocytic calcium stores favors extrasynaptic neuron-astrocyte communication. Cell Calcium 2013, 54, 343–349. [Google Scholar] [CrossRef]
- Bergles, D.E.; Jahr, C.E. Glial contribution to glutamate uptake at Schaffer collateral- commissural synapses in the hippocampus. J. Neurosci. 1998, 18, 7709–7716. [Google Scholar] [CrossRef] [PubMed]
- Danbolt, N.C. Glutamate uptake. Progr. Neurobiol. 2001, 65, 1–105. [Google Scholar] [CrossRef]
- Murphy-Royal, C.; Dupuis, J.; Groc, L.; Oliet, S.H.R. Astroglial glutamate transporters in the brain: Regulating neurotransmitter homeostasis and synaptic transmission. J. Neurosci. Res. 2017, 95, 2140–2151. [Google Scholar] [CrossRef]
- Verkhratsky, A.; Nedergaard, M. Physiology of Astroglia. Physiol. Rev. 2018, 98, 239–389. [Google Scholar] [CrossRef]
- Bazargani, N.; Attwell, D. Astrocyte calcium signaling: The third wave. Nat. Neurosci. 2016, 19, 182–189. [Google Scholar] [CrossRef]
- Lozovaya, N.A.; Kopanitsa, M.V.; Boychuk, Y.A.; Krishtal, O.A. Enhancement of glutamate release uncovers spillover-mediated transmission by N-methyl-D-aspartate receptors in the rat hippocampus. Neuroscience 1999, 91, 1321–1330. [Google Scholar] [CrossRef]
- Diamond, J.S. Neuronal glutamate transporters limit activation of NMDA receptors by neurotransmitter spillover on CA1 pyramidal cells. J. Neurosci. 2001, 21, 8328–8338. [Google Scholar] [CrossRef]
- Zheng, K.; Rusakov, D.A. Efficient integration of synaptic events by NMDA receptors in three-dimensional neuropil. Biophys. J. 2015, 108, 2457–2464. [Google Scholar] [CrossRef] [PubMed]
- Scimemi, A.; Fine, A.; Kullmann, D.M.; Rusakov, D.A. NR2B-containing receptors mediate cross talk among hippocampal synapses. J. Neurosci. 2004, 24, 4767–4777. [Google Scholar] [CrossRef] [PubMed]
- Scimemi, A.; Tian, H.; Diamond, J.S. Neuronal transporters regulate glutamate clearance, NMDA receptor activation, and synaptic plasticity in the hippocampus. J. Neurosci. 2009, 29, 14581–14595. [Google Scholar] [CrossRef]
- Szapiro, G.; Barbour, B. Multiple climbing fibers signal to molecular layer interneurons exclusively via glutamate spillover. Nat. Neurosci. 2007, 10, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Rose, C.R.; Felix, L.; Zeug, A.; Dietrich, D.; Reiner, A.; Henneberger, C. Astroglial Glutamate Signaling and Uptake in the Hippocampus. Front. Mol. Neurosci. 2017, 10, 451. [Google Scholar] [CrossRef]
- Tanaka, K.; Watase, K.; Manabe, T.; Yamada, K.; Watanabe, M.; Takahashi, K.; Iwama, H.; Nishikawa, T.; Ichihara, N.; Kikuchi, T.; et al. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science 1997, 276, 1699–1702. [Google Scholar] [CrossRef]
- Chu, K.; Lee, S.T.; Sinn, D.I.; Ko, S.Y.; Kim, E.H.; Kim, J.M.; Kim, S.J.; Park, D.K.; Jung, K.H.; Song, E.C.; et al. Pharmacological Induction of Ischemic Tolerance by Glutamate Transporter-1 (EAAT2) Upregulation. Stroke 2007, 38, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Sykova, E.; Nicholson, C. Diffusion in brain extracellular space. Physiol. Rev. 2008, 88, 1277–1340. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.R.; Binder, D.K. Post-translational Regulation of GLT-1 in Neurological Diseases and Its Potential as an Effective Therapeutic Target. Front. Mol. Neurosci. 2019, 12, 164. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.C.; Lambert, H.K.; Grossman, Y.S.; Dumitriu, D.; Waldman, R.; Jannetty, S.K.; Calakos, K.; Janssen, W.G.; McEwen, B.S.; Morrison, J.H. Glutamatergic regulation prevents hippocampal-dependent age-related cognitive decline through dendritic spine clustering. Proc. Natl. Acad. Sci. USA 2014, 111, 18733–18738. [Google Scholar] [CrossRef]
