Exosomal MicroRNAs as Novel Cell-Free Therapeutics in Tissue Engineering and Regenerative Medicine
Abstract
:1. Introduction
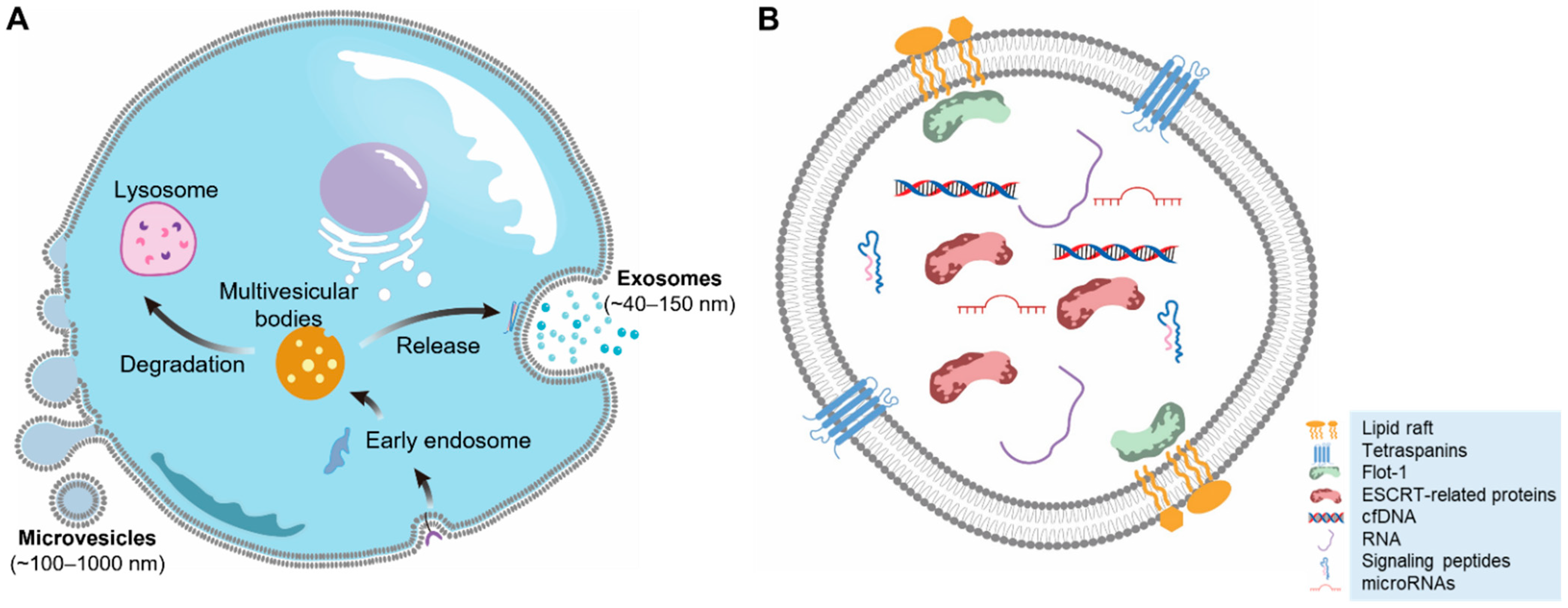
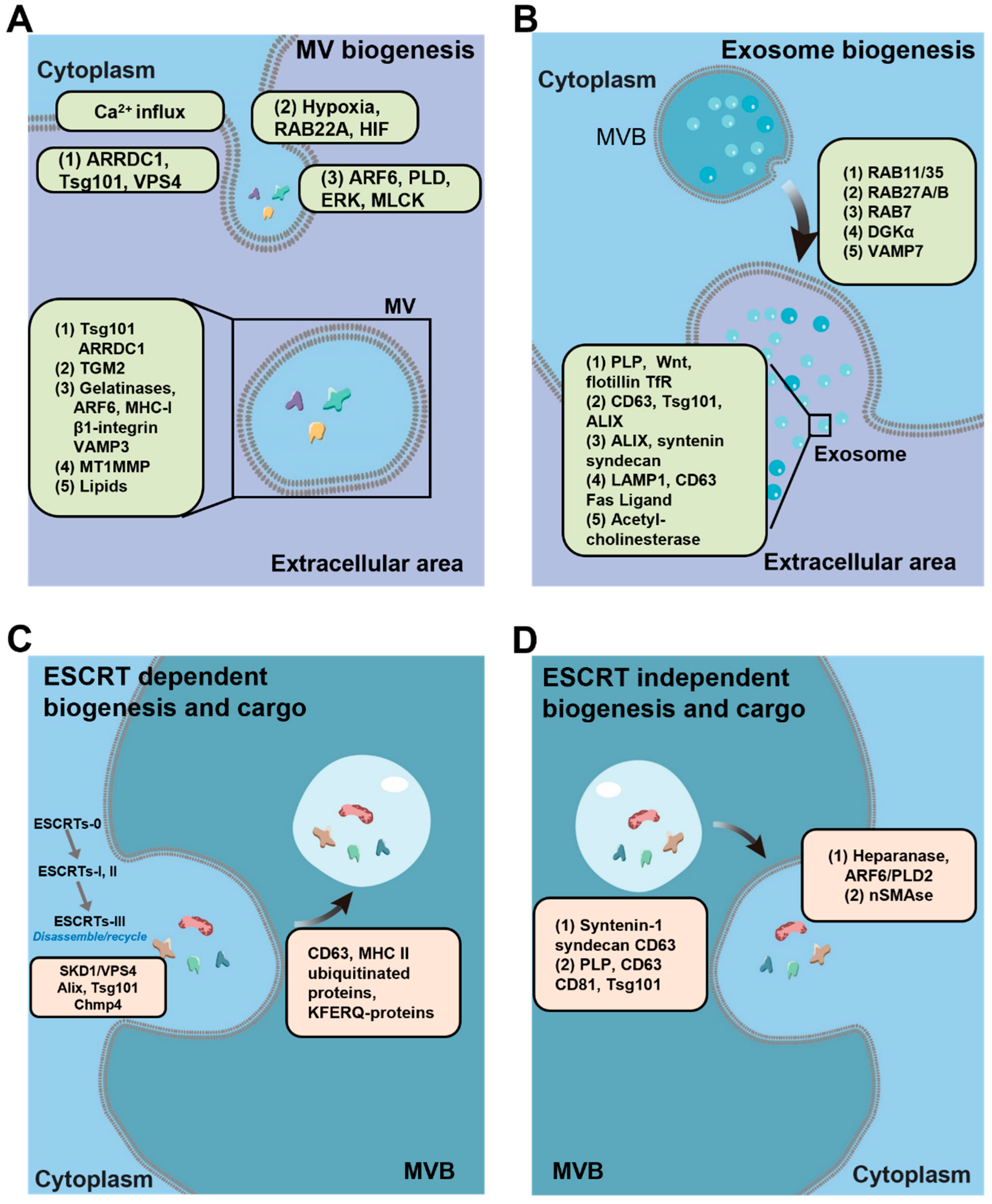
2. EV Biogenesis
2.1. Microvesicle Biogenesis
2.2. Exosome Biogenesis
2.2.1. ESCRT-Dependent Pathways
2.2.2. ESCRT-Independent Pathways
3. Exo-miRNA Loading and Sorting in EVs
4. Mechanism for EV Uptake by Recipient Cells and Exosomal miRNA Functions
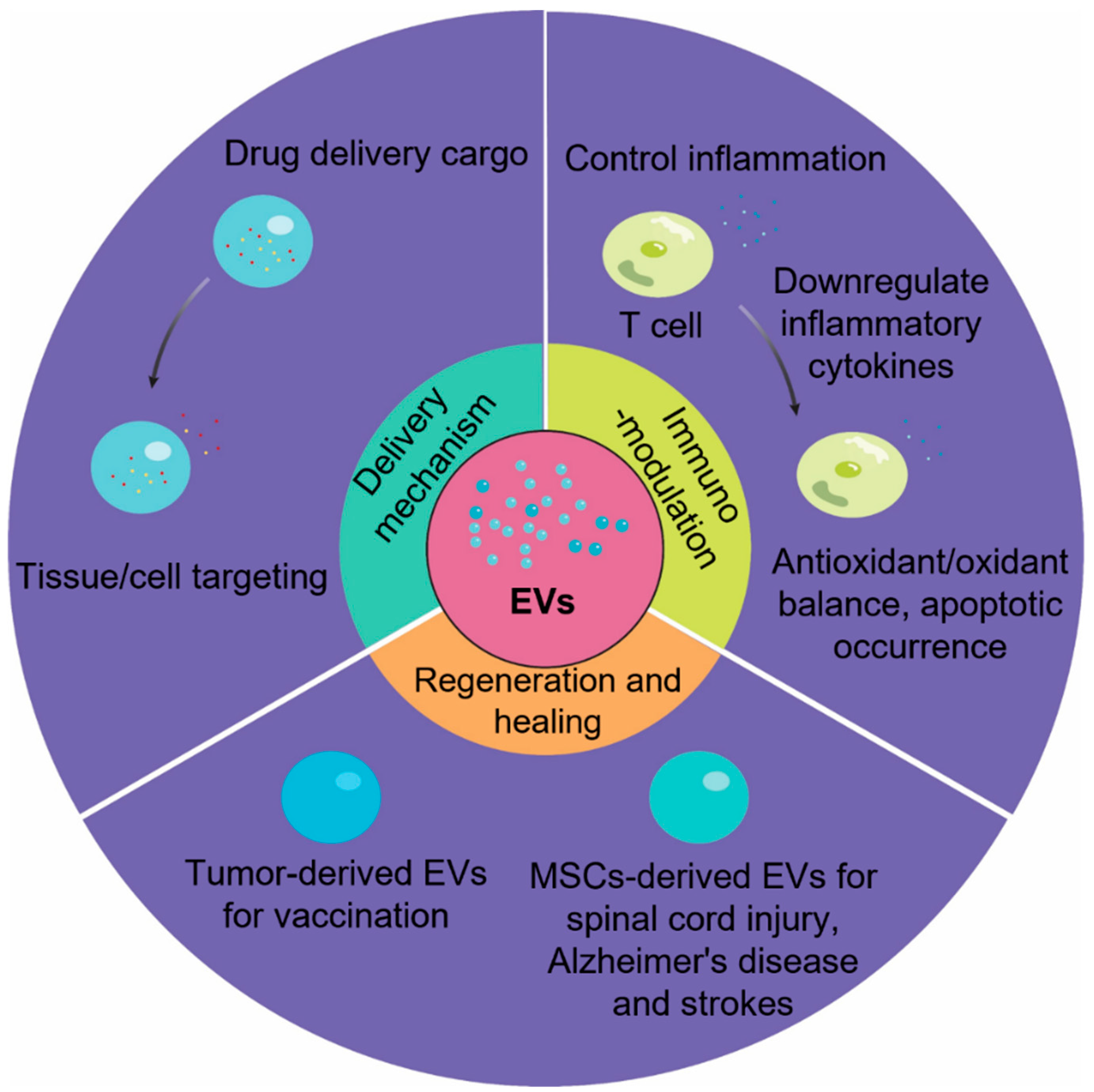
5. Engineering and Therapeutic Strategies with Exosomal miRs in Regenerative Medicine
5.1. Exo-miR from Mesenchymal Stem Cells (MSCs) in Bone-Associated Regeneration
5.2. Exo-miR from MSCs in Cancer Treatment
5.3. Exo-miR in Alzheimer’s Disease Pathology and Treatment
5.4. Exo-miR in Spinal Cord Injury and Treatment
5.5. Exo-miR from MSCs in Ischemic Diseases
5.6. Exo-miR Detection and EV Biomanufacturing
6. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Battistelli, M.; Falcieri, E. Apoptotic bodies: Particular extracellular vesicles involved in intercellular communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, T.K.; Ozkocak, D.C.; Poon, I.K.H. Unleashing the therapeutic potential of apoptotic bodies. Biochem. Soc. Trans. 2020, 48, 2079–2088. [Google Scholar] [CrossRef]
- Meehan, B.; Rak, J.; Di Vizio, D. Oncosomes-large and small: What are they, where they came from? J. Extracell Vesicles 2016, 5, 33109. [Google Scholar] [CrossRef]
- Rich, J.N. Cancer stem cells in radiation resistance. Cancer Res. 2007, 67, 8980–8984. [Google Scholar] [CrossRef] [Green Version]
- Hauser, P.; Wang, S.; Didenko, V.V. Apoptotic bodies: Selective detection in extracellular vesicles. In Signal Transduction Immunohistochemistry; Springer: Berlin/Heidelberg, Germany, 2017; pp. 193–200. [Google Scholar]
- Muralidharan-Chari, V.; Clancy, J.; Plou, C.; Romao, M.; Chavrier, P.; Raposo, G.; D’Souza-Schorey, C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. 2009, 19, 1875–1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Antonyak, M.A.; Zhang, J.; Cerione, R.A. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene 2012, 31, 4740–4749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neuro-Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Schlienger, S.; Campbell, S.; Claing, A. ARF1 regulates the Rho/MLC pathway to control EGF-dependent breast cancer cell invasion. Mol. Biol. Cell 2014, 25, 17–29. [Google Scholar] [CrossRef]
- Sedgwick, A.E.; Clancy, J.W.; Olivia Balmert, M.; D’Souza-Schorey, C. Extracellular microvesicles and invadopodia mediate non-overlapping modes of tumor cell invasion. Sci. Rep. 2015, 5, 14748. [Google Scholar] [CrossRef]
- Pap, E.; Pallinger, E.; Pasztoi, M.; Falus, A. Highlights of a new type of intercellular communication: Microvesicle-based information transfer. Inflamm. Res. 2009, 58, 1–8. [Google Scholar] [CrossRef]
- Bianco, F.; Perrotta, C.; Novellino, L.; Francolini, M.; Riganti, L.; Menna, E.; Saglietti, L.; Schuchman, E.H.; Furlan, R.; Clementi, E. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009, 28, 1043–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Pendurthi, U.R.; Rao, L.V.M. Sphingomyelin encrypts tissue factor: ATP-induced activation of A-SMase leads to tissue factor decryption and microvesicle shedding. Blood Adv. 2017, 1, 849–862. [Google Scholar] [CrossRef]
- Marrone, M.C.; Morabito, A.; Giustizieri, M.; Chiurchiù, V.; Leuti, A.; Mattioli, M.; Marinelli, S.; Riganti, L.; Lombardi, M.; Murana, E. TRPV1 channels are critical brain inflammation detectors and neuropathic pain biomarkers in mice. Nat. Commun. 2017, 8, 15292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Conde, I.; Shrimpton, C.N.; Thiagarajan, P.; López, J.A. Tissue-factor–bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood 2005, 106, 1604–1611. [Google Scholar] [CrossRef] [PubMed]
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Raymond, C.K.; Howald-Stevenson, I.; Vater, C.; Stevens, T. Morphological classification of the yeast vacuolar protein sorting mutants: Evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 1992, 3, 1389–1402. [Google Scholar] [CrossRef] [Green Version]
- McCullough, J.; Frost, A.; Sundquist, W.I. Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu. Rev. Cell Dev. Biol. 2018, 34, 85–109. [Google Scholar] [CrossRef] [PubMed]
- Garrus, J.E.; von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Côté, M.; Rich, R.L. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef]
