Large-Scale Functional Genomics Screen to Identify Modulators of Human β-Cell Insulin Secretion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Text Mining
2.2. EndoC-βH1 Cell Culture
2.3. EndoC-βH5 Cell Culture
2.4. Glucose Stimulated Insulin Secretion (GSIS)
2.5. Insulin Quantification
2.6. siRNA Transfection in 96-Well and 384-Well Format
2.7. siRNA Randomisation on the Screening Plates
2.8. siRNA Assay Plates Preparation
2.9. High-Throughput siRNA Screen
2.10. Strictly Standardized Mean Difference SSMD
2.11. RNA Extraction and qPCR
2.12. Statistical Analysis
2.13. Viability Assay
2.14. Western Blot
3. Results
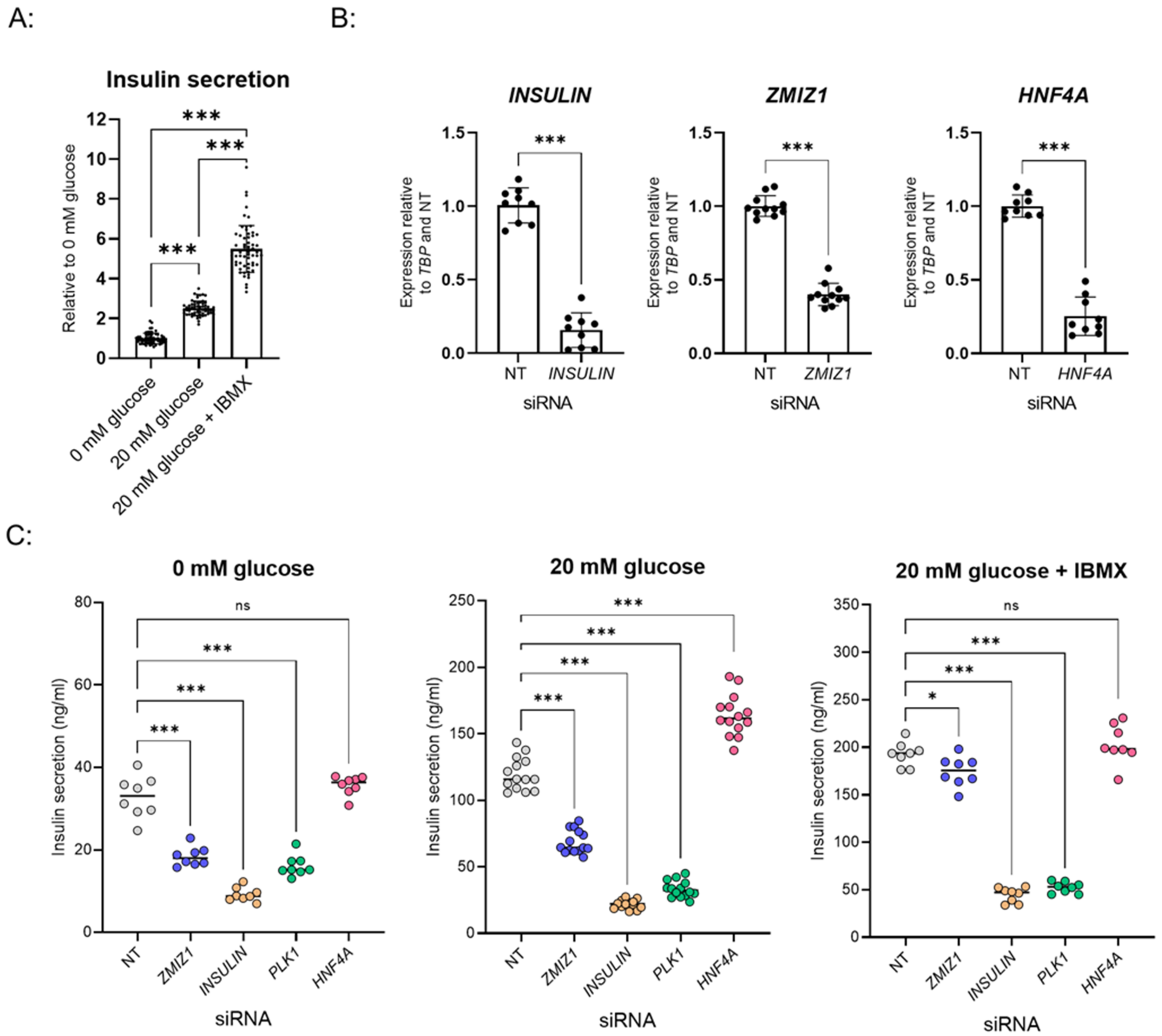
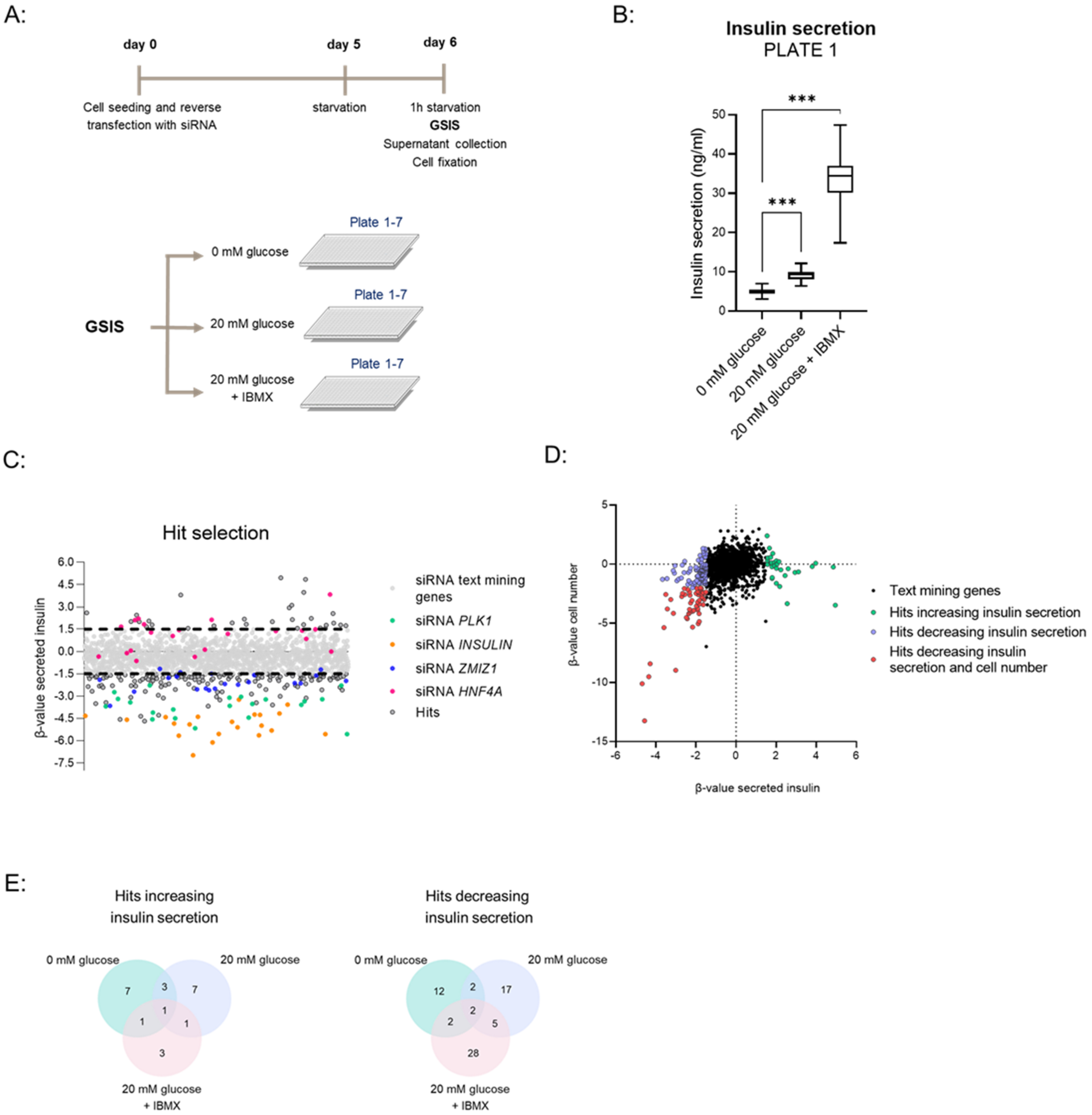
3.1. Establishment of GSIS Assay and siRNA Transfection in 384-Well Plate Format
3.2. High-Throughput siRNA Screen with GSIS as Readout
3.3. Validation of Hits
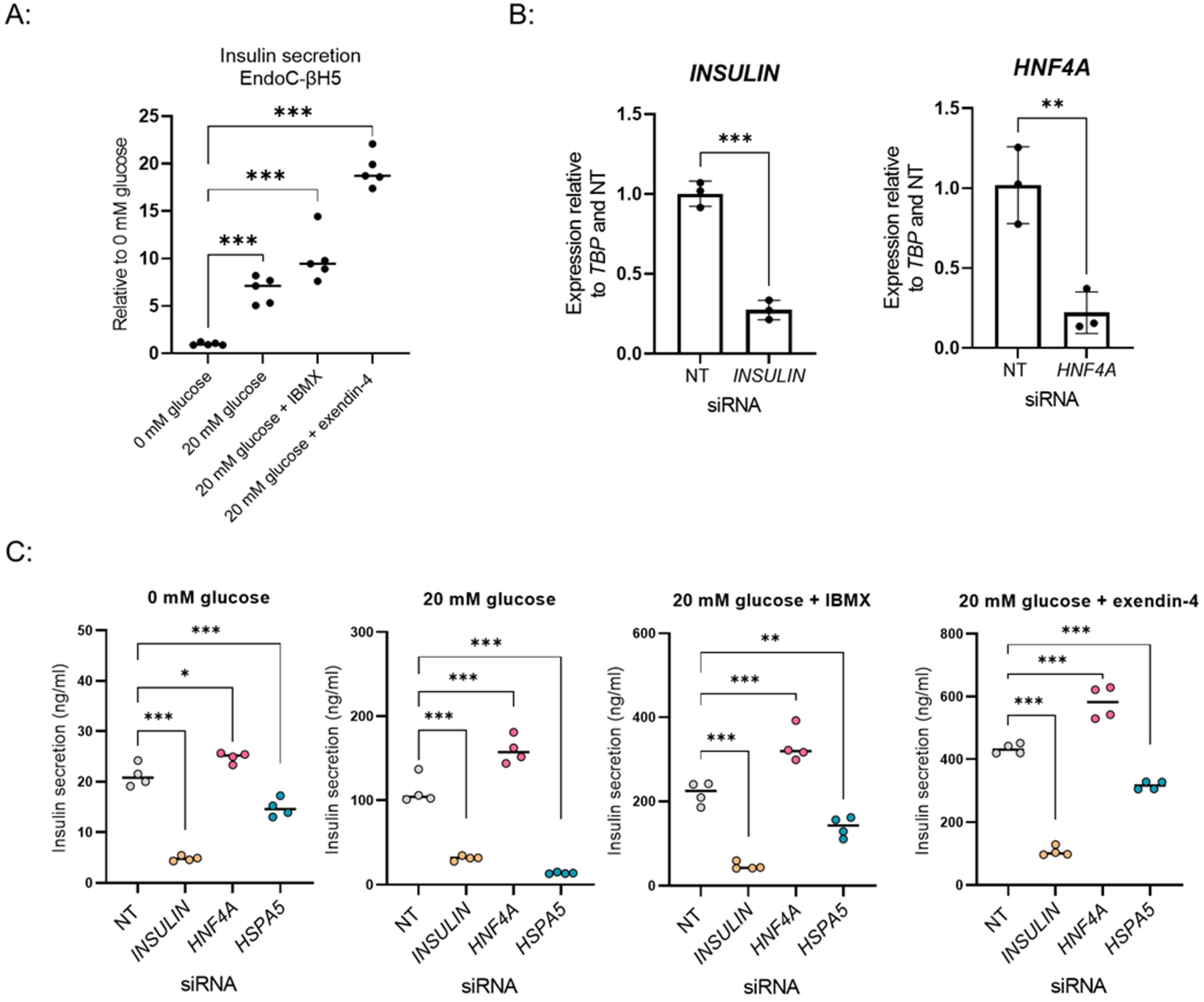
3.4. Adaptation of the Screening Setup to EndoC-βH5, a New in Vitro β-Cell Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [Green Version]
- Boyle, J.P.; Thompson, T.J.; Gregg, E.W.; Barker, L.E.; Williamson, D.F. Projection of the year 2050 burden of diabetes in the US adult population: Dynamic modeling of incidence, mortality, and prediabetes prevalence. Popul. Health Metr. 2010, 8, 29. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, V.A. Defining and characterizing the progression of type 2 diabetes. Diabetes Care 2009, 32 (Suppl. 2), S151–S156. [Google Scholar] [CrossRef] [Green Version]
- King, P.; Peacock, I.; Donnelly, R. The UK Prospective Diabetes Study (UKPDS) clinical and therapeutic implications for type 2 diabetes. Br. J. Clin. Pharmacol. 1999, 48, 643–648. [Google Scholar] [CrossRef]