- Gass, J.T.; Sinclair, C.M.; Cleva, R.M.; Widholm, J.J.; Olive, M.F. Alcohol-seeking behavior is associated with increased glutamate transmission in basolateral amygdala and nucleus accumbens as measured by glutamate-oxidase-coated biosensors. Addict. Biol. 2011, 16, 215–228. [Google Scholar] [CrossRef]
- Gipson, C.D.; Reissner, K.J.; Kupchik, Y.M.; Smith, A.C.W.; Stankeviciute, N.; Hensley-Simon, M.E.; Kalivas, P.W. Reinstatement of nicotine seeking is mediated by glutamatergic plasticity. Proc. Natl. Acad. Sci. USA 2013, 110, 9124–9129. [Google Scholar] [CrossRef]
- McFarland, K.; Lapish, C.C.; Kalivas, P.W. Prefrontal glutamate release into the core of the nucleus accumbens mediates cocaine-induced reinstatement of drug-seeking behavior. J. Neurosci. 2003, 23, 3531–3537. [Google Scholar] [CrossRef]
- Shen, H.W.; Scofield, M.D.; Boger, H.; Hensley, M.; Kalivas, P.W. Synaptic glutamate spillover due to impaired glutamate uptake mediates heroin relapse. J. Neurosci. 2014, 34, 5649–5657. [Google Scholar] [CrossRef]
- Kruyer, A.; Scofield, M.D.; Wood, D.; Reissner, K.J.; Kalivas, P.W. Heroin Cue-Evoked Astrocytic Structural Plasticity at Nucleus Accumbens Synapses Inhibits Heroin Seeking. Biol. Psychiat. 2019, 86, 811–819. [Google Scholar] [CrossRef]
- Kruyer, A.; Angelis, A.; Garcia-Keller, C.; Li, H.; Kalivas, P.W. Plasticity in astrocyte subpopulations regulates heroin relapse. Sci. Adv. 2022, 8, eabo7044. [Google Scholar] [CrossRef]
- Marvin, J.S.; Borghuis, B.G.; Tian, L.; Cichon, J.; Harnett, M.T.; Akerboom, J.; Gordus, A.; Renninger, S.L.; Chen, T.W.; Bargmann, C.I.; et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 2013, 10, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.P.; Zheng, K.Y.; Cole, N.; Marvin, J.S.; Looger, L.L.; Rusakov, D.A. Multiplex imaging relates quantal glutamate release to presynaptic Ca2+ homeostasis at multiple synapses in situ. Nat. Commun. 2019, 10, 1414. [Google Scholar] [CrossRef] [PubMed]
- Arizono, M.; Inavalli, V.; Panatier, A.; Pfeiffer, T.; Angibaud, J.; Levet, F.; Ter Veer, M.J.T.; Stobart, J.; Bellocchio, L.; Mikoshiba, K.; et al. Structural basis of astrocytic Ca2+ signals at tripartite synapses. Nat. Commun. 2020, 11, 1906. [Google Scholar] [CrossRef] [PubMed]
- Barbour, B. An evaluation of synapse independence. J. Neurosci. 2001, 21, 7969–7984. [Google Scholar] [CrossRef]
- Franks, K.M.; Bartol, T.M., Jr.; Sejnowski, T.J. A Monte Carlo model reveals independent signaling at central glutamatergic synapses. Biophys. J. 2002, 83, 2333–2348. [Google Scholar] [CrossRef]
- Kinney, J.P.; Spacek, J.; Bartol, T.M.; Bajaj, C.L.; Harris, K.M.; Sejnowski, T.J. Extracellular sheets and tunnels modulate glutamate diffusion in hippocampal neuropil. J. Comp. Neurol. 2013, 521, 448–464. [Google Scholar] [CrossRef]
- Armbruster, M.; Dulla, C.G.; Diamond, J.S. Effects of fluorescent glutamate indicators on neurotransmitter diffusion and uptake. eLife 2020, 9, e54441. [Google Scholar] [CrossRef]
- Thorne, R.G.; Nicholson, C. In vivo diffusion analysis with quantum dots and dextrans predicts the width of brain extracellular space. Proc. Natl. Acad. Sci. USA 2006, 103, 5567–5572. [Google Scholar] [CrossRef]
- Tonnesen, J.; Inavalli, V.V.G.K.; Nagerl, U.V. Super-resolution imaging of the extracellular space in living brain tissue. Cell 2018, 172, 1108–1121. [Google Scholar] [CrossRef]
