Recent Achievements in Electrochemical and Surface Plasmon Resonance Aptasensors for Mycotoxins Detection
Abstract
:1. Introduction
2. General Characterization of Mycotoxins
3. Conventional Methods of Mycotoxin Determination
4. Aptamers Utilized in Aptasensor Assembly
5. Aptasensing Strategies
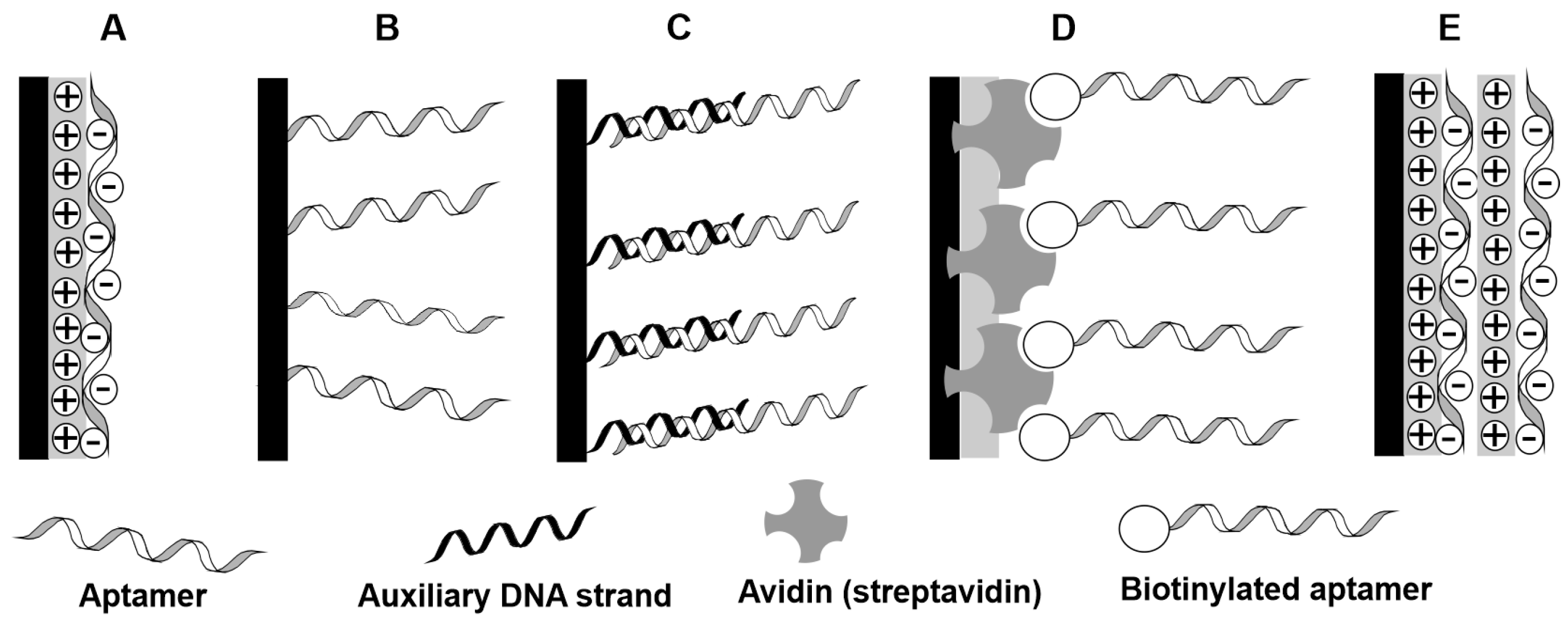
5.1. Immobiization of Aptamers
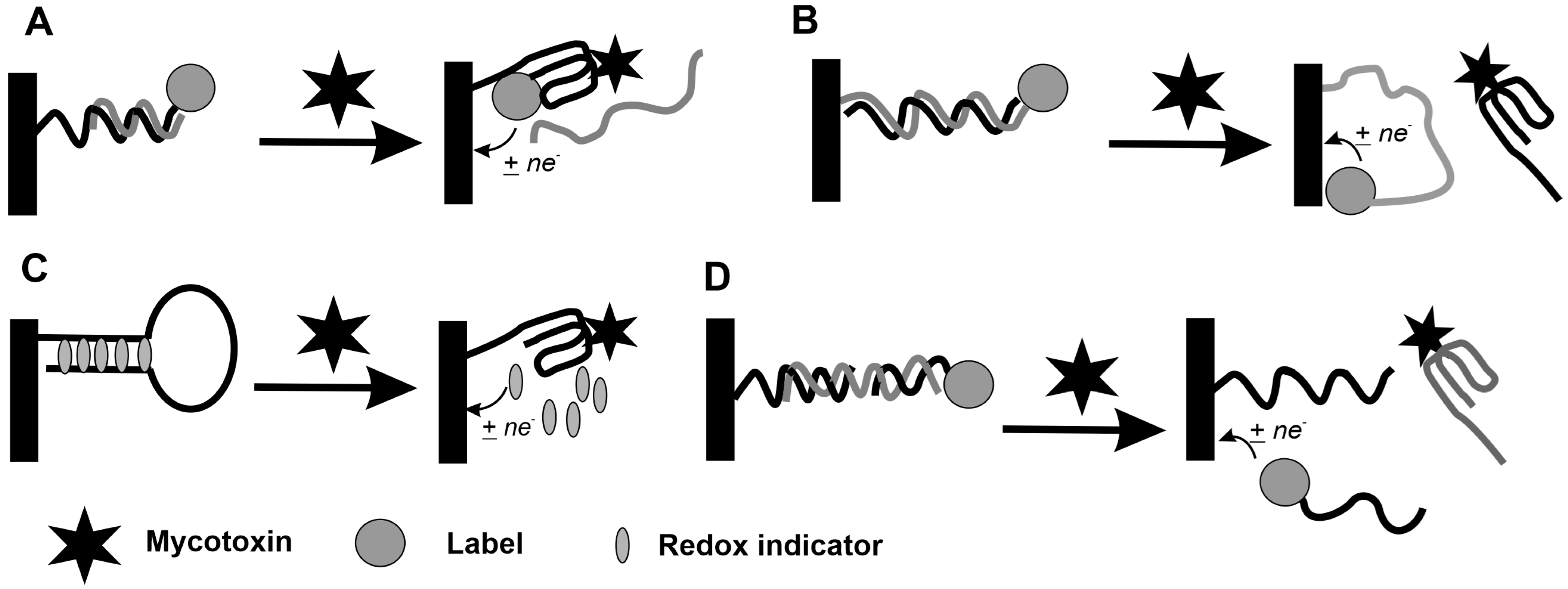
5.2. Assembling of Electrochemical Aptasensors
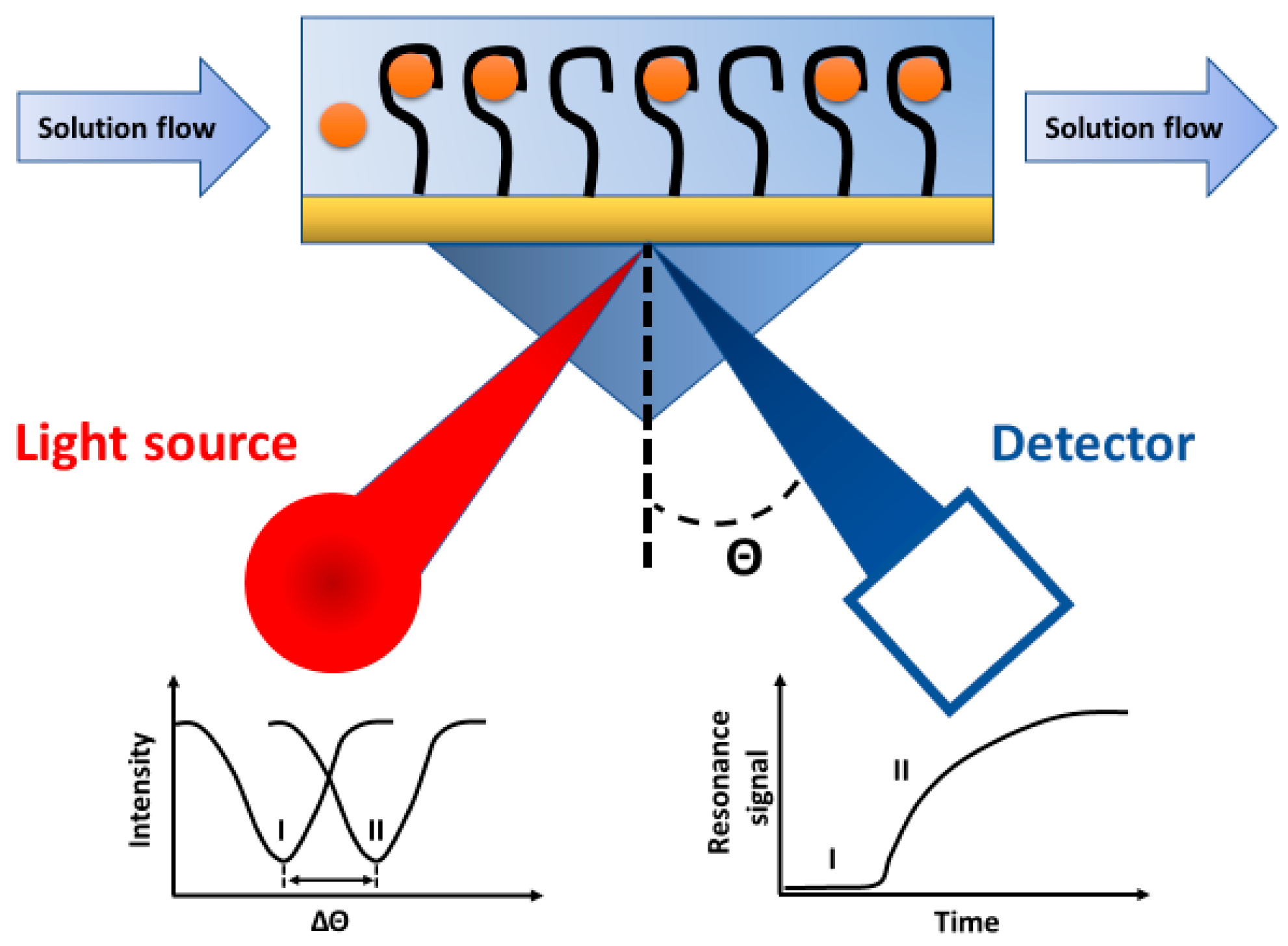
5.3. Surface Plasmon Resonance Aptasensors
6. Conclusions
- Although the review covers only recent publications in the area of aptasensors for mycotoxin determination, some trends in further progress can be mentioned;
- More attention will be paid to the combination of aptasensing with microfluidics and simplified biosensor formats, e.g., paper-based biosensors [211] and colorimetric devices based on SERS principles;
- The focus on the development of new measurement formats will be shifted to the signal-on (switch-on) aptasensors offering better metrological characteristics, especially in real sample assay;
- The interest in the selection of new aptamer structures and their derivatization in favor of aptasensor assembling will improve both the operational and analytical characteristics of aptasensors and result in the formation of chimeric materials combining aptasensing with the artificial 3D structures of synthetic materials;
- In general, further efforts in aptasensor design will extend to the area of monitoring the environment and foodstuffs to establish safer and more comfortable life for the population.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brack, W.; Dulio, V.; Ågerstrand, M.; Allan, I.; Altenburger, R.; Brinkmann, M.; Bunke, D.; Burgess, R.M.; Cousins, I.; Escher, B.I.; et al. Towards the review of the European Union Water Framework Directive: Recommendations for more efficient assessment and management of chemical contamination in European surface water resources. Sci. Total Environ. 2017, 576, 720–737. [Google Scholar] [CrossRef]
- Uwizeyimana, H.; Wang, M.; Chen, W.; Khan, K. The eco-toxic effects of pesticide and heavy metal mixtures towards earthworms in soil. Environ. Toxicol. Pharm. 2017, 55, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Grandjean, P.; Bellanger, M. Calculation of the disease burden associated with environmental chemical exposures: Application of toxicological information in health economic estimation. Environ. Health 2017, 16, 123. [Google Scholar] [CrossRef] [Green Version]
- Schmeller, D.S.; Loyau, A.; Bao, K.; Brack, W.; Chatzinotas, A.; De Vleeschouwer, F.; Friesen, J.; Gandois, L.; Hansson, S.V.; Haver, M.; et al. People, pollution and pathogens—Global change impacts in mountain freshwater ecosystems. Sci. Total Environ. 2018, 622–623, 756–763. [Google Scholar] [CrossRef] [Green Version]
- Cimbalo, A.; Alonso-Garrido, M.; Font, G.; Manyes, L. Toxicity of mycotoxins in vivo on vertebrate organisms: A review. Food Chem. Toxicol. 2020, 137, 111161. [Google Scholar] [CrossRef]
- Lee, H.J.; Ryu, D. Worldwide occurrence of mycotoxins in cereals and cereal-derived food products: Public health perspectives of their co-occurrence. J. Agric. Food Chem. 2017, 65, 7034–7051. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Thrane, U.; Samson, R.A.; Pitt, J.I. Important mycotoxins and the fungi which produce them. In Advances in Food Mycology. Advances in Experimental Medicine and Biology; Hocking, A.D., Pitt, J.I., Samson, R.A., Thrane, U., Eds.; Springer: Boston, MA, USA, 2006; Volume 571, pp. 3–31. [Google Scholar] [CrossRef]
- Omotayo, O.P.; Omotayo, A.O.; Mwanza, M.; Babalola, O.O. Prevalence of mycotoxins and their consequences on human health. Toxicol. Res. 2019, 35, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zinedine, A.; Soriano, J.M.; Moltó, J.C.; Mañes, J. Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: An oestrogenic mycotoxin. Food Chem. Toxicol. 2007, 45, 1–18. [Google Scholar] [CrossRef]
- Tammer, B.; Lehmann, I.; Nieber, K.; Altenburger, R. Combined effects of mycotoxin mixtures on human T cell function. Toxicol. Lett. 2007, 170, 124–133. [Google Scholar] [CrossRef]
- Ülger, T.G.; Uçar, A.; Çakıroğlu, F.P.; Yilmaz, S. Genotoxic effects of mycotoxins. Toxicon 2020, 185, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Dou, X.-W.; Zhang, C.; Logrieco, A.F.; Yang, M.-H. A review of current methods for analysis of mycotoxins in herbal medicines. Toxins 2018, 10, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alshannaq, A.; Yu, J.-H. Occurrence, toxicity, and analysis of major mycotoxins in food. Int. J. Environ. Res. Public Health 2017, 14, 632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tittlemier, S.A.; Cramer, B.; Dall’Asta, C.; Iha, M.H.; Lattanzio, V.M.T.; Maragos, C.; Solfrizzo, M.; Stranska, M.; Stroka, J.; Sumarah, M. Developments in mycotoxin analysis: An update for 2018–2019. World Mycotoxin J. 2020, 13, 3–24. [Google Scholar] [CrossRef] [Green Version]
- Alahi, M.E.E.; Mukhopadhyay, S.C. Detection methodologies for pathogen and toxins: A review. Sensors 2017, 17, 1885. [Google Scholar] [CrossRef] [Green Version]
- Lieberzeit, P.A.; Dickert, F.L. Sensor technology and its application in environmental analysis. Anal. Bioanal. Chem. 2007, 387, 237–247. [Google Scholar] [CrossRef]
- Thevenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Pure Appl. Chem. 1999, 71, 2333–2348. [Google Scholar] [CrossRef] [Green Version]
- Justino, C.I.L.; Duarte, A.C.; Rocha-Santos, T.A.P. Recent progress in biosensors for environmental monitoring: A review. Sensors 2017, 17, 2918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gevaerd, A.; Banks, C.S.; Bergamini, M.F.; Marcolino-Junior, L.H. Nanomodified screen-printed electrode for direct determination of aflatoxin B1 in malted barley samples. Sens. Actuators B 2020, 307, 127547. [Google Scholar] [CrossRef]
- Arduini, F.; Amine, A.; Moscone, D.; Palleschi, G. Biosensors based on cholinesterase inhibition for insecticides, nerve agents and aflatoxin B1 detection (review). Microchim. Acta 2010, 170, 193–214. [Google Scholar] [CrossRef] [Green Version]
- Soldatkin, O.O.; Stepurska, K.V.; Arkhypova, V.M.; Soldatkin, A.P.; El’skaya, A.V.; Lagarde, F.; Dzyadevych, S.V. Conductometric enzyme biosensor for patulin determination. Sens. Actuators B 2017, 239, 1010–1015. [Google Scholar] [CrossRef]
- Dunn, M.R.; Jimenez, R.M.; Chaput, J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017, 1, 0076. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Jayasen, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Liu, B.; Cui, X.; Li, Y.; Tang, J.; Wang, X.; Zhang, D.; Li, Z. Recent advances in aptasensors for mycotoxin detection: On the surface and in the colloid. Talanta 2021, 223, 121729. [Google Scholar] [CrossRef]
- Shkembi, S.; Svobodova, M.; Skouridou, V.; Bashammakh, A.S.; Alyoubi, A.O.; O’Sullivan, C.K. Aptasensors for mycotoxin detection: A review. Anal. Biochem. 2021, 114156, in press. [Google Scholar] [CrossRef]