- Wang, T.; Gilkes, D.M.; Takano, N.; Xiang, L.; Luo, W.; Bishop, C.J.; Chaturvedi, P.; Green, J.J.; Semenza, G.L. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2014, 111, E3234–E3242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.; Roulson, J.-A.; Hart, C.A.; Tawadros, T.; Clarke, N.W. Arachidonic acid induction of Rho-mediated transendothelial migration in prostate cancer. Br. J. Cancer 2014, 110, 2099–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Morley, S.; Le, M.; Bedoret, D.; Umetsu, D.T.; Di Vizio, D.; Freeman, M.R. Enhanced shedding of extracellular vesicles from amoeboid prostate cancer cells: Potential effects on the tumor microenvironment. Cancer Biol. Ther. 2014, 15, 409–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConnell, R.E.; Higginbotham, J.N.; Shifrin, D.A., Jr.; Tabb, D.L.; Coffey, R.J.; Tyska, M.J. The enterocyte microvillus is a vesicle-generating organelle. J. Cell Biol. 2009, 185, 1285–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H. The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 463–487. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; Van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Mayers, J.R.; Wang, L.; Edwardson, J.M.; Audhya, A. Hrs and STAM function synergistically to bind ubiquitin-modified cargoes in vitro. Biophys. J. 2015, 108, 76–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katzmann, D.J.; Babst, M.; Emr, S.D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001, 106, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Bilodeau, P.S.; Winistorfer, S.C.; Kearney, W.R.; Robertson, A.D.; Piper, R.C. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 2003, 163, 237–243. [Google Scholar] [CrossRef]
- Gill, D.J.; Teo, H.; Sun, J.; Perisic, O.; Veprintsev, D.B.; Emr, S.D.; Williams, R.L. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. EMBO J. 2007, 26, 600–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostelansky, M.S.; Schluter, C.; Tam, Y.Y.C.; Lee, S.; Ghirlando, R.; Beach, B.; Conibear, E.; Hurley, J.H. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell 2007, 129, 485–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Souza, R.F.; Aravind, L. UMA and MABP domains throw light on receptor endocytosis and selection of endosomal cargoes. Bioinformatics 2010, 26, 1477–1480. [Google Scholar] [CrossRef] [Green Version]
- Im, Y.J.; Hurley, J.H. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev. Cell 2008, 14, 902–913. [Google Scholar] [CrossRef] [Green Version]
- Hierro, A.; Sun, J.; Rusnak, A.S.; Kim, J.; Prag, G.; Emr, S.D.; Hurley, J.H. Structure of the ESCRT-II endosomal trafficking complex. Nature 2004, 431, 221–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, H.; Perisic, O.; González, B.; Williams, R.L. ESCRT-II, an endosome-associated complex required for protein sorting: Crystal structure and interactions with ESCRT-III and membranes. Dev. Cell 2004, 7, 559–569. [Google Scholar] [CrossRef] [Green Version]
- Teis, D.; Saksena, S.; Emr, S.D. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 2008, 15, 578–589. [Google Scholar] [CrossRef]
- Wollert, T.; Wunder, C.; Lippincott-Schwartz, J.; Hurley, J.H. Membrane scission by the ESCRT-III complex. Nature 2009, 458, 172–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonciarz, M.D.; Whitby, F.G.; Eckert, D.M.; Kieffer, C.; Heroux, A.; Sundquist, W.I.; Hill, C.P. Biochemical and structural studies of yeast Vps4 oligomerization. J. Mol. Biol. 2008, 384, 878–895. [Google Scholar] [CrossRef] [Green Version]
- Yeo, S.C.; Xu, L.; Ren, J.; Boulton, V.J.; Wagle, M.D.; Liu, C.; Ren, G.; Wong, P.; Zahn, R.; Sasajala, P. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 2003, 116, 3957–3970. [Google Scholar] [CrossRef] [Green Version]
- Raiborg, C.; Bremnes, B.; Mehlum, A.; Gillooly, D.J.; D’Arrigo, A.; Stang, E.; Stenmark, H. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 2001, 114, 2255–2263. [Google Scholar] [CrossRef]
- Razi, M.; Futter, C. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol. Biol. Cell 2006, 17, 3469–3483. [Google Scholar] [CrossRef] [Green Version]
- Slagsvold, T.; Aasland, R.; Hirano, S.; Bache, K.G.; Raiborg, C.; Trambaiolo, D.; Wakatsuki, S.; Stenmark, H. Eap45 in Mammalian ESCRT-II Binds Ubiquitin via a Phosphoinositide-interacting GLUE Domain*♦. J. Biol. Chem. 2005, 280, 19600–19606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böker, K.O.; Lemus-Diaz, N.; Ferreira, R.R.; Schiller, L.; Schneider, S.; Gruber, J. The impact of the CD9 tetraspanin on lentivirus infectivity and exosome secretion. Mol. Ther. 2018, 26, 634–647. [Google Scholar] [CrossRef] [Green Version]
- Adell, M.A.Y.; Migliano, S.M.; Upadhyayula, S.; Bykov, Y.S.; Sprenger, S.; Pakdel, M.; Vogel, G.F.; Jih, G.; Skillern, W.; Behrouzi, R. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife 2017, 6, e31652. [Google Scholar] [CrossRef]
- Chiaruttini, N.; Redondo-Morata, L.; Colom, A.; Humbert, F.; Lenz, M.; Scheuring, S.; Roux, A. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 2015, 163, 866–879. [Google Scholar] [CrossRef] [Green Version]