- Wu, W.; Shang, J.; Feng, Y.; Thompson, C.M.; Horwitz, S.; Thompson, J.R.; Macintyre, E.D.; Thornberry, N.A.; Chapman, K.; Zhou, Y.P.; et al. Identification of glucose-dependant insulin secretion targets in pancreatic beta cells by combining defined-mechanism compound library screening and siRNA gene silencing. J. Biomol. Screen. 2008, 13, 128–134. [Google Scholar]
- Burns, S.M.; Vetere, A.; Walpita, D.; Dančík, V.; Khodier, C.; Perez, J.; Clemons, P.A.; Wagner, B.K.; Altshuler, D. High-throughput luminescent reporter of insulin secretion for discovering regulators of pancreatic Beta-cell function. Cell Metab. 2015, 21, 126–137. [Google Scholar] [CrossRef] [Green Version]
- Ku, G.M.; Pappalardo, Z.; Luo, C.C.; German, M.S.; McManus, M.T. An siRNA screen in pancreatic beta cells reveals a role for Gpr27 in insulin production. PLoS Genet. 2012, 8, e1002449. [Google Scholar] [CrossRef] [Green Version]
- Rorsman, P.; Ashcroft, F.M. Pancreatic beta-Cell Electrical Activity and Insulin Secretion: Of Mice and Men. Physiol. Rev. 2018, 98, 117–214. [Google Scholar] [CrossRef]
- Benner, C.; Van der Meulen, T.; Cacéres, E.; Tigyi, K.; Donaldson, C.J.; Huising, M.O. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genom. 2014, 15, 620. [Google Scholar] [CrossRef] [Green Version]
- Ravassard, P.; Hazhouz, Y.; Pechberty, S.; Bricout-Neveu, E.; Armanet, M.; Czernichow, P.; Scharfmann, R. A genetically engineered human pancreatic beta cell line exhibiting glucose-inducible insulin secretion. J. Clin. Investig. 2011, 121, 3589–3597. [Google Scholar] [CrossRef]
- Swinney, D.C.; Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011, 10, 507–519. [Google Scholar] [CrossRef]