- Ngea, G.L.N.; Yang, Q.; Castoria, R.; Zhang, X.; Routledge, M.N.; Zhang, H. Recent trends in detecting, controlling, and detoxifying of patulin mycotoxin using biotechnology methods. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2447–2472. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, M.J.; Dobson, A.D.W. Mycotoxin production by aspergillus, fusarium and penicillium species. Int. J. Food Microbiol. 1998, 43, 141–158. [Google Scholar] [CrossRef]
- Thanushree, M.P.; Sailendri, D.; Yoha, K.S.; Moses, J.A.; Anandharamakrishna, C. Mycotoxin contamination in food: An exposition on spices. Trends Food Sci. Technol. 2019, 93, 69–80. [Google Scholar] [CrossRef]
- Pinotti, L.; Ottoboni, M.; Giromini, C.; Dell’Orto, V.; Cheli, F. Mycotoxin contamination in the EU feed supply chain: A focus on cereal byproducts. Toxins 2016, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.I. Toxigenic fungi: Which are important? Med. Mycol. 2000, 38, 17–22. [Google Scholar] [CrossRef]
- Gurban, A.-M.; Epure, P.; Oancea, F.; Doni, M. Achievements and prospects in electrochemical-based biosensing platforms for aflatoxinM1 detection in milk and dairy products. Sensors 2017, 17, 2951. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.V. Aflatoxins: Properties, toxicity and detoxification. Nutr. Food Sci. Int. J. 2018, 6, 555696. [Google Scholar] [CrossRef]
- Do, J.H.; Choi, D.-K. Aflatoxins: Detection, toxicity and biosynthesis. Biotechnol. Bioprocess Eng. 2007, 12, 585–593. [Google Scholar] [CrossRef]
- Parkin, D.M.; Pisani, P.; Ferlay, J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int. J. Cancer 1999, 80, 827–841. [Google Scholar] [CrossRef]
- Koszegi, T.; Poor, M. Ochratoxin A: Molecular interactions, mechanisms of toxicity and prevention at the molecular level. Toxins 2016, 15, 111. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.I. Penicillium viridicatum, Penicillium verrucosum, and production of ochratoxin A. Appl. Environ. Microbiol. 1987, 53, 266–269. [Google Scholar] [CrossRef] [Green Version]
- Varga, J.; Kevei, E.; Rinyu, E.; Teren, J.; Kozakiewicz, Z. Ochratoxin production by Aspergillus species. Appl. Environ. Microbiol. 1996, 62, 4461–4464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serra, R.; Abrunhosa, L.; Kozakiewicz, Z.; Venancio, A. Black Aspergillus species as ochratoxin A producers in Portuguese wine grapes. Int. J. Food Microbiol. 2003, 88, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Sage, L.; Krivobok, S.; Delbos, E.; Seigle-Murandi, F.; Creppy, E.E. Fungal flora and ochratoxin A production in grapes and musts from France. J. Agric. Food Chem. 2002, 50, 1306–1311. [Google Scholar] [CrossRef]
- Palumbo, J.D.; O’Keeffe, T.L.; Ho, Y.S.; Santillan, C.J. Occurrence of ochratoxin a contamination and detection of ochratoxigenic Aspergillus species in retail samples of dried fruits and nuts. J. Food Prot. 2015, 78, 836–842. [Google Scholar] [CrossRef]
- O’Brien, E.; Dietrich, D.R. Ochratoxin A: The continuing enigma. Crit. Rev. Toxicol. 2005, 35, 33–60. [Google Scholar] [CrossRef]
- Pfohl-Leszkowicz, A.; Manderville, R.A. Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 2007, 51, 1192. [Google Scholar] [CrossRef]
- Ringot, D.; Chango, A.; Schneider, Y.J.; Larondelle, Y. Toxicokinetics and toxicodynamics of ochratoxin A, an update. Chem. Biol. Interact. 2006, 5, 18–46. [Google Scholar] [CrossRef]
- Boesch-Saadatmandi, C.; Loboda, A.; Jozkowicz, A.; Huebbe, P.; Blank, R.; Wolffram, S.; Dulak, J.; Rimbach, G. Effect of ochratoxin A on redox-regulated transcription factors, antioxidant enzymes and glutathione-S-transferase in cultured kidney tubulus cells. Food Chem. Toxicol. Int. J. Br. Ind. Biol. Res. Assoc. 2008, 46, 2665–2671. [Google Scholar] [CrossRef]
- Malir, F.; Ostry, V.; Pfohl-Leszkowicz, A.; Malir, J.; Toman, J. Ochratoxin a: 50 Years of research. Toxins 2016, 8, 191. [Google Scholar] [CrossRef] [Green Version]
- Rheeder, J.P.; Marasas, W.F.; Vismer, H.F. Production of fumonisin analogs by Fusarium species. Appl. Environ. Microbiol. 2002, 68, 2101–2105. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Mahato, D.K.; Kamle, M.; Mohanta, T.K.; Kang, S.G. Review aflatoxins: A global concern for food safety, human health and their management. Front. Microbiol. 2016, 7, 2170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, F.S.; Li, G.Y. Simultaneous occurrence of fumonisin B1 and other mycotoxins in moldy corn collected from the People’s Republic of China in regions with high incidences of esophageal cancer. Appl. Environ. Microbiol. 1994, 60, 847–852. [Google Scholar] [CrossRef] [Green Version]
- Kamle, M.; Mahato, D.K.; Devi, S.; Lee, K.E.; Kang, S.G.; Kumar, P. Fumonisins: Impact on agriculture, food, and human health and their management strategies. Toxins 2019, 11, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Mahunu, G.K.; Castoria, R.; Yang, Q.; Apaliya, M.T. Recent developments in the enhancement of some postharvest biocontrol agents with unconventional chemicals compounds. Trends Food Sci. Technol. 2018, 78, 180–187. [Google Scholar] [CrossRef]
- Solairaj, D.; Legrand, N.N.G.; Yang, Q.; Zhang, H. Isolation of pathogenic fungi causing postharvest decay in table grapes and in vivo biocontrol activity of selected yeasts against them. Physiol. Mol. Plant Pathol. 2020, 110, 101478. [Google Scholar] [CrossRef]
- Biango-Daniels, M.N.; Snyder, A.B.; Worobo, R.W.; Hodge, K.T. Fruit infected with Paecilomyces niveus: A source of spoilage inoculum and patulin in apple juice concentrate. Food Control 2019, 97, 81–86. [Google Scholar] [CrossRef]
- Saleh, I.; Goktepe, I. The characteristics, occurrence, and toxicological effects of patulin. Food Chem. Toxicol. 2019, 129, 301–311. [Google Scholar] [CrossRef]
- Pal, S.; Singh, N.; Ansari, K.M. Toxicological effects of patulin mycotoxin on the mammalian system: An overview. Toxicol. Res. 2017, 6, 764–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudakin, D.L. Trichothecenes in the environment: Relevance to human health. Toxicol. Lett. 2003, 143, 97–107. [Google Scholar] [CrossRef]
- McCormick, S.P.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From simple to complex mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Foroud, N.A.; Baines, D.; Gagkaeva, T.Y.; Thakor, N.; Badea, A.; Steiner, B.; Bürstmayr, M.; Bürstmayr, H. Trichothecenes in cereal grains—An update. Toxins 2019, 11, 634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arunachalam, C.; Doohan, F.M. Trichothecene toxicity in eukaryotes: Cellular and molecular mechanisms in plants and animals. Toxicol. Lett. 2013, 217, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J. Mechanisms of deoxynivalenol-induced gene expression and apoptosis. Food Addit. Contam. 2008, 22, 1128–1140. [Google Scholar] [CrossRef] [PubMed]
- Ueno, Y. The toxicology of mycotoxins. Crit. Rev. Toxicol. 1985, 14, 99–132. [Google Scholar] [CrossRef]