- Coulter, M.E.; Dorobantu, C.M.; Lodewijk, G.A.; Delalande, F.; Cianférani, S.; Ganesh, V.S.; Smith, R.S.; Lim, E.T.; Xu, C.S.; Pang, S. The ESCRT-III protein CHMP1A mediates secretion of sonic hedgehog on a distinctive subtype of extracellular vesicles. Cell Rep. 2018, 24, 973–986.e8. [Google Scholar] [CrossRef]
- Bishop, N.; Woodman, P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell 2000, 11, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Fujita, H.; Yamanaka, M.; Imamura, K.; Tanaka, Y.; Nara, A.; Yoshimori, T.; Yokota, S.; Himeno, M. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: Class E vps phenotype in mammalian cells. J. Cell Sci. 2003, 116, 401–414. [Google Scholar] [CrossRef] [Green Version]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428. [Google Scholar] [CrossRef] [Green Version]
- Laulagnier, K.; Grand, D.; Dujardin, A.; Hamdi, S.; Vincent-Schneider, H.; Lankar, D.; Salles, J.-P.; Bonnerot, C.; Perret, B.; Record, M. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 2004, 572, 11–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghossoub, R.; Lembo, F.; Rubio, A.; Gaillard, C.B.; Bouchet, J.; Vitale, N.; Slavík, J.; Machala, M.; Zimmermann, P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014, 5, 3477. [Google Scholar] [CrossRef] [Green Version]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Edgar, J.R.; Eden, E.R.; Futter, C.E. Hrs-and CD63-dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic 2014, 15, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Chen, T.; Guo, J.; Yang, M.; Zhu, X.; Cao, X. Chemokine-containing exosomes are released from heat-stressed tumor cells via lipid raft-dependent pathway and act as efficient tumor vaccine. J. Immunol. 2011, 186, 2219–2228. [Google Scholar] [CrossRef]
- Möbius, W.; Van Donselaar, E.; Ohno-Iwashita, Y.; Shimada, Y.; Heijnen, H.; Slot, J.; Geuze, H. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic 2003, 4, 222–231. [Google Scholar] [CrossRef]
- Kajimoto, T.; Okada, T.; Miya, S.; Zhang, L.; Nakamura, S. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat. Commun. 2013, 4, 2712. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and-dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; Bergam, P.; Di Cicco, A.; Hurbain, I.; Cicero, A.L.; Dingli, F.; Palmulli, R.; Fort, C.; Potier, M.C.; Schurgers, L.J. Apolipoprotein E regulates amyloid formation within endosomes of pigment cells. Cell Rep. 2015, 13, 43–51. [Google Scholar] [CrossRef]
- Hurwitz, S.N.; Nkosi, D.; Conlon, M.M.; York, S.B.; Liu, X.; Tremblay, D.C.; Meckes, D.G., Jr. CD63 regulates epstein-barr virus LMP1 exosomal packaging, enhancement of vesicle production, and noncanonical NF-κB signaling. J. Virol. 2017, 91, e02251-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurwitz, S.N.; Cheerathodi, M.R.; Nkosi, D.; York, S.B.; Meckes, D.G., Jr. Tetraspanin CD63 bridges autophagic and endosomal processes to regulate exosomal secretion and intracellular signaling of Epstein-Barr virus LMP1. J. Virol. 2018, 92, e01969-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauthier, S.A.; Pérez-González, R.; Sharma, A.; Huang, F.-K.; Alldred, M.J.; Pawlik, M.; Kaur, G.; Ginsberg, S.D.; Neubert, T.A.; Levy, E. Enhanced exosome secretion in Down syndrome brain-a protective mechanism to alleviate neuronal endosomal abnormalities. Acta Neuropathol. Commun. 2017, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Mazurov, D.; Barbashova, L.; Filatov, A. Tetraspanin protein CD 9 interacts with metalloprotease CD 10 and enhances its release via exosomes. FEBS J. 2013, 280, 1200–1213. [Google Scholar] [CrossRef]
- Buschow, S.I.; Nolte-‘T Hoen, E.N.; Van Niel, G.; Pols, M.S.; Ten Broeke, T.; Lauwen, M.; Ossendorp, F.; Melief, C.J.; Raposo, G.; Wubbolts, R. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic 2009, 10, 1528–1542. [Google Scholar] [CrossRef]
- Chairoungdua, A.; Smith, D.L.; Pochard, P.; Hull, M.; Caplan, M.J. Exosome release of β-catenin: A novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 2010, 190, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Guix, F.X.; Sannerud, R.; Berditchevski, F.; Arranz, A.M.; Horré, K.; Snellinx, A.; Thathiah, A.; Saido, T.; Saito, T.; Rajesh, S. Tetraspanin 6: A pivotal protein of the multiple vesicular body determining exosome release and lysosomal degradation of amyloid precursor protein fragments. Mol. Neurodegener. 2017, 12, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zöller, M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678. [Google Scholar] [CrossRef] [Green Version]
- Hirano, S.; Kawasaki, M.; Ura, H.; Kato, R.; Raiborg, C.; Stenmark, H.; Wakatsuki, S. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat. Struct. Mol. Biol. 2006, 13, 272–277. [Google Scholar] [CrossRef]