- Paviolo, C.; Soria, F.N.; Ferreira, J.S.; Lee, A.; Groc, L.; Bezard, E.; Cognet, L. Nanoscale exploration of the extracellular space in the live brain by combining single carbon nanotube tracking and super-resolution imaging analysis. Methods 2020, 174, 91–99. [Google Scholar] [CrossRef]
- Savtchenko, L.P.; Zheng, K.Y.; Rusakov, D.A. Buffering by Transporters Can Spare Geometric Hindrance in Controlling Glutamate Escape. Front. Cell. Neurosci. 2021, 15, 707813. [Google Scholar] [CrossRef] [PubMed]
- Grosche, J.; Matyash, V.; Moller, T.; Verkhratsky, A.; Reichenbach, A.; Kettenmann, H. Microdomains for neuron-glia interaction: Parallel fiber signaling to Bergmann glial cells. Nat. Neurosci. 1999, 2, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Borst, J.G.; Soria van Hoeve, J. The calyx of Held synapse: From model synapse to auditory relay. Annu. Rev. Physiol. 2012, 74, 199–224. [Google Scholar] [CrossRef] [PubMed]
- DiGregorio, D.A.; Nusser, Z.; Silver, R.A. Spillover of glutamate onto synaptic AMPA receptors enhances fast transmission at a cerebellar synapse. Neuron 2002, 35, 521–533. [Google Scholar] [CrossRef]
- Harris, K.M.; Jensen, F.E.; Tsao, B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: Implications for the maturation of synaptic physiology and long-term potentiation. J. Neurosci. 1992, 12, 2685–2705. [Google Scholar] [CrossRef]
- Vorisek, I.; Sykova, E. Ischemia-induced changes in the extracellular space diffusion parameters, K+, and pH in the developing rat cortex and corpus callosum. J. Cereb. Blood Flow Metab. 1997, 17, 191–203. [Google Scholar] [CrossRef]
- Thorne, R.G.; Lakkaraju, A.; Rodriguez-Boulan, E.; Nicholson, C. In vivo diffusion of lactoferrin in brain extracellular space is regulated by interactions with heparan sulfate. Proc. Natl. Acad. Sci. USA 2008, 105, 8416–8421. [Google Scholar] [CrossRef]
- Hrabetova, S.; Hrabe, J.; Nicholson, C. Dead-space microdomains hinder extracellular diffusion in rat neocortex during ischemia. J. Neurosci. 2003, 23, 8351–8359. [Google Scholar] [CrossRef]
- Savtchenko, L.P.; Bard, L.; Jensen, T.P.; Reynolds, J.P.; Kraev, I.; Medvedev, N.; Stewart, M.G.; Henneberger, C.; Rusakov, D.A. Disentangling astroglial physiology with a realistic cell model in silico. Nat. Commun. 2018, 9, 3554. [Google Scholar] [CrossRef]
- Bergles, D.E.; Dzubay, J.A.; Jahr, C.E. Glutamate transporter currents in Bergmann glial cells follow the time course of extrasynaptic glutamate. Proc. Natl. Acad. Sci. USA 1997, 94, 14821–14825. [Google Scholar] [CrossRef]
- Wadiche, J.I.; Arriza, J.L.; Amara, S.G.; Kavanaugh, M.P. Kinetics of a human glutamate transporter. Neuron 1995, 14, 1019–1027. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savtchenko, L.P.; Rusakov, D.A. Increased Extrasynaptic Glutamate Escape in Stochastically Shaped Probabilistic Synaptic Environment. Biomedicines 2022, 10, 2406. https://doi.org/10.3390/biomedicines10102406
Savtchenko LP, Rusakov DA. Increased Extrasynaptic Glutamate Escape in Stochastically Shaped Probabilistic Synaptic Environment. Biomedicines. 2022; 10(10):2406. https://doi.org/10.3390/biomedicines10102406
Chicago/Turabian StyleSavtchenko, Leonid P., and Dmitri A. Rusakov. 2022. "Increased Extrasynaptic Glutamate Escape in Stochastically Shaped Probabilistic Synaptic Environment" Biomedicines 10, no. 10: 2406. https://doi.org/10.3390/biomedicines10102406
APA StyleSavtchenko, L. P., & Rusakov, D. A. (2022). Increased Extrasynaptic Glutamate Escape in Stochastically Shaped Probabilistic Synaptic Environment. Biomedicines, 10(10), 2406. https://doi.org/10.3390/biomedicines10102406