- McLean, M. The phytotoxicity of Fusarium metabolites: An update since 1989. Mycopathologia 1996, 133, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Applegate, T. Zearalenone (ZEN) in livestock and poultry: Dose toxicokinetics, toxicity and estrogenicity. Toxins 2020, 12, 377. [Google Scholar] [CrossRef] [PubMed]
- Rogowska, A.; Pomastowski, P.; Sagandykova, G.; Buszewski, B. Zearalenone and its metabolites: Effect on human health, metabolism and neutralisation methods. Toxicon 2019, 162, 46–56. [Google Scholar] [CrossRef]
- Carlos, O.G.; Elena, C.L.; Vytautas, T.; Carsten, M.; Stefanka, B.; Joerg, S. Determination of regulated mycotoxins and enniatins and beauvericin in cereals. In Report on the 2016 Proficiency Test of the European Union Reference Laboratory for Mycotoxins; Cat. No KJ-NA-28790-EN-N; European Union: Luxembourg, 2017. [Google Scholar] [CrossRef]
- Sulyok, M.; Krska, R.; Schuhmacher, R. A liquid chromatography/tandem mass spectrometric multi-mycotoxin method for the quantification of 87 analytes and its application to semi-quantitative screening of moldy food samples. Anal. Bioanal. Chem. 2007, 389, 1505–1523. [Google Scholar] [CrossRef] [PubMed]
- Sulyok, M.; Krska, R.; Schuhmacher, R. Application of a liquid chromatography–tandem mass spectrometric method to multi-mycotoxin determination in raw cereals and evaluation of matrix effects. Food Additiv. Contamin. 2007, 24, 1184–1195. [Google Scholar] [CrossRef]
- Spanjer, M.C. Mass spectrometry in multi-mycotoxin and fungal spore analysis. In Determining Mycotoxins and Mycotoxigenic Fungi in Food and Feed. Woodhead Publishing Series in Food Science, Technology and Nutrition; De Saeger, S., Ed.; Woodhead Publishing Limited: Cambridge, UK, 2011; pp. 90–134. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Hu, X.F.; Zhang, Q.; Li, P.W. Determination for multiple mycotoxins in agricultural products using HPLC-MS/MS via a multiple antibody immunoaffinity column. J. Chromatogr. B 2016, 1021, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.F.; Jia, B.Y.; Sun, C.N.; Shi, L.C.; Liu, X.; Zhou, L.D.; Kong, W.J. Reuse of regenerated immunoaffinity column for excellent clean-up and low-cost detection of trace aflatoxins in malt. Microchem. J. 2020, 157, 105007. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, N.; Gao, H.L.; Guo, Z.X.; Lu, A.X.; Guo, X.J.; Lu, J.H.; Luan, Y.X. Determination of aflatoxin B1 in lotus seed by high performance liquid chromatography with aptamer affinity column for purification and enrichment. Chin. J. Anal. Chem. 2020, 48, 662–669. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Chen, M.L.; Chi, J.X.; Yu, X.; Chen, Y.X.; Lin, X.C.; Xie, Z.H. Aptamer-based polyhedral oligomeric silsesquioxane (POSS)-containing hybrid affinity monolith prepared via a “one-pot” process for selective extraction of ochratoxin A. J. Chromatogr. A 2018, 1563, 37–46. [Google Scholar] [CrossRef]
- Moez, E.; Noel, D.; Brice, S.; Benjamin, G.; Pascaline, A.; Didier, M. Aptamer assisted ultrafiltration cleanup with high performance liquid chromatography-fluorescence detector for the determination of OTA in green coffee. Food Chem. 2020, 310, 125851. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, Y.; Zhang, C.; Zheng, Y.; Wu, Y.; Wang, X. Development of an aptamer-functionalized capillary monolithic column for the highly-selective and highly-efficient recognition of patulin. Food Control 2021, 119, 107461. [Google Scholar] [CrossRef]
- He, D.; Wu, Z.; Cui, B.; Xu, E. Aptamer and gold nanorod-based fumonisin B1 assay using both fluorometry and SERS. Microchim. Acta 2020, 187, 215. [Google Scholar] [CrossRef]
- Singh, G.; Velasquez, L.; Brady, B.; Koerner, T.; Huet, A.-C.; Delahaut, P. Development of an indirect competitive ELISA for analysis of alternariol in bread and bran samples. Food Anal. Methods 1018, 11, 1444–1450. [Google Scholar] [CrossRef]
- Xiao, Z.-L.; Wang, Y.-L.; Shen, Y.-D.; Xu, Z.-L.; Dong, J.-X.; Wang, H.; Situ, C.; Wang, F.; Yang, J.-Y.; Lei, H.T.; et al. Specific monoclonal antibody-based enzyme immunoassay for sensitive and reliable detection of Alternaria mycotoxin iso-tenuazonic acid in food droducts. Food Anal. Methods 2018, 11, 635–645. [Google Scholar] [CrossRef]
- Xiong, Y.; Pei, K.; Wu, Y.Q.; Xiong, Y.H. Colorimetric ELISA based on glucose oxidase-regulated the color of acid-base indicator for sensitive detection of aflatoxin B1 in corn. Food Control 2017, 78, 317–323. [Google Scholar] [CrossRef]
- Hendrickson, O.D.; Chertovich, J.O.; Zherdeva, A.V.; Sveshnikov, P.G.; Dzantiev, B.B. Ultrasensitive magnetic ELISA of zearalenone with pre-concentration and chemiluminescent detection. Food Control 2018, 84, 330–338. [Google Scholar] [CrossRef]
- Maragos, C.M.; Sieve, K.K.; Bobell, J. Detection of cyclopiazonic acid (CPA) in maize by immunoassay. Mycotoxin Res. 2017, 33, 157–165. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, M.; Wang, Y.S.; Liu, J.M. Nanozyme and aptamer-based immunosorbent assay for aflatoxin B1. J. Hazard. Mater. 2020, 399, 123154. [Google Scholar] [CrossRef]
- Xiong, Y.; Pei, K.; Wu, Y.Q.; Duan, H.; Lai, W.H.; Xiong, Y.H. Plasmonic ELISA based on enzyme-assisted etching of Au nanorods for the highly sensitive detection of aflatoxin B1in corn samples. Sens. Actuators B 2018, 267, 320–327. [Google Scholar] [CrossRef]
- Zhao, F.; Tian, Y.; Shen, Q.; Liu, R.; Shi, R.; Wang, H.; Yang, Z. A novel nanobody and mimotope based immunoassay for rapid analysis of aflatoxin B1. Talanta 2019, 195, 55–61. [Google Scholar] [CrossRef]
- Liu, N.; Wu, A. Chromogenic platform-based lateral flow immunoassay. In Food Safety & Mycotoxins; Wu, A., Ed.; Springer: Singapore, 2010; pp. 3–11. [Google Scholar] [CrossRef]
- Xing, K.Y.; Shan, S.; Liu, D.F.; Lai, W.H. Recent advances of lateral flow immunoassay for mycotoxins detection. TrAC Trends Anal. Chem. 2020, 133, 116087. [Google Scholar] [CrossRef]
- Famulok, M.; Mayer, G. Aptamers as tools in molecular biology and immunology. Curr. Top. Microb. Immun. 1999, 243, 123–136. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Vorobyeva, M.A.; Davydova, A.S.; Vorobjev, P.E.; Venyaminova, A.G. Key aspects of nucleic acid library design for in vitro selection. Int. J. Mol. Sci. 2018, 19, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randrianjatovo-Gbalou, I.; Rosario, S.; Sismeiro, O.; Varet, H.; Legendre, R.; Coppée, J.Y.; Huteau, V.; Pochet, S.; Delarue, M. Enzymatic synthesis of random sequences of RNA and RNA analogues by DNA polymerase theta mutants for the generation of aptamer libraries. Nucleic Acids Res. 2018, 46, 6271–6284. [Google Scholar] [CrossRef]
- Yan, J.; Xiong, H.; Cai, S.; Wen, N.; He, Q.; Liu, Y.; Peng, D.; Liu, Z. Advances in aptamer screening technologies. Talanta 2019, 200, 124–144. [Google Scholar] [CrossRef]
- Chang, Y.C.; Kao, W.C.; Wang, W.Y.; Yang, R.B.; Peck, K. Identification and characterization of oligonucleotides that inhibit Toll-like receptor 2-associated immune responses. FASEB J. 2009, 23, 3078–3088. [Google Scholar] [CrossRef] [PubMed]