- Hansson, L.K.; Hansen, R.B.; Pletscher-Frankild, S.; Berzins, R.; Hansen, D.H.; Madsen, D.; Christensen, S.B.; Christiansen, M.R.; Boulund, U.; Wolf, X.A.; et al. Semantic text mining in early drug discovery for type 2 diabetes. PLoS ONE 2020, 15, e0233956. [Google Scholar] [CrossRef]
- Thomsen, S.K.; Ceroni, A.; Van de Bunt, M.; Burrows, C.; Barrett, A.; Scharfmann, R.; Ebner, D.; McCarthy, M.I.; Gloyn, A.L. Systematic Functional Characterization of Candidate Causal Genes for Type 2 Diabetes Risk Variants. Diabetes 2016, 65, 3805–3811. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.D. A new method with flexible and balanced control of false negatives and false positives for hit selection in RNA interference high-throughput screening assays. J. Biomol. Screen. 2007, 12, 645–655. [Google Scholar] [CrossRef] [Green Version]
- Van de Bunt, M.; Manning Fox, J.E.; Dai, X.; Barrett, A.; Grey, C.; Li, L.; Bennett, A.J.; Johnson, P.R.; Rajotte, R.V.; Gaulton, K.J.; et al. Transcript Expression Data from Human Islets Links Regulatory Signals from Genome-Wide Association Studies for Type 2 Diabetes and Glycemic Traits to Their Downstream Effectors. PLoS Genet. 2015, 11, e1005694. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Saito, M.; Kadokura, H.; Miyazaki, J.I.; Tashiro, F.; Imagawa, Y.; Iwawaki, T.; Kohno, K. IRE1-XBP1 pathway regulates oxidative proinsulin folding in pancreatic beta cells. J. Cell Biol. 2018, 217, 1287–1301. [Google Scholar] [CrossRef]
- Roche, T.E.; Hiromasa, Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell. Mol. Life Sci. 2007, 64, 830–849. [Google Scholar] [CrossRef]
- Nellemann, B.; Vendelbo, M.H.; Nielsen, T.S.; Bak, A.M.; Høgild, M.; Pedersen, S.B.; Biensø, R.S.; Pilegaard, H.; Moller, N.; Jessen, N.; et al. Growth hormone-induced insulin resistance in human subjects involves reduced pyruvate dehydrogenase activity. Acta Physiol. 2014, 210, 392–402. [Google Scholar] [CrossRef]