- Ren, X.; Hurley, J.H. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010, 29, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Raiborg, C.; Wesche, J.; Malerød, L.; Stenmark, H. Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains. J. Cell Sci. 2006, 119, 2414–2424. [Google Scholar] [CrossRef] [Green Version]
- Shields, S.B.; Oestreich, A.J.; Winistorfer, S.; Nguyen, D.; Payne, J.A.; Katzmann, D.J.; Piper, R. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 2009, 185, 213–224. [Google Scholar] [CrossRef]
- Gangalum, R.K.; Atanasov, I.C.; Zhou, Z.H.; Bhat, S.P. αB-crystallin is found in detergent-resistant membrane microdomains and is secreted via exosomes from human retinal pigment epithelial cells. J. Biol. Chem. 2011, 286, 3261–3269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Gassart, A.; Géminard, C.; Février, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells*♦. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [Green Version]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.-A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.; Taunk, N.K. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.J.; Le Bert, N.; Chitre, A.A.; Koo, C.X.E.; Nga, X.H.; Ho, S.S.; Khatoo, M.; Tan, N.Y.; Ishii, K.J.; Gasser, S. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep. 2015, 11, 460–473. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. [Google Scholar] [CrossRef] [Green Version]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; LeBleu, V.S. Discovery of double-stranded genomic DNA in circulating exosomes. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 275–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitai, Y.; Kawasaki, T.; Sueyoshi, T.; Kobiyama, K.; Ishii, K.J.; Zou, J.; Akira, S.; Matsuda, T.; Kawai, T. DNA-containing exosomes derived from cancer cells treated with topotecan activate a STING-dependent pathway and reinforce antitumor immunity. J. Immunol. 2017, 198, 1649–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, Q.; Xu, J.; Yan, S.; Huang, M.; Ding, H.; Sun, X.; Bi, A.; Ding, J.; Sun, B.; Geng, M. Chemotherapy-induced intestinal inflammatory responses are mediated by exosome secretion of double-strand DNA via AIM2 inflammasome activation. Cell Res. 2017, 27, 784–800. [Google Scholar] [CrossRef] [PubMed]
- Lázaro-Ibáñez, E.; Sanz-Garcia, A.; Visakorpi, T.; Escobedo-Lucea, C.; Siljander, P.; Ayuso-Sacido, Á.; Yliperttula, M. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: Apoptotic bodies, microvesicles, and exosomes. Prostate 2014, 74, 1379–1390. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [Green Version]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef]
- Xiao, D.; Ohlendorf, J.; Chen, Y.; Taylor, D.D.; Rai, S.N.; Waigel, S.; Zacharias, W.; Hao, H.; McMasters, K.M. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE 2012, 7, e46874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Goldie, B.J.; Dun, M.D.; Lin, M.; Smith, N.D.; Verrills, N.M.; Dayas, C.V.; Cairns, M.J. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014, 42, 9195–9208. [Google Scholar] [CrossRef] [Green Version]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [Green Version]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [Green Version]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 2016, 5, e19276. [Google Scholar] [CrossRef]
- Shurtleff, M.J.; Yao, J.; Qin, Y.; Nottingham, R.M.; Temoche-Diaz, M.M.; Schekman, R.; Lambowitz, A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E8987–E8995. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.S.; Pressman, S.; Andress, A.P.; Kim, K.; White, J.L.; Cassidy, J.J.; Li, X.; Lubell, K.; Lim, D.H.; Cho, I.S. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009, 11, 1150–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Pan, X.; Fu, R. Role and Function of T Cell-Derived Exosomes and Their Therapeutic Value. Mediat. Inflamm. 2021, 2021, 8481013. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Tkach, M.; Kowal, J.; Zucchetti, A.E.; Enserink, L.; Jouve, M.; Lankar, D.; Saitakis, M.; Martin-Jaular, L.; Théry, C. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. EMBO J. 2017, 36, 3012–3028. [Google Scholar] [CrossRef]
- Hemler, M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003, 19, 397–422. [Google Scholar] [CrossRef]
- Nolte-‘t Hoen, E.N.; Buschow, S.I.; Anderton, S.M.; Stoorvogel, W.; Wauben, M.H. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood J. Am. Soc. Hematol. 2009, 113, 1977–1981. [Google Scholar] [CrossRef] [Green Version]
- Rana, S.; Yue, S.; Stadel, D.; Zöller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef]