- Nutiu, R.; Li, Y. In vitro selection of structure-switching signaling aptamers. Angew. Chem. Int. Ed. Engl. 2005, 44, 1061–1065. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Schubert, T.; Strehlitz, B. In vitro selection and interaction studies of a DNA aptamer targeting protein A. PLoS ONE. 2015, 10, e0134403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef] [PubMed]
- Mosing, R.K.; Bowser, M.T. Isolating aptamers using capillary electrophoresis-SELEX (CE-SELEX). Methods Mol. Biol. 2009, 535, 33–43. [Google Scholar] [CrossRef]
- Sinha, A.; Gopinathan, P.; Chung, Y.D.; Lin, H.Y.; Li, K.H.; Ma, H.P.; Huang, P.C.; Shiesh, S.C.; Lee, G.B. An integrated microfluidic platform to perform uninterrupted SELEX cycles to screen affinity reagents specific to cardiovascular biomarkers. Biosens. Bioelectron. 2018, 122, 104–112. [Google Scholar] [CrossRef]
- Takenaka, M.; Okumura, Y.; Amino, T.; Miyachi, Y.; Ogino, C.; Kondo, A. DNA-duplex linker for AFM-SELEX of DNA aptamer against human serum albumin. Bioorg. Med. Chem. Lett. 2017, 27, 954–957. [Google Scholar] [CrossRef]
- Hybarger, G.; Bynum, J.; Williams, R.F.; Valdes, J.J.; Chambers, J.P. A microfluidic SELEX prototype. Anal. Bioanal. Chem. 2006, 384, 191–198. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef] [Green Version]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Al Bawab, A.; Ismail, S.I. Aptamers chemistry: Chemical modifications and conjugation strategies. Molecules 2020, 25, 3. [Google Scholar] [CrossRef] [Green Version]
- Shuichiro, U.; Naohiko, S.; Yoichi, T.; Yoshikazu, K.; Yoshifumi, T.; Hironori, A.; Susumu, S.; Atsushi, U.; Kazuo, S. 3′ Poly(dA)-tailed thrombin DNA aptamer to increase DNase-resistance and clotting inhibitory activity. Bull. Chem. Soc. Jpn. 2008, 81, 1485–1491. [Google Scholar] [CrossRef]
- Prodeus, A.; Abdul-Wahid, A.; Fischer, N.W.; Huang, E.H.-B.; Cydzik, M.; Gariépy, J. Targeting the PD-1/PD-L1 immune evasion axis with DNA aptamers as a novel therapeutic strategy for the treatment of disseminated cancers. Mol. Ther. Nucleic Acids 2015, 28, e237. [Google Scholar] [CrossRef]
- Willis, M.C.; Collins, B.D.; Zhang, T.; Green, L.S.; Sebesta, D.P.; Bell, C.; Kellogg, E.; Gill, S.C.; Magallanez, A.; Knauer, S.; et al. Liposome-anchored vascular endothelial growth factor aptamers. Bioconjug. Chem. 1998, 9, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Musumeci, D.; Krauss, I.R.; Piccolo, M.; Irace, C.; Paduano, L.; Montesarchio, D. Exploring the conformational behaviour and aggregation properties of lipid-conjugated AS1411 aptamers. Int. J. Biol. Macromol. 2018, 118 Pt B, 1384–1399. [Google Scholar] [CrossRef] [PubMed]
- Gatto, B.; Palumbo, M.; Sissi, C. Nucleic acid aptamers based on the G-quadruplex structure: Therapeutic and diagnostic potential. Curr. Med. Chem. 2009, 16, 1248–1265. [Google Scholar] [CrossRef]
- Ilgu, M.; Nilsen-Hamilton, M. Aptamers in Analytics. Analyst 2016, 141, 1551–1568. [Google Scholar] [CrossRef] [Green Version]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS mapper: A web based server for predicting G-quadruplexes in nucleic acid sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef] [PubMed]
- Markham, N.R.; Zuker, M. UNAFold: Software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008, 453, 3–31. [Google Scholar] [CrossRef] [PubMed]
- Evtugyn, G.; Hianik, T. Electrochemical immuno- and aptasensors for mycotoxin determination. Chemosensors 2019, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, R.; Adams, M.C.; Naik, R.R.; Milam, V.T. Analyzing secondary structure patterns in DNA aptamers identified via CompELS. Molecules 2019, 24, 1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekhon, S.S.; Lee, S.H.; Lee, K.A.; Min, J.; Lee, B.T.; Kim, K.W.; Ahn, J.Y.; Kim, Y.H. Defining the copper binding aptamotif and aptamer integrated recovery platform (AIRP). Nanoscale 2017, 9, 2883–2894. [Google Scholar] [CrossRef]
- Cai, S.; Yan, I.; Xiong, H.; Liu, Y.; Peng, D.; Liu, Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 2018, 143, 5317–5338. [Google Scholar] [CrossRef]
- Hamaguchi, N.; Ellington, E.; Stanton, M. Aptamer beacons for the direct detection of proteins. Anal. Biochem. 2001, 294, 126–131. [Google Scholar] [CrossRef] [Green Version]
- He, J.-L.; Wu, Z.-S.; Zhou, H.; Wang, H.-Q.; Jiang, J.-H.; Shen, G.-L.; Yu, R.Q. Fluorescence aptameric sensor for strand displacement amplification detection of cocaine. Anal. Chem. 2010, 82, 1358–1364. [Google Scholar] [CrossRef]
- Deng, L.; Du, J.; Xu, J.-J.; Chen, H.-Y. An off-on-off electrochemiluminescence approach for ultrasensitive detection of thrombin. Biosens. Bioelectron. 2014, 59, 58–63. [Google Scholar] [CrossRef]
- Alyamani, B.J.; Alsager, O.A.; Zourob, M. Label-free fluorescent aptasensor for small targets via displacement of groove bound curcumin molecules. Sensors 2019, 19, 4181. [Google Scholar] [CrossRef] [Green Version]
- Castillo, G.; Spinella, K.; Poturnayova, A.; Snejdarkova, M.; Mosiello, L.; Hianik, T. Detection of aflatoxin B1 by aptamer-based biosensor using PAMAM dendrimers as immobilization platform. Food Control 2015, 52, 9–18. [Google Scholar] [CrossRef]
- Ma, X.; Wang, W.; Chen, X.; Xia, Y.; Wu, S.; Duan, N.; Wang, Z. Selection, identification, and application of aflatoxin B1 aptamer. Eur. Food Res. Technol. 2014, 238, 919–925. [Google Scholar] [CrossRef]
- Nguyen, B.H.; Tran, L.D.; Do, Q.P.; Nguyen, H.L.; Tran, N.H.; Nguyen, P.X. Label-free detection of aflatoxin M1 with electrochemical Fe3O4/polyaniline-based aptasensor. Mat. Sci. Eng. C 2013, 33, 2229–2234. [Google Scholar] [CrossRef] [PubMed]
- Hamula, C.L.A.; Guthrie, J.W.; Zhang, H.; Li, X.F.; Le, X.C. Selection and analytical applications of aptamers. TrAC Trends Anal. Chem. 2006, 25, 681–691. [Google Scholar] [CrossRef]
- Malhotra, S.; Pandey, A.K.; Rajput, Y.S.; Sharma, R. Selection of aptamers for aflatoxin M1 and their characterization. J. Mol. Recognit. 2014, 27, 493–500. [Google Scholar] [CrossRef]
- Chen, X.; Huang, Y.; Duan, N.; Wu, S.; Xia, Y.; Ma, X.; Zhu, C.; Jiang, Y.; Ding, Z.; Wang, Z. Selection and characterization of single stranded DNA aptamers recognizing fumonisin B1. Microchim. Acta 2014, 181, 1317–1324. [Google Scholar] [CrossRef]
- Chen, X.; Bai, X.; Li, H.; Zhang, B. Aptamer-based microcantilever array biosensor for detection of fumonisin B-1. RSC Adv. 2015, 5, 35448–35452. [Google Scholar] [CrossRef]
- Frost, N.R.; McKeague, M.; Falcioni, D.; DeRosa, M.C. An in solution assay for interrogation of affinity and rational minimer design for small molecule-binding aptamers. Analyst 2015, 140, 6643–6651. [Google Scholar] [CrossRef]
- McKeague, M.; Bradley, C.R.; De Girolamo, A.; Visconti, A.; Miller, L.D.; De Rosa, M.C. Screening and initial binding assessment of fumonisin B1 aptamers. Int. J. Mol. Sci. 2010, 11, 4864–4881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Aguado, J.A.; Penner, G. Determination of ochratoxin A with a DNA aptamer. J. Agric. Food Chem. 2008, 56, 10456. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Duan, N.; Zhang, W.; Zhao, S.; Wang, Z. Screening and development of DNA aptamers as capture probes for colorimetric detection of patulin. Anal. Biochem. 2016, 508, 58–64. [Google Scholar] [CrossRef]