- Zhang, S.; Hulver, M.W.; McMillan, R.P.; Cline, M.A.; Gilbert, E.R. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. 2014, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Fu, A.; Robitaille, K.; Faubert, B.; Reeks, C.; Dai, X.-Q.; Hardy, A.B.; Sankar, K.S.; Ogrel, S.; Al-Dirbashi, O.Y.; Rocheleau, J.V.; et al. LKB1 couples glucose metabolism to insulin secretion in mice. Diabetologia 2015, 58, 1513–1522. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, G.; Wu, C.S.; Villarreal, D.; Lee, J.H.; Han, H.W.; Gaharwar, A.; Tian, Y.; Fu, W.; Guo, S.; Smith, R.G.; et al. Beta Cell GHS-R Regulates Insulin Secretion and Sensitivity. Int. J. Mol. Sci. 2021, 22, 3950. [Google Scholar] [CrossRef]
- Kurashina, T.; Dezaki, K.; Yoshida, M.; Rita, R.S.; Ito, K.; Taguchi, M.; Miura, R.; Tominaga, M.; Ishibashi, S.; Kakei, M.; et al. The beta-cell GHSR and downstream cAMP/TRPM2 signaling account for insulinostatic and glycemic effects of ghrelin. Sci. Rep. 2015, 5, 14041. [Google Scholar] [CrossRef]
- Fusakio, M.E.; Willy, J.A.; Wang, Y.; Mirek, E.T.; Al-Baghdadi, R.; Adams, C.; Anthony, T.G.; Wek, R.C. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol. Biol. Cell 2016, 27, 1536–1551. [Google Scholar] [CrossRef]
- Laybutt, D.R.; Preston, A.M.; Åkerfeldt, M.C.; Kench, J.; Busch, A.K.; Biankin, A.; Biden, T.J. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia 2007, 50, 752–763. [Google Scholar] [CrossRef]