- Klibi, J.; Niki, T.; Riedel, A.; Pioche-Durieu, C.; Souquere, S.; Rubinstein, E.; Le Moulec, S.; Guigay, J.; Hirashima, M.; Guemira, F. Blood diffusion and Th1-suppressive effects of galectin-9–containing exosomes released by Epstein-Barr virus–infected nasopharyngeal carcinoma cells. Blood J. Am. Soc. Hematol. 2009, 113, 1957–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Céspedes, P.F.; Jainarayanan, A.; Fernández-Messina, L.; Valvo, S.; Saliba, D.G.; Kurz, E.; Kvalvaag, A.; Chen, L.; Ganskow, C.; Colin-York, H. T-cell trans-synaptic vesicles are distinct and carry greater effector content than constitutive extracellular vesicles. Nat. Commun. 2022, 13, 3460. [Google Scholar] [CrossRef]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Tian, T.; Zhu, Y.-L.; Zhou, Y.-Y.; Liang, G.-F.; Wang, Y.-Y.; Hu, F.-H.; Xiao, Z.-D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [Green Version]
- Feng, D.; Zhao, W.L.; Ye, Y.Y.; Bai, X.C.; Liu, R.Q.; Chang, L.F.; Zhou, Q.; Sui, S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef]
- Laulagnier, K.; Javalet, C.; Hemming, F.J.; Chivet, M.; Lachenal, G.; Blot, B.; Chatellard, C.; Sadoul, R. Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell. Mol. Life Sci. 2018, 75, 757–773. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [Green Version]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Coleman, B.M.; Hill, A.F. Extracellular vesicles–Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin. Cell Dev. Biol. 2015, 40, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, H.M.; Masoumi, N.; Witwer, K.W.; Pegtel, D.M. Extracellular vesicles exploit viral entry routes for cargo delivery. Microbiol. Mol. Biol. Rev. 2016, 80, 369–386. [Google Scholar] [CrossRef] [PubMed]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Beer Stolz, D.; Sullivan, M.L.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood J. Am. Soc. Hematol. 2012, 119, 756–766. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. eLife 2016, 5, e10250. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Zhai, S.; Fu, Q.; Liu, D. Bone marrow mesenchymal stem cells-derived exosomal microRNA-150-3p promotes osteoblast proliferation and differentiation in osteoporosis. Hum. Gene Ther. 2021, 32, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Omar, O.; Vazirisani, F.; Thomsen, P.; Ekström, K. Mesenchymal stem cell-derived exosomes have altered microRNA profiles and induce osteogenic differentiation depending on the stage of differentiation. PLoS ONE 2018, 13, e0193059. [Google Scholar] [CrossRef] [Green Version]
- Mao, G.; Zhang, Z.; Hu, S.; Zhang, Z.; Chang, Z.; Huang, Z.; Liao, W.; Kang, Y. Exosomes derived from miR-92a-3p-overexpressing human mesenchymal stem cells enhance chondrogenesis and suppress cartilage degradation via targeting WNT5A. Stem Cell Res. Ther. 2018, 9, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.; Tian, G.; Yang, Z.; Gao, X.; Wang, F.; Li, J.; Tian, Z.; Huang, B.; Wei, F.; Sang, X. Enhancement of acellular cartilage matrix scaffold by Wharton’s jelly mesenchymal stem cell-derived exosomes to promote osteochondral regeneration. Bioact. Mater. 2021, 6, 2711–2728. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ding, X.; Zhou, L.; Zhang, L.; Yang, X. Mesenchymal stem cells-derived exosomal microRNA-139-5p restrains tumorigenesis in bladder cancer by targeting PRC1. Oncogene 2021, 40, 246–261. [Google Scholar] [CrossRef]
- Ma, Y.-S.; Liu, J.-B.; Lin, L.; Zhang, H.; Wu, J.-J.; Shi, Y.; Jia, C.-Y.; Zhang, D.-D.; Yu, F.; Wang, H.-M. Exosomal microRNA-15a from mesenchymal stem cells impedes hepatocellular carcinoma progression via downregulation of SALL4. Cell Death Discov. 2021, 7, 224. [Google Scholar] [CrossRef]
- Jeong, K.; Yu, Y.J.; You, J.Y.; Rhee, W.J.; Kim, J.A. Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model. Lab Chip 2020, 20, 548–557. [Google Scholar] [CrossRef]
- Hou, C.; Sun, N.; Han, W.; Meng, Y.; Wang, C.; Zhu, Q.; Tang, Y.; Ye, J. Exosomal microRNA-23b-3p promotes tumor angiogenesis and metastasis by targeting PTEN in Salivary adenoid cystic carcinoma. Carcinogenesis 2022, 43, 682–692. [Google Scholar] [CrossRef]
- Ma, Y.-S.; Wu, T.-M.; Ling, C.-C.; Yu, F.; Zhang, J.; Cao, P.-S.; Gu, L.-P.; Wang, H.-M.; Xu, H.; Li, L. M2 macrophage-derived exosomal microRNA-155-5p promotes the immune escape of colon cancer by downregulating ZC3H12B. Mol. Ther.-Oncolytics 2021, 20, 484–498. [Google Scholar] [CrossRef]
- Yang, C.; Dou, R.; Wei, C.; Liu, K.; Shi, D.; Zhang, C.; Liu, Q.; Wang, S.; Xiong, B. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol. Ther. 2021, 29, 2088–2107. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Liu, Y.; Fang, X.; Liu, Y.; Fang, L.; Lin, L.; Liu, X.; Wang, N. Tumor-derived microRNA-494 promotes angiogenesis in non-small cell lung cancer. Angiogenesis 2015, 18, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-A.; Lu, C.-H.; Ke, C.-C.; Chiu, S.-J.; Jeng, F.-S.; Chang, C.-W.; Yang, B.-H.; Liu, R.-S. Mesenchymal stem cell-derived exosomes ameliorate Alzheimer’s disease pathology and improve cognitive deficits. Biomedicines 2021, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Jäkel, L.; Bruinsma, I.B.; Claassen, J.A.; Kuiperij, H.B.; Verbeek, M.M. MicroRNA-29a is a candidate biomarker for Alzheimer’s disease in cell-free cerebrospinal fluid. Mol. Neurobiol. 2016, 53, 2894–2899. [Google Scholar] [CrossRef] [Green Version]