- He, B.; Dong, X. Aptamer based voltammetric patulin assay based on the use of ZnO nanorods. Microchim. Acta 2018, 185, 462. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Y.; Duan, N.; Wu, S.; Xia, Y.; Ma, X.; Zhu, C.; Jiang, Y.; Wang, Z. Screening and identification of DNA aptamers against T-2 toxin assisted by graphene oxide. J. Agric. Food Chem. 2014, 62, 10368–10374. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Y.; Duan, N.; Wu, S.; Ma, X.; Xia, Y.; Zhu, C.; Jiang, Y.; Wang, Z. Selection and identification of ssDNA aptamers recognizing zearalenone. Anal. Bioanal. Chem. 2013, 405, 6573–6581. [Google Scholar] [CrossRef]
- Liu, X.; Wen, Y.; Wang, W.; Zhao, Z.; Han, Y.; Tang, K.; Wang, D. Nanobody-based electrochemical competitive immunosensor for the detection of AFB1 through AFB1-HCR as signal amplifier. Microchim. Acta 2020, 187, 352. [Google Scholar] [CrossRef] [PubMed]
- Romanazzo, D.; Ricci, F.; Volpe, G.; Elliott, C.T.; Vesco, S.; Kroeger, K.; Moscone, D.; Stroka, J.; Van Egmond, H.; Vehniäinen, M.; et al. Development of a recombinant Fab-fragment based electrochemical immunosensor for deoxynivalenol detection in food samples. Biosens. Bioelectron. 2010, 25, 2615–2621. [Google Scholar] [CrossRef]
- Arshavsky-Graham, S.; Urmann, K.; Salama, R.; Massad-Ivanir, N.; Walter, J.-G.; Scheper, T.; Segal, E. Aptamers vs. antibodies as capture probes in optical porous silicon biosensors. Analyst 2020, 145, 4991–5003. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gopinath, S.C.B.; Wang, Z.; Li, Y.; Anbu, P.; Zhang, W. Zeolite-iron oxide nanocomposite from fly ash formed a ‘clubbell’ structure: Integration of cardiac biocapture macromolecules in serum on microelectrodes. Microchim. Acta 2021, 188, 187. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; He, B. Ultrasensitive sandwich-type electrochemical biosensor based on octahedral gold nanoparticles modified poly (ethylenimine) functionalized graphitic carbon nitride nanosheets for the determination of sulfamethazine. Sens. Actuators B 2021, 329, 129158. [Google Scholar] [CrossRef]
- Rashid, J.I.A.; Yusof, N.A. The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: A review. Sens. Bio-Sens. Res. 2017, 16, 19–31. [Google Scholar] [CrossRef]
- Shi, Z.-Y.; Zheng, Y.-T.; Zhang, H.-B.; He, C.-H.; Wu, W.-D.; Zhang, H.-B. DNA electrochemical aptasensor for detecting fumonisins B1 based on graphene and thionine nanocomposite. Electroananlysis 2015, 27, 1097–1103. [Google Scholar] [CrossRef]
- Rosy Goyal, R.N.; Shim, Y.-B. Glutaraldehyde sandwiched amino functionalized polymer based aptasensor for the determination and quantification of chloramphenicol. RSC Adv. 2015, 5, 69356–69364. [Google Scholar] [CrossRef]
- Tian, J.; Wei, W.; Wang, J.; Ji, S.; Chen, G.; Lu, J. Fluorescence resonance energy transfer aptasensor between nanoceria and graphene quantum dots for the determination of ochratoxin A. Anal. Chim. Acta 2018, 1000, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Vasilescu, A.; Wang, Q.; Coffinier, Y.; Li, M.; Boukherroub, R.; Szunerits, S. Electrophoretic approach for the simultaneous deposition and functionalization of reduced graphene oxide nanosheets with diazonium compounds: Application for lysozyme sensing in serum. ACS Appl. Mater. Interfaces 2017, 9, 12823–12831. [Google Scholar] [CrossRef] [PubMed]
- Rabai, S.; Benounis, M.; Catanante, G.; Baraket, A.; Errachid, A.; Jaffrezic Renault, N.; Marty, J.-L.; Rhouati, A. Development of a label-free electrochemical aptasensor based on diazonium electrodeposition: Application to cadmium detection in water. Anal. Biochem. 2021, 612, 113956. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Y.; Ma, X.; Jia, F.; Guo, X.; Wang, Z. Impedimetric aptamer-based determination of the mold toxin fumonisin B1. Microchim. Acta 2015, 182, 1709–1714. [Google Scholar] [CrossRef]
- Dupont-Filliard, A.; Billon, M.; Livache, T.; Guillerez, S. Biotin/avidin system for the generation of fully renewable DNA sensor based on biotinylated polypyrrole film. Anal. Chim. Acta 2004, 515, 271–277. [Google Scholar] [CrossRef]
- Yang, Z.; Kasprzyk-Hordern, B.; Goggins, S.; Frost, C.G.; Estrela, P. A novel immobilization strategy for electrochemical detection of cancer biomarkers: DNA-directed immobilization of aptamer sensors for sensitive detection of prostate specific antigens. Analyst 2015, 140, 2628–2633. [Google Scholar] [CrossRef] [Green Version]
- Kong, T.; Zhou, R.; Zhang, Y.; Hao, L.; Cai, X.; Zhu, B. AS1411 aptamer modified carbon dots via polyethylenimine-assisted strategy for efficient targeted cancer cell imaging. Cell Prolif. 2020, 53, 312713. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Gao, R.; Chen, Q.; Jia, L. Dual-aptamers labeled polydopamine-polyethyleneimine copolymer dots assisted engineering a fluorescence biosensor for sensitive detection of Pseudomonas aeruginosa in food samples. Spectrochim. Acta Part A 2020, 224, 117417. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Ma, X.; Ding, C.; Jia, L. Colorimetric determination of lysozyme based on the aggregation of gold nanoparticles controlled by a cationic polymer and an aptamer. Microchim. Acta 2016, 183, 2353–2359. [Google Scholar] [CrossRef]
- Malile, B.; Chen, J.I.L. Factors influencing polyelectrolyte-aptamer multilayered films with target-controlled permeability for sensing applications. Analyst 2016, 141, 3794–3802. [Google Scholar] [CrossRef]
- Oberhaus, F.W.; Frense, D.; Beckmann, D. Immobilization techniques for aptamers on gold electrodes for the electrochemical detection of proteins: A review. Biosensors 2020, 10, 45. [Google Scholar] [CrossRef]
- Liu, J.; Jasim, I.; Shen, Z.; Zhao, L.; Dweik, M.; Zhang, S.; Almasri, M. A microfluidic based biosensor for rapid detection of Salmonella in food products. PLoS ONE 2019, 14, e0216873. [Google Scholar] [CrossRef] [Green Version]
- Chaubey, A.; Malhotra, B.D. Mediated biosensors. Biosens. Bioelectron. 2002, 17, 441–456. [Google Scholar] [CrossRef]
- Rai, M.; Jogee, P.S.; Ingle, A.P. Emerging nanotechnology for detection of mycotoxins in food and feed. Int. J. Food Sci. Nutr. 2015, 66, 363–370. [Google Scholar] [CrossRef]
- Ruscito, A.; Smith, M.; Goudreau, D.N.; De Rosa, M.C. Current status and future prospects for aptamer-based mycotoxin detection. J. AOAC Inetrn. 2016, 99, 865–877. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Z.; Zhang, Q.; Li, P. Mycotoxin determination in foods using advanced sensors based on antibodies or aptamers. Toxins 2016, 8, 239. [Google Scholar] [CrossRef] [Green Version]
- Geleta, G.S.; Zhao, Z.; Wang, Z. A novel reduced graphene oxide/molybdenum disulfide/polyaniline nanocomposite-based electrochemical aptasensor for detection of aflatoxin B1. Analyst 2018, 143, 1644–1649. [Google Scholar] [CrossRef]