- Fonseca, S.G.; Burcin, M.; Gromada, J.; Urano, F. Endoplasmic reticulum stress in beta-cells and development of diabetes. Curr. Opin. Pharmacol. 2009, 9, 763–770. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, N.; De Franco, E.; Arvan, P.; Cnop, M. Pathological beta-Cell Endoplasmic Reticulum Stress in Type 2 Diabetes: Current Evidence. Front. Endocrinol. 2021, 12, 650158. [Google Scholar] [CrossRef]
- Kitakaze, K.; Oyadomari, M.; Zhang, J.; Hamada, Y.; Takenouchi, Y.; Tsuboi, K.; Inagaki, M.; Tachikawa, M.; Fujitani, Y.; Okamoto, Y.; et al. ATF4-mediated transcriptional regulation protects against beta-cell loss during endoplasmic reticulum stress in a mouse model. Mol. Metab. 2021, 54, 101338. [Google Scholar] [CrossRef]
- Wang, J.; Lee, J.; Liem, D.; Ping, P. HSPA5 Gene encoding Hsp70 chaperone BiP in the endoplasmic reticulum. Gene 2017, 618, 14–23. [Google Scholar] [CrossRef]
- Polonsky, K.S.; Burant, C.F. Type 2 Diabetes Mellitus. In Williams Textbook of Endocrinology; Elsevier/Saunders: Philadelphia, PA, USA, 2016; pp. 1385–1450. [Google Scholar]
- Njølstad, P.R.; Søvik, O.; Cuesta-Muñoz, A.; Bjørkhaug, L.; Massa, O.; Barbetti, F.; Undlien, D.E.; Shiota, C.; Magnuson, M.A.; Molven, A.; et al. Neonatal diabetes mellitus due to complete glucokinase deficiency. N. Engl. J. Med. 2001, 344, 1588–1592. [Google Scholar] [CrossRef]
- Grotz, A.K.; Navarro-Guerrero, E.; Bevacqua, R.J.; Baronio, R.; Thomsen, S.K.; Nawaaz, S.; Rajesh, V.; Wesolowska-Andersen, A.; Kem, S.K.; Ebner, D.; et al. A genome-wide CRISPR screen identifies regulators of beta cell function involved in type 2 diabetes risk. bioRxiv 2021. [Google Scholar] [CrossRef]
- Diedisheim, M.; Oshima, M.; Albagli, O.; Huldt, C.W.; Ahlstedt, I.; Clausen, M.; Menon, S.; Aivazidis, A.; Andreasson, A.-C.; Haynes, W.G.; et al. Modeling human pancreatic beta cell dedifferentiation. Mol. Metab. 2018, 10, 74–86. [Google Scholar] [CrossRef]