- Jahangard, Y.; Monfared, H.; Moradi, A.; Zare, M.; Mirnajafi-Zadeh, J.; Mowla, S.J. Therapeutic effects of transplanted exosomes containing miR-29b to a rat model of Alzheimer’s disease. Front. Neurosci. 2020, 14, 564. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.H.; Wu, J.; Mou, F.F.; Xie, W.H.; Wang, F.B.; Wang, Q.L.; Fang, J.; Xu, Y.W.; Dong, Y.R.; Liu, J.R. Exosomes derived from hypoxia-preconditioned mesenchymal stromal cells ameliorate cognitive decline by rescuing synaptic dysfunction and regulating inflammatory responses in APP/PS1 mice. FASEB J. 2018, 32, 654–668. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Sun, L.; Jeske, R.; Nkosi, D.; York, S.B.; Liu, Y.; Grant, S.C.; Meckes, D.G., Jr.; Li, Y. Engineering extracellular vesicles by three-dimensional dynamic culture of human mesenchymal stem cells. J. Extracell. Vesicles 2022, 11, e12235. [Google Scholar] [CrossRef] [PubMed]
- Cone, A.S.; Yuan, X.; Sun, L.; Duke, L.C.; Vreones, M.P.; Carrier, A.N.; Kenyon, S.M.; Carver, S.R.; Benthem, S.D.; Stimmell, A.C. Mesenchymal stem cell-derived extracellular vesicles ameliorate Alzheimer’s disease-like phenotypes in a preclinical mouse model. Theranostics 2021, 11, 8129. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Xu, X.; Zhang, M.; Song, X. Human urine-derived stem cell-derived exosomal miR-21-5p promotes neurogenesis to attenuate Rett syndrome via the EPha4/TEK axis. Lab. Investig. 2021, 101, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Huda, M.N.; Nurunnabi, M. Potential application of exosomes in vaccine development and delivery. Pharm. Res. 2022, 1–37. [Google Scholar] [CrossRef]
- Naseri, M.; Bozorgmehr, M.; Zoller, M.; Pirmardan, E.R.; Madjd, Z. Tumor-derived exosomes: The next generation of promising cell-free vaccines in cancer immunotherapy. Oncoimmunology 2020, 9, 1779991. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Liu, W.; Zhu, S.; Fan, B.; Yu, N.; Ning, G.; Feng, S. Potential of different cells-derived exosomal microRNA cargos for treating spinal cord injury. J. Orthop. Transl. 2021, 31, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Ashmwe, M.; Posa, K.; Rührnößl, A.; Heinzel, J.C.; Heimel, P.; Mock, M.; Schädl, B.; Keibl, C.; Couillard-Despres, S.; Redl, H. Effects of Extracorporeal Shockwave Therapy on Functional Recovery and Circulating miR-375 and miR-382-5p after Subacute and Chronic Spinal Cord Contusion Injury in Rats. Biomedicines 2022, 10, 1630. [Google Scholar] [CrossRef]
- Kang, J.; Li, Z.; Zhi, Z.; Wang, S.; Xu, G. MiR-21 derived from the exosomes of MSCs regulates the death and differentiation of neurons in patients with spinal cord injury. Gene Ther. 2019, 26, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zeng, L.; Huang, J.; Wang, G.; Lu, H. miR-126 promotes angiogenesis and attenuates inflammation after contusion spinal cord injury in rats. Brain Res. 2015, 1608, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Xia, C.; Cai, Y.; Lin, Y.; Guan, R.; Xiao, G.; Yang, J. MiR-133b-5p regulates the expression of the heat shock protein 70 during rat neuronal cell apoptosis induced by the gp120 V3 loop peptide. J. Med. Virol. 2016, 88, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Rong, Y.; Wang, J.; Zhou, Z.; Ge, X.; Ji, C.; Jiang, D.; Gong, F.; Li, L.; Chen, J. Exosome-shuttled miR-216a-5p from hypoxic preconditioned mesenchymal stem cells repair traumatic spinal cord injury by shifting microglial M1/M2 polarization. J. Neuroinflamm. 2020, 17, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Gong, F.; Ge, X.; Lv, C.; Huang, C.; Feng, S.; Zhou, Z.; Rong, Y.; Wang, J.; Ji, C. Neuron-derived exosomes-transmitted miR-124-3p protect traumatically injured spinal cord by suppressing the activation of neurotoxic microglia and astrocytes. J. Nanobiotechnol. 2020, 18, 105. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, P. A novel cell-cell communication mechanism in the nervous system: Exosomes. J. Neurosci. Res. 2018, 96, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, D.; Kuo, W.P.; Frühbeis, C.; Sun, J.-J.; Zehendner, C.M.; Luhmann, H.J.; Pinto, S.; Toedling, J.; Trotter, J.; Krämer-Albers, E.-M. Multifaceted effects of oligodendroglial exosomes on neurons: Impact on neuronal firing rate, signal transduction and gene regulation. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130510. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Xue, X.; Yin, Z.; Cao, L.; Li, M.; Huang, J. Engineered Cell Membrane-Derived Nanocarriers: The Enhanced Delivery System for Therapeutic Applications. Front. Cell Dev. Biol. 2022, 10, 844050. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, D.; Gao, F.; Lv, H.; Zhang, G.; Sun, X.; Liu, L.; Mo, D.; Ma, N.; Song, L. Exosomes derived from microRNA-138-5p-overexpressing bone marrow-derived mesenchymal stem cells confer neuroprotection to astrocytes following ischemic stroke via inhibition of LCN2. J. Biol. Eng. 2019, 13, 71. [Google Scholar] [CrossRef]