- Yagati, A.K.; Chavan, S.G.; Baek, C.; Lee, M.-H.; Min, J. Label-free impedance sensing of aflatoxin B1 with polyaniline nanofibers/Au nanoparticle electrode array. Sensors 2018, 18, 1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, G.; Li, X.; Cui, F.; Qiu, Q.; Chen, X.; Huang, H. Aflatoxin B1 electrochemical aptasensor based on tetrahedral DNA nanostructures functionalized three dimensionally ordered macroporous MoS2–AuNPs film. ACS Appl. Mater. Interfaces 2018, 10, 17551–17559. [Google Scholar] [CrossRef]
- Wang, C.; Qian, J.; An, K.; Ren, C.; Lu, X.; Hao, N.; Liu, Q.; Li, H.; Huang, X.; Wang, K. Fabrication of magnetically assembled aptasensing device for label-free determination of aflatoxin B1 based on EIS. Biosens. Bioelectron. 2018, 108, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.S.; Wei, M.; Wei, W.; Liu, Y.; Liu, S. Electrochemical aptasensor for aflatoxin B1 based on smart host-guest recognition of β-cyclodextrin polymer. Biosens. Bioelectron. 2019, 129, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, D.; Zhu, C.; Shen, X.; Liu, Y.; You, T. Sensitivity programmable ratiometric electrochemical aptasensor based on signal engineering for the detection of aflatoxin B1 in peanut. J. Hazard. Mater. 2020, 387, 122001. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Zhao, Q. A signal-on electrochemical aptasensor for rapid detection of aflatoxin B1 based on competition with complementary DNA. Biosens. Bioelectron. 2019, 144, 111641. [Google Scholar] [CrossRef]
- Selvolini, G.; Lettieria, M.; Tassoni, L.; Gastaldello, S.; Grillo, M.; Maran, C.; Marrazza, G. Electrochemical enzyme-linked oligonucleotide array for aflatoxin B1 detection. Talanta 2019, 203, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, Q. A reagentless electrochemical sensor for aflatoxin B1 with sensitive signal-on responses using aptamer with methylene blue label at specific internal thymine. Biosens. Bioelectron. 2020, 167, 112478. [Google Scholar] [CrossRef]
- Smolko, V.; Shurpik, D.; Porfireva, A.; Evtugyn, G.; Stoikov, I.; Hianik, T. Electrochemical aptasensor based on poly(Neutral red) and carboxylated pillar[5]arene for sensitive determination of aflatoxin M1. Electroanalysis 2018, 30, 486–496. [Google Scholar] [CrossRef]
- Lin, T.; Shen, Y. Fabricating electrochemical aptasensors for detecting aflatoxin B1 via layer-by-layer self-assembly. J. Electroanal. Chem. 2020, 870, 114247. [Google Scholar] [CrossRef]
- Jalalian, S.H.; Ramezani, M.; Danesh, N.M.; Alibolandi, M.; Abnous, K.H.; Taghdisi, S.M. A novel electrochemical aptasensor for detection of aflatoxin M1 based on target-induced immobilization of gold nanoparticles on the surface of electrode. Biosens. Bioelectron. 2018, 117, 487–492. [Google Scholar] [CrossRef]
- Ong, C.C.; Sangu, S.S.; Illias, N.M.; Gopinath, S.C.B.; Saheed, M.S.M. Iron nanoflorets on 3D-graphene-nickel: A ‘Dandelion’ nanostructure for selective deoxynivalenol detection. Biosens. Bioelectron. 2020, 154, 112088. [Google Scholar] [CrossRef]
- Li, D.; Zhang, X.; Ma, Y.; Deng, Y.; Hu, R.; Yang, Y. Preparation of an OTA aptasensor based on a metal-organic framework. Anal. Methods 2018, 10, 3273–3279. [Google Scholar] [CrossRef]
- Zejli, K.; Goud, K.Y.; Marty, J.-L. Label free aptasensor for ochratoxin A detection using polythiophene-3-carboxylic acid. Talanta 2018, 185, 513–519. [Google Scholar] [CrossRef]
- Wang, Y.; Ning, G.; Bi, H.; Wu, Y.; Liu, G.; Zhao, Y. A novel ratiometric electrochemical assay for ochratoxin A coupling Au nanoparticles decorated MoS2 nanosheets with aptamer. Electrochim. Acta 2018, 285, 120–127. [Google Scholar] [CrossRef]
- Suea-Ngam, A.; Howes, P.D.; Stanley, C.E.; de Mello, A.J. An exonuclease I-assisted silver-metallized electrochemical aptasensor for ochratoxin A detection. ACS Sens. 2019, 28, 1560–1568. [Google Scholar] [CrossRef]
- Gökçe, G.; Aissa, S.B.; Nemčeková, K.; Catanante, G.; Raouafi, N.; Marty, J.-L. Aptamer-modified pencil graphite electrodes for the impedimetric determination of ochratoxin A. Food Control 2020, 115, 107271. [Google Scholar] [CrossRef]
- Qiao, X.; Wang, Y.; Arsalan, M.; Yang, S.; Wang, Y.; Sheng, Q.; Yue, T. A label-free aptasensor for the detection of Ochratoxin A based on competitive molecule-level interactions. J. Electrochem. Soc. 2020, 167, 147518. [Google Scholar] [CrossRef]
- Chen, W.; Yan, C.; Cheng, L.; Yao, L.; Xue, F.; Xu, J. An ultrasensitive signal-on electrochemical aptasensor for ochratoxin A determination based on DNA controlled layer-by-layer assembly of dual gold nanoparticle conjugates. Biosens. Bioelectron. 2018, 117, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhi, H.; Zhu, M.; Wang, F.; Meng, H.; Feng, L. Electrochemical/visual dual-readout aptasensor for Ochratoxin A detection integrated into a miniaturized paper-based analytical device. Biosens. Bioelectron. 2021, 180, 113146. [Google Scholar] [CrossRef] [PubMed]
- Mejri-Omrani, N.; Miodek, A.; Zribi, B.; Marrakchi, M.; Hamdi, M.; Marty, J.-L.; Korri-Youssouf, H. Direct detection of OTA by impedimetric aptasensor based on modified polypyrrole-dendrimers. Anal. Chim. Acta 2016, 920, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Aissa, S.B.; Sherazi, T.A.; Catanante, G.; Hayat, A.; Marty, J.-L. Development of an impedimetric aptasensor for label free detection of patulin in apple juice. Molecules 2019, 24, 1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, T.; Yu, C.; Gao, R.; Chen, J.; Yu, Y.; Geng, Y.; Wen, Y.; He, J. A novel sandwich aptasensor for detecting T-2 toxin based on rGO-TEPAAu@Pt nanorods with a dual signal amplification strategy. Biosens. Bioelectron. 2019, 144, 111635. [Google Scholar] [CrossRef] [PubMed]
- Mu, Z.; Ma, L.; Wang, J.; Zhou, J.; Yuan, Y.; Bai, L. A target-induced amperometic aptasensor for sensitive zearalenone detection by CS@AB-MWCNTs nanocomposite as enhancers. Food Chem. 2021, 340, 128128. [Google Scholar] [CrossRef] [PubMed]
- Azri, F.A.; Eissa, S.; Zourob, M.; Chinnappan, R.; Sukor, R.; Yusof, N.A.; Raston, N.H.A.; Alhoshani, A.; Jinap, S. Electrochemical determination of zearalenone using a label-free competitive aptasensor. Microchim. Acta 2020, 187, 266. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Tang, Z.; Jiang, K.; Huang, Q.; Meng, J.; Nie, D.; Zhao, Z. Dual-target electrochemical aptasensor based on co-reduced molybdenum disulfide and Au NPs (rMoS2-Au) for multiplex detection of mycotoxins. Biosens. Bioelectron. 2020, 150, 111894. [Google Scholar] [CrossRef]
- Nogueira, J.J.; González, L. Molecular dynamics simulations of binding modes between Methylene blue and DNA with alternating GC and AT sequences. Biochemistry 2014, 53, 2391–2412. [Google Scholar] [CrossRef]
- Lucarelli, F.; Marrazza, G.; Turner, A.P.F.; Mascini, M. Carbon and gold electrodes as electrochemical transducers for DNA hybridisation sensors. Biosens. Bioelectron. 2004, 19, 515–530. [Google Scholar] [CrossRef]