- Oshima, M.; Pechberty, S.; Bellini, L.; Göpel, S.O.; Campana, M.; Rouch, C.; Dairou, J.; Cosentino, C.; Fantuzzi, F.; Toivonen, S.; et al. Stearoyl CoA desaturase is a gatekeeper that protects human beta cells against lipotoxicity and maintains their identity. Diabetologia 2020, 63, 395–409. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szczerbinska, I.; Tessitore, A.; Hansson, L.K.; Agrawal, A.; Ragel Lopez, A.; Helenius, M.; Malinowski, A.R.; Gilboa, B.; Ruby, M.A.; Gupta, R.; et al. Large-Scale Functional Genomics Screen to Identify Modulators of Human β-Cell Insulin Secretion. Biomedicines 2022, 10, 103. https://doi.org/10.3390/biomedicines10010103
Szczerbinska I, Tessitore A, Hansson LK, Agrawal A, Ragel Lopez A, Helenius M, Malinowski AR, Gilboa B, Ruby MA, Gupta R, et al. Large-Scale Functional Genomics Screen to Identify Modulators of Human β-Cell Insulin Secretion. Biomedicines. 2022; 10(1):103. https://doi.org/10.3390/biomedicines10010103
Chicago/Turabian StyleSzczerbinska, Iwona, Annamaria Tessitore, Lena Kristina Hansson, Asmita Agrawal, Alejandro Ragel Lopez, Marianne Helenius, Andrzej R. Malinowski, Barak Gilboa, Maxwell A. Ruby, Ramneek Gupta, and et al. 2022. "Large-Scale Functional Genomics Screen to Identify Modulators of Human β-Cell Insulin Secretion" Biomedicines 10, no. 1: 103. https://doi.org/10.3390/biomedicines10010103
APA StyleSzczerbinska, I., Tessitore, A., Hansson, L. K., Agrawal, A., Ragel Lopez, A., Helenius, M., Malinowski, A. R., Gilboa, B., Ruby, M. A., Gupta, R., & Ämmälä, C. (2022). Large-Scale Functional Genomics Screen to Identify Modulators of Human β-Cell Insulin Secretion. Biomedicines, 10(1), 103. https://doi.org/10.3390/biomedicines10010103