- Ai, Z.; Cheng, C.; Zhou, L.; Yin, S.; Wang, L.; Liu, Y. Bone marrow mesenchymal stem cells-derived extracellular vesicles carrying microRNA-221-3p protect against ischemic stroke via ATF3. Brain Res. Bull. 2021, 172, 220–228. [Google Scholar] [CrossRef]
- Xin, H.; Li, Y.; Liu, Z.; Wang, X.; Shang, X.; Cui, Y.; Zhang, Z.G.; Chopp, M. MiR-133b promotes neural plasticity and functional recovery after treatment of stroke with multipotent mesenchymal stromal cells in rats via transfer of exosome-enriched extracellular particles. Stem Cells 2013, 31, 2737–2746. [Google Scholar] [CrossRef] [Green Version]
- Xin, H.; Li, Y.; Buller, B.; Katakowski, M.; Zhang, Y.; Wang, X.; Shang, X.; Zhang, Z.G.; Chopp, M. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells 2012, 30, 1556–1564. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.-H.; Wang, X.; Zhang, Y.; Hui, J. Exosomes of bone-marrow stromal cells inhibit cardiomyocyte apoptosis under ischemic and hypoxic conditions via miR-486-5p targeting the PTEN/PI3K/AKT signaling pathway. Thromb. Res. 2019, 177, 23–32. [Google Scholar] [CrossRef]
- Wang, W.; Zheng, Y.; Wang, M.; Yan, M.; Jiang, J.; Li, Z. Exosomes derived miR-126 attenuates oxidative stress and apoptosis from ischemia and reperfusion injury by targeting ERRFI1. Gene 2019, 690, 75–80. [Google Scholar] [CrossRef]
- Li, Q.; Jin, Y.; Ye, X.; Wang, W.; Deng, G.; Zhang, X. Bone marrow mesenchymal stem cell-derived exosomal MicroRNA-133a restrains myocardial fibrosis and epithelial–mesenchymal transition in viral myocarditis rats through suppressing MAML1. Nanoscale Res. Lett. 2021, 16, 111. [Google Scholar] [CrossRef]
- Pei, Y.; Xie, S.; Li, J.; Jia, B. Bone marrow-mesenchymal stem cell-derived exosomal microRNA-141 targets PTEN and activates β-catenin to alleviate myocardial injury in septic mice. Immunopharmacol. Immunotoxicol. 2021, 43, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Xiao, Y.; Wang, J.; Li, Y.; Li, S.; Wei, B.; Du, W.; Feng, X.; Chen, P.; Liu, B.-F. Rapid exosomes concentration and in situ detection of exosomal microRNA on agarose-based microfluidic chip. Sens. Actuators B Chem. 2021, 333, 129559. [Google Scholar] [CrossRef]
- Xiao, P.-P.; Wan, Q.-Q.; Liao, T.; Tu, J.-Y.; Zhang, G.-J.; Sun, Z.-Y. Peptide nucleic acid-functionalized nanochannel biosensor for the highly sensitive detection of tumor exosomal microRNA. Anal. Chem. 2021, 93, 10966–10973. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Lu, H.; Liu, L.; Ni, W.; Yao, Q.; Zhang, G.-J.; Yang, F. Ultrasensitive exosomal MicroRNA detection with a supercharged DNA framework nanolabel. Anal. Chem. 2021, 93, 5917–5923. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Anand, S.; Samuel, M.; Mathivanan, S. Exomeres: A new member of extracellular vesicles family. In New Frontiers: Extracellular Vesicles; Springer: Berlin/Heidelberg, Germany, 2021; pp. 89–97. [Google Scholar]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183. [Google Scholar] [CrossRef]
- Staufer, O.; Dietrich, F.; Rimal, R.; Schroter, M.; Fabritz, S.; Boehm, H.; Singh, S.; Moller, M.; Platzman, I.; Spatz, J.P. Bottom-up assembly of biomedical relevant fully synthetic extracellular vesicles. Sci. Adv. 2021, 7, eabg6666. [Google Scholar] [CrossRef]
- Jeske, R.; Liu, C.; Duke, L.; Castro, M.L.C.; Muok, L.; Arthur, P.; Singh, M.; Jung, S.; Sun, L.; Li, Y. Upscaling human mesenchymal stromal cell production in a novel vertical-wheel bioreactor enhances extracellular vesicle secretion and cargo profile. Bioact. Mater. 2022; in press. [Google Scholar] [CrossRef]
- Cao, J.; Wang, B.; Tang, T.; Lv, L.; Ding, Z.; Li, Z.; Hu, R.; Wei, Q.; Shen, A.; Fu, Y. Three-dimensional culture of MSCs produces exosomes with improved yield and enhanced therapeutic efficacy for cisplatin-induced acute kidney injury. Stem Cell Res. Ther. 2020, 11, 206. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, J.; Fu, H.; Kuang, S.; He, F.; Zhang, M.; Shen, Z.; Qin, W.; Lin, Z.; Huang, S. Exosomes derived from 3D-cultured MSCs improve therapeutic effects in periodontitis and experimental colitis and restore the Th17 cell/Treg balance in inflamed periodontium. Int. J. Oral Sci. 2021, 13, 43. [Google Scholar] [CrossRef]
- Kim, M.; Yun, H.-W.; Park, D.Y.; Choi, B.H.; Min, B.-H. Three-dimensional spheroid culture increases exosome secretion from mesenchymal stem cells. Tissue Eng. Regen. Med. 2018, 15, 427–436. [Google Scholar] [CrossRef]