- Wagenknecht, H.-A. Electron transfer processes in DNA: Mechanisms, biological relevance and applications in DNA analytics. Nat. Prod. Rep. 2006, 23, 973–1006. [Google Scholar] [CrossRef]
- Pheeney, C.G.; Barton, J.K. DNA electrochemistry with tethered Methylene blue. Langmuir 2012, 28, 7063–7070. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.; Zuo, X.; Yang, R.; Xia, F.; Plaxco, K.W.; White, R.J. Comparing the properties of electrochemical-based DNA sensors employing different redox tags. Anal. Chem. 2009, 81, 9109–9113. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Lim, B.J.; Li, B.; Jiang, Y.S.; Sessler, J.L.; Ellington, A.D. Reagentless, ratiometric electrochemical DNA sensors with improved robustness and reproducibility. Anal. Chem. 2014, 86, 8010–8016. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Q.; Chen, S.; Cheng, F.; Wang, H.; Peng, W. Surface plasmon resonance biosensor based on smart phone platforms. Sci. Rep. 2015, 5, 12864. [Google Scholar] [CrossRef] [Green Version]
- Bano, A.; Olivero, M.; Vallan, A.; Perrone, G. Portable SPR refractometer for contamination analysis in a flowing liquid. In Proceedings of the 19th Italian National Conference on Photonic Technologies (Fotonica 2017), Padua, Italy, 3–5 May 2017; pp. 1–3. [Google Scholar] [CrossRef]
- Brulé, T.; Granger, G.; Bukar, N.; Deschênes-Rancourt, C.; Havard, T.; Schmitzer, A.R.; Martel, R.; Masson, J.-F. A field-deployed surface plasmon resonance (SPR) sensor for RDX quantification in environmental waters. Analyst 2017, 142, 2161–2168. [Google Scholar] [CrossRef] [Green Version]
- Evtugyn, G.; Hianik, T. Aptamer-based biosensors for mycotoxin detection. In Nanomycotoxicology. Treating Mycotoxins in the Nano Way; Rai, M., Abd-Elsalam, K.A., Eds.; Academic Press: San Diego, CA, USA, 2020; pp. 35–70. [Google Scholar]
- Mahmoudpour, M.; Dolatabadi, J.E.N.; Torbati, M.; Tazehkand, A.P.; Homayouni-Rad, A.; de la Guardia, M. Nanomaterials and new biorecognition molecules based surface plasmon resonance biosensors for mycotoxin detection. Biosens. Bioelectron. 2019, 143, 111603. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, I.S.; da Silva, A.G., Jr.; de Andrade, C.A.S.; Oliveira, M.D.L. Biosensors for early detection of fungi spoilage and toxigenic and mycotoxins in food. Curr. Opin. Food Sci. 2019, 29, 64–79. [Google Scholar] [CrossRef]
- Mirón-Mérida, V.A.; Gong, Y.Y.; Goycoolea, F.M. Aptamer-based detection of fumonisin B1: A critical review. Anal. Chim. Acta 2021, 1160, 338395. [Google Scholar] [CrossRef]
- Hill, R.T. Plasmonic biosensors. Wiley Interdiscip. Rev. Nanomed. Nanobiotech. 2015, 7, 152–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Liu, X.; Lin, Z. Recent developments and applications of surface plasmon resonance biosensors for the detection of mycotoxins in foodstuffs. Food Chem. 2012, 132, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Feng, M.; Zuo, L.; Zhu, Z.; Wang, F.; Chen, L.; Li, J.; Shan, G.; Luo, S.-Z. An aptamer based surface plasmon resonance biosensor for the de- tection of ochratoxin A in wine and peanut oil. Biosens. Bioelectron. 2015, 65, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Bianco, M.; Sonato, A.; De Girolamo, A.; Pascale, M.; Romanato, F.; Rinaldi, R.; Arima, V. An aptamer-based SPR-polarization platform for high sensitive OTA detection. Sens. Actuators B 2017, 241, 314–320. [Google Scholar] [CrossRef]
- Sun, L.; Wu, L.; Zhao, Q. Aptamer based surface plasmon resonance sensor for aflatoxin B1. Microchim. Acta 2017, 184, 2605–2610. [Google Scholar] [CrossRef]
- Gnedenko, O.V.; Mezentsev, Y.V.; Molnar, A.A.; Lisitsa, A.V.; Ivanov, A.S.; Archakov, A.I. Highly sensitive detection of human cardiac myoglobin using a reverse sandwich immunoassay with a gold nanoparticle-enhanced surface plasmon resonance biosensor. Anal. Chim. Acta 2013, 759, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Homola, J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, Z.; Li, B.; Liu, Z.; Jia, L.; Zuo, L.; Chen, L.; Zhu, Z.; Shan, G.; Luo, S.-Z. A direct determination of AFBs in vinegar by aptamer-based surface plasmon resonance biosensor. Toxicon 2018, 146, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Nabok, A.; Al-Rubaye, A.G.; Al-Jawdah, A.M.; Tsargorodska, A.; Marty, J.-L.; Catanante, G.; Szekacs, A.; Takacs, E. Novel optical biosensing technologies for detection of mycotoxins. Opt. Laser Technol. 2019, 109, 212–221. [Google Scholar] [CrossRef]
- Al-Rubaye, A.; Nabok, A.; Catanante, G.; Marty, J.-L.; Takacs, E.; Szekacs, A. Detection of ochratoxin A in aptamer assay using total internalreflection ellipsometryA. Sens. Actuators B 2018, 263, 248–251. [Google Scholar] [CrossRef] [Green Version]
- Caglayan, M.O.; Üstündağ, Z. Detection of zearalenone in an aptamer assay using attenuated internal reflection ellipsometry and it’s cereal sample applications. Food Chem. Toxicol. 2020, 136, 111081. [Google Scholar] [CrossRef] [PubMed]
- Evtugyn, G.; Belyakova, S.; Porfireva, A.; Hianik, T. Electrochemical aptasensors based on hybrid metal-organic frameworks. Sensors 2020, 20, 6963. [Google Scholar] [CrossRef]
- Evtugyn, G.; Porfireva, A.; Shamagsumova, R.; Hianik, T. Advances in electrochemical aptasensors based on carbon nanomaterials. Chemosensors 2020, 8, 96. [Google Scholar] [CrossRef]
- Le, V.T.; Vasseghian, Y.; Dragoi, E.-N.; Moradi, M.; Khaneghah, A.M. A review on graphene-based electrochemical sensor for mycotoxins detection. Food Chem. Toxicol. 2021, 148, 111931. [Google Scholar] [CrossRef]
- Masson, J.-F. Portable and field-deployed surface plasmon resonance and plasmonic sensors. Analyst 2020, 145, 3778–3800. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Tian, S.; Zhao, W.; Liu, K.; Ma, X.; Guo, J. Aptamer-based lateral flow assay on-site biosensors. Biosens. Bioelectron. 2021, 186, 113279. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-X.; Zhao, Y.-W.; Li, Z.; Liu, B.-S.; Zhang, D. Development and application of aptamer-based surface-enhanced Raman spectroscopy sensors in quantitative analysis and biotherapy. Sensors 2019, 19, 3806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
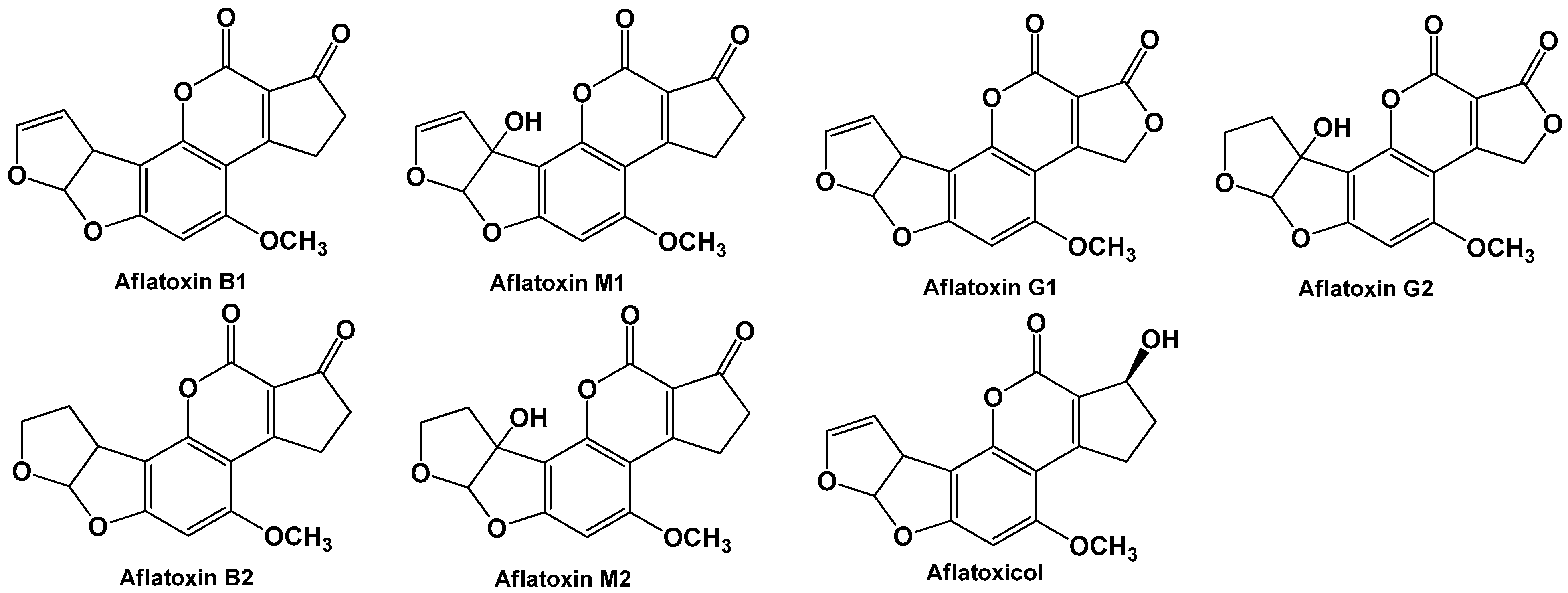
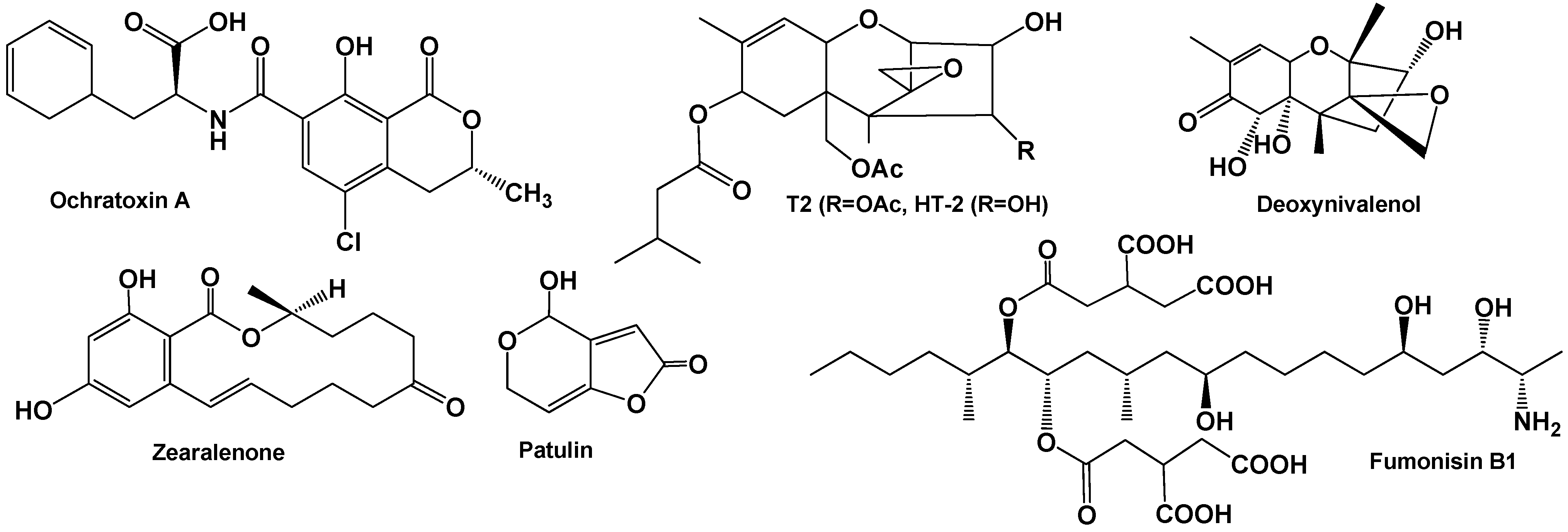
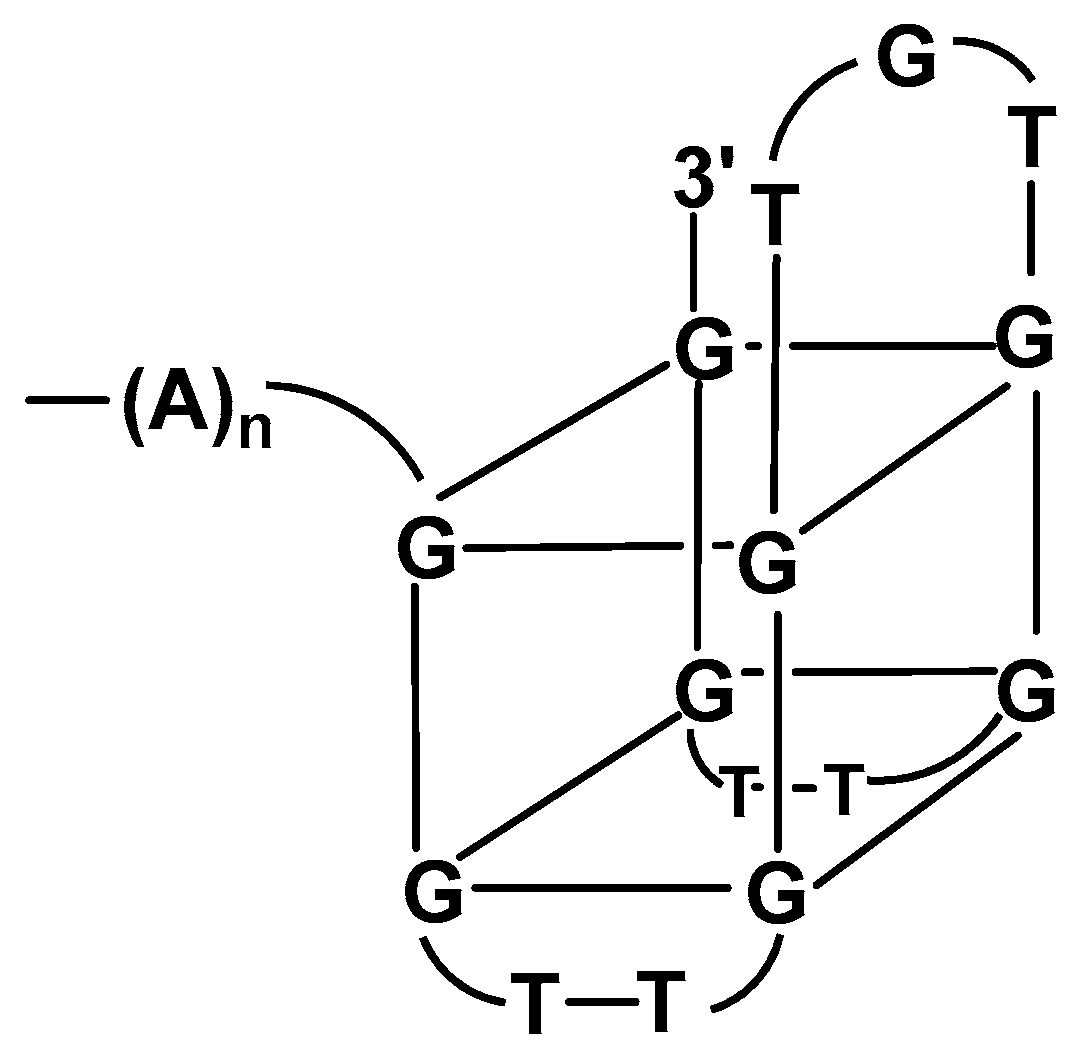
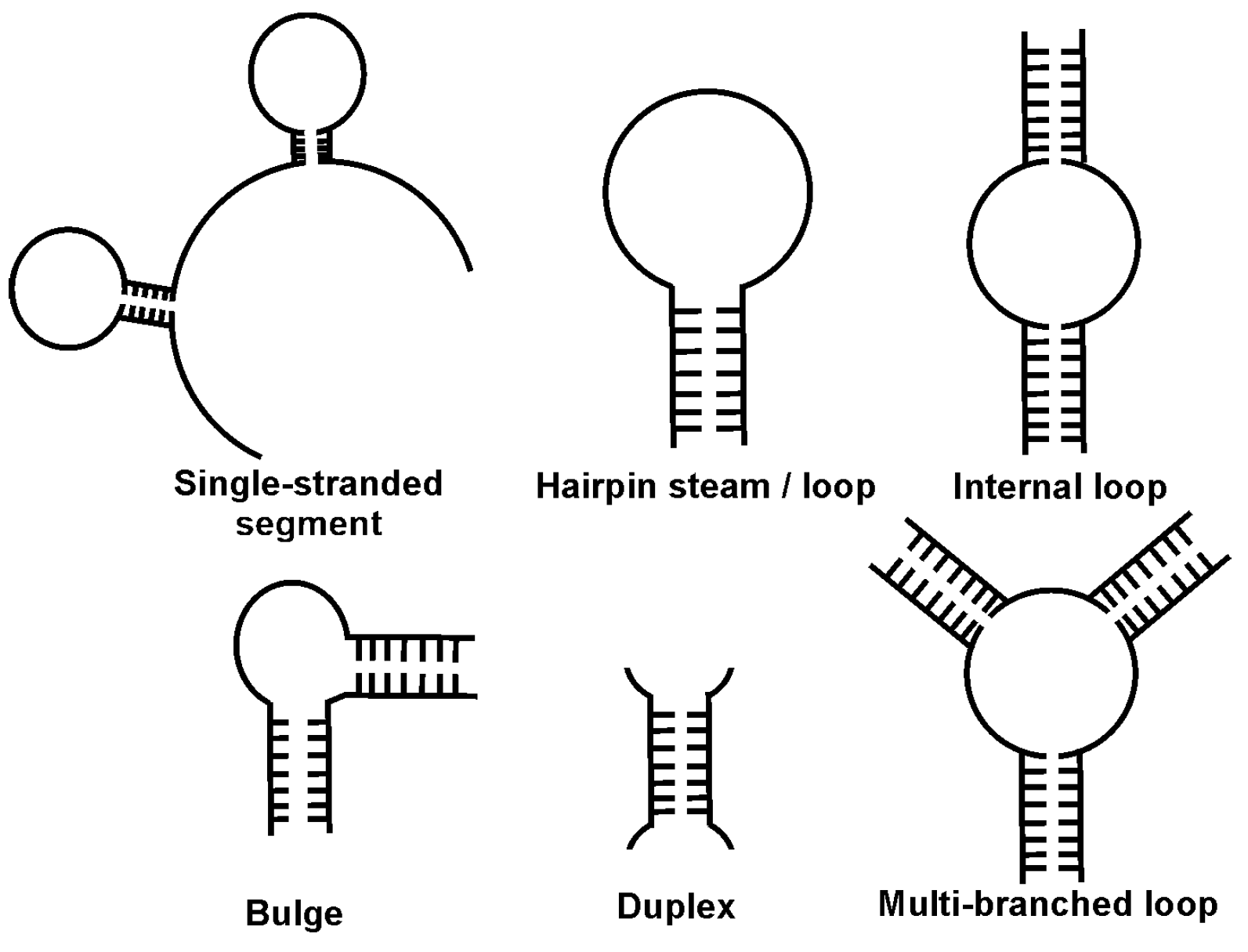

| Mycotoxin | Fungal Source | IARC Group | Contaminated Food | Maximal Admissible Levels (μg/kg) | |
|---|---|---|---|---|---|
| USA Food and Drug Administration | European Food Safety Authority | ||||
| Aflatoxins (B1, B2, G1, G2) | Aspergillus flavus Aspergillus parasiticus | 1 | Wheat, maize, rice, peanut, pistachio, almond, hazelnut, ground nuts, tree nuts, figs, cottonseed | 20 | 4–10 for total 2–5 for B1 0.1 for B1 in baby food |
| Aflatoxin M1 | Metabolite of aflatoxin B1 | 2B | Milk and dairy products | 0.5 | 0.05 0.025 baby milk |
| Fumonisin B1, B2, B3 | Fusarium verticillionides Fusarium proliferatum | 2B | Maize, asparagus, corn-based food, white and yellow popcorn, sweet corn | 2000–4000 | 800–1000 200 baby food |