- Cha, J.M.; Shin, E.K.; Sung, J.H.; Moon, G.J.; Kim, E.H.; Cho, Y.H.; Park, H.D.; Bae, H.; Kim, J.; Bang, O.Y. Efficient scalable production of therapeutic microvesicles derived from human mesenchymal stem cells. Sci. Rep. 2018, 8, 1171. [Google Scholar] [CrossRef] [PubMed]

| Proteins | Location | Functions | Ref |
|---|---|---|---|
| ARF1 | Golgi apparatus and shedding microvesicles | Regulation of matrix degradation by directly acting on the structures associated with invasiveness—invadopodia maturation and the shedding of membrane-derived microvesicles | [10] |
| ARF6 | Plasma membrane, cytosol, and endosomal membranes | Regulating the actomyosin-based membrane abscission mechanism to control the shedding of microvesicle in tumor cells | [7] |
| Rab22a | Nonclathrin-derived Endosomes, budding microvesicles | Increasing microvesicle shedding in human breast cancer under hypoxic conditions and knockdown of RAB22A impairs breast cancer metastasis | [22] |
| RhoA | Membrane and cytosol | Involved in microvesicle biogenesis through regulation of myosin light chain phosphatase. required for microvesicle shedding | [11] |
| ARRDC1 | Plasma membrane | ARRDC1-mediated relocalization of TSG101 may alter endosomal trafficking and sorting and signal transduction by receptors subjected to endosomal sorting mechanisms | [23] |
| DIAPH3 | Plasma membrane, Microtubules/microvilli | DIAPH3 silencing also promotes shedding of extracellular vesicles (EV) containing bioactive cargo and increases proliferation of recipient tumor cells, and suppresses proliferation of human macrophages and peripheral blood mononuclear cells | [24,25] |
| Myosin-1a | Plasma membrane | Enterocyte microvilli containing Myosin-1a are active vesicle-generating organelles | [26] |
| Complex | Location | Cargo Sorting | Functions | Ref | |
|---|---|---|---|---|---|
| ESCRTs-0 | HRS | VHS, FYVE, P(S/T)XP, GAT domain and coiled-coil core, clathrin-binding | Binding to/clustering with ubiquitinated cargo for delivery into MVBs, and recruits clathrin, ubiquitin ligases, and deubiquitinating enzymes, and almost certainly has other functions as well | Clustering of Ub cargo, MVB biogenesis | [31] |
| STAM1/2 | VHS, UIM, SH3, GAT domain and coiled-coil core | [32] | |||
| ESCRTs-I | TSG101 | UEV, PRD, stalk, headpiece | Binding ubiquitinated cargo, ESCRT-0, ESCRT1, BRO1 and viral proteins | Membrane budding, MVB biogenesis, viral budding, replication and cytolinesis | [33,34] |
| HVPS28 | headpiece, Vps28 CTD | ESCRT-0, ESCRT1, BRO1 and viral proteins | [35] | ||
| VPS37 | basic helix, stalk, headpiece | Membrane binding | [36] | ||
| hMVB12 | stalk, headpiece (“UMA domain”), MAPB | N/A | [37] | ||
| ESCRTs-II | EAP20/VPS25 | Winged-helix | Binding ubiquitinated cargo, binding to human ESCRT-I | The essential partner of ESCRT-I in MVB biogenesis and budding formation, membrane budding | [38] |
| EAP30/VPS22 | basic helix, Winged-helix | Forming nearly equivalent interactions with the two Vps25 molecules | [39] | ||
| EAP45/VPS36 | Winged-helix, GLUE, | Binding PI containing membranes, ubiquitinated cargo and ESCRT-1-i | [40] | ||
| ESCRTs-III | CHMP2/VPS2 | MIM1 | Recruits VPS4, initiates ESCRT disassembly | Membrane scission | [41] |
| CHMP3/VPS24 | weak MIM1 | Caps Snf7 polymer, recruits VPS2 | [41] | ||
| CHMP4/SNF7 | weak MIM2 | Main driver of membrane scission, bind Bro1 | [42] | ||
| CHMP6/VPS20 | MIM2 | Binding ESCRT-II and Doa4, acts as nucleator of Snf7 polymer | [41] | ||
| VPS4 | SKD1/VPS4 | MIT, AAA | AAA ATPase disassembles ESCRT-III, active function in MVB membrane scission | Vps4 solubilizes ESCRT-III subunits at the cost of ATP hydrolysis. LIP5 binds to Vps4 and promotes its oligomerization, activity, and ESCRT-III binding | [43] |
| LIP5 | MIT | Binding vps4 to promote ESCRT-III recycling | [44] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, E.Z.; Chen, I.; Chen, X.; Yuan, X. Exosomal MicroRNAs as Novel Cell-Free Therapeutics in Tissue Engineering and Regenerative Medicine. Biomedicines 2022, 10, 2485. https://doi.org/10.3390/biomedicines10102485
Zeng EZ, Chen I, Chen X, Yuan X. Exosomal MicroRNAs as Novel Cell-Free Therapeutics in Tissue Engineering and Regenerative Medicine. Biomedicines. 2022; 10(10):2485. https://doi.org/10.3390/biomedicines10102485
Chicago/Turabian StyleZeng, Eric Z., Isabelle Chen, Xingchi Chen, and Xuegang Yuan. 2022. "Exosomal MicroRNAs as Novel Cell-Free Therapeutics in Tissue Engineering and Regenerative Medicine" Biomedicines 10, no. 10: 2485. https://doi.org/10.3390/biomedicines10102485
APA StyleZeng, E. Z., Chen, I., Chen, X., & Yuan, X. (2022). Exosomal MicroRNAs as Novel Cell-Free Therapeutics in Tissue Engineering and Regenerative Medicine. Biomedicines, 10(10), 2485. https://doi.org/10.3390/biomedicines10102485