| Ochratoxin A | Aspergillus ochraceus Penicillium verrucosum Aspergillus carbonarius | 2B | Cereals, coffee, cocoa, wine, beer, dried fruits, grapes, pig kidney | Not set | 3–10, 0.5 baby food |
| Patulin | Penicillium expansum | 3 | Maize, asparagus, apple, pears, grapes, vegetables, cereals and cheese. | 50 | 25–50 10 baby food |
| Zearalenone | Fusarium graminearum Fusarium culmorum | 2A | Wheat, corn, barley, oats, sorghum and sesame seeds, hay and corn silage. | Not set | 50–100 20 baby food |
| Deoxynivalenol | Fusarium graminearum Fusarium culmorum | 3 | Corn, wheat, oats, barley, rice, grains, beer, animal’s kidney and liver, milk, eggs | 1000 | 750–1250 200 baby food |
| Nivalenol | Fusarium graminearum Fusarium culmorum | 3 | Oats, barley, maize, wheat, bread and fine bakery wares, pasta, cereals | Not set | 1.2 |
| T-2 toxin | Fusarium sporotrichioides | 3 | Maize, wheat, corn gluten feed, corn, gluten meal, barley, bran | Not set | 0.012–0.043 |
| Method | Key Aspects | Advantages | Disadvantages |
|---|---|---|---|
| IP-SELEX | Includes immunoprecipitation. | Selects aptamers against proteins under normal physiological conditions. Increased affinity and specificity. | More time-consuming than standard SELEX. |
| Capture-SELEX | Oligonucleotide library immobilized on a support instead of the targets to identify aptamers against small molecules. | Suitable for the selection of aptamers against small molecules. Immobilization of the target not required. Used for the discovery of structure-switching aptamers. | Some oligonucleotides from the library might be not released/selected. |
| Cell-SELEX | Utilizes whole live cells as targets for selection of aptamers. | Prior knowledge of the target not required. Aptamers are selected against molecules in their native state. Many potential targets available on the cell surface. Protein purification is not required. | Suitable for cell surface targets. Requires high level of technical expertise. Costly. Time consuming. Post SELEX identification of the target is required. |
| CE-SELEX | Involves separation of ions based on electrophoretic mobility. | Fast. Only few (1–4) rounds of selection required. Reduced non-specific binding. Target immobilization is not required. | Not suitable for small molecules. Expensive equipment. |
| M-SELEX | Combines SELEX with a microfluidic system. | Rapid. Very efficient (only small amounts of reagents needed).Applicable to small molecules. | Low purity/recovery of aptamers. Target immobilization required. |
| AFM-SELEX | Employs AFM to create 3D image of the sample surface. | Able to isolate high affinity aptamers. Fast (only 3–4 rounds are required). | Expensive equipment is required. Immobilization of target and aptamers are required |
| Mycotoxin | Sequence 5′-3′ | Ref. |
|---|---|---|
| Aflatoxin B1 | GTT GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CCT TCG CTA GGC CCA CA | [106,117] |
| AGC AGC ACA GAG GTC AGA TGG TGC TAT CAT GCG CTC AAT GGG AGA CTT TAG CTG CCC CCA CCT ATG CGT GCT ACC GTG AA | [118] | |
| Aflatoxin M1 | ACT GCT AGA GAT TTT CCA CAT | [119] |
| GTT GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CCT TCG CTA GGC CCA CA | [120] | |
| ATC CGT CAC ACC TGC TCT GAC GCT GGG GTC GAC CCG GAG AAA TGC ATT CCC CTG TGG TGT TGG CTC CCG TAT | [121] | |
| Fumonisin B1 | CGA TCT GGA TAT TAT TTT TGA TAC CCC TTT GGG GAG ACA T | [122] |
| ATA CCA GCT TAT TCA ATT AAT CGC ATT ACC TTA TAC CAG CTT ATT CAA TTA CGT CTG CAC ATA CCA GCT TAT TCA ATT AGA TAG TAA GTG CAA TCT | [123] | |
| ATA CCA GCT TAT TCA ATT AAT CGC ATT ACC TTA TAC CAG CTT ATT CAA TTA CGT CTG CAC ATA CCA GCT TAT TCA ATT | [124] | |
| AAT CGC ATT ACC TTA TAC CAG CTT ATT CAA TTA CGT CTG CAC ATA CCA GCT TAT TCA ATT | [125] | |
| Ochratoxin A | GAT CGG GTG TGG GTG GCG TAA AGG GAG CAT CGG ACA; TGG TGG CTG TAG GTC AGC ATC TGA TCG GGT GTG GGT GGC GTA AAG GGA GCA TCG GAC AAC G | [126] |
| Patulin | GGC CCG CCA ACC CGC ATC ATC TAC ACT GAT ATT TTA CCT T | [127] |
| SH-CAGCTCAGAAGCTTGATCCT-GGCC CGC CAA CCC GCA TCA TCT ACA CTG ATA TTT TAC CTT GAC TCG AAG TCG TGC ATC TG | [128] | |
| T-2 Toxin | GTA TAT CAA GCA TCG CGT GTT TAC ACA TGC GAG AGG TGA A | [129] |
| Zearalenone | TCA TCT ATC TAT GGT ACA TTA CTA TCT GTA ATG TGA TAT | [130] |
| Transducer | Transduction Principles | Samples Analyzed | LOD, Linearity Range | Ref. |
|---|---|---|---|---|
| Aflatoxin B1 | ||||
| Glassy carbon electrode (GCE) covered with reduced graphene oxide (rGO) polyaniline/nanoAu/MoS2 composite | Differential pulse voltammetry (DPV), electrochemical impedance spectroscopy (EIS) with [Fe(CN)6]3−/4− redox indicator. | Wine | LOD 0.003 fg/mL, 0.01–1.0 fg/mL (DPV) | [155] |
| Indium-tin oxide (ITO) electrode covered with nanoAu/polyaniline | EIS with [Fe(CN)6]3−/4− redox indicator. | Corn | LOD 0.05 ng/mL, 0.1–100 ng/mL | [156] |
| Au electrode with immobilized tetrahedral DNAs bearing auxiliary DNA sequences complementary to aptamers. | Mycotoxin binding releases aptamers from the surface, auxiliary DNA binds to complementary sequences bearing Au nanoparticles modified with peroxidase. Enzyme activity measured by redox current thionine utilized as a substrate | Rice, wheat powder | LOD 0.01 fg/mL, 0.1 fg/mL–0.1 μg/mL | [157] |
| Screen-printed electrode modified with magnetically collected Fe3O4@Au nanoparticles with aptamer immobilized via Au-SH bonds | EIS with [Fe(CN)6]3−/4− redox indicator. | Peanut | LOD 15 pg/mL, 20 pg/mL–50 ng/mL | [158] |
| GCE modified with Au nanoparticles and β-cyclodextrin. | Aptamer is first hybridized with complementary DNA sequence with terminal ferrocene label. Mycotoxin binding releases auxiliary DNA. Ferrocene group is involved in the inclusion complex with the macrocycle; charge transfer resistance increases. DPV signal of ferrocene increases with the analyte concentration. | Peanut oil | EIS: LOD 0.049 ng/mL (0.147 pM), 0.1–10 ng/mL | [159] |
| GCE modified with rGO-thionine composite followed by electrodeposition of Au nanoparticles and immobilization of auxiliary DNA sequence complementary to aptamer bearing ferrocene label | Analyte binding results in release of aptamer form the electrode interface. Rational signal measurement based on simultaneous monitoring of ferrocene and thionine signals with alternating current voltammetry. | Peanut | LOD 0.016 ng/mL, 0.05–20 ng/mL | [160] |
| Au electrode modified with thiolated stem-loop aptamer with methylene blue on the opposite end | Square wave voltammetry (SWV) signal of methylene blue changing with target reaction resulted in transformation of the initial aptamer structure. Reaction is amplified by addition of short DNA sequence complementary to the aptamer. | Beer, white wine | LOD 8 nM, 8 nM–4 μM | [161] |
| Screen-printed electrode with electropolymerized poly(aniline-anthranilic acid) film and covalently attached BSA-aflatoxin conjugate | Reaction with biotinylated aptamer is followed by attachment of streptavidin-alkaline phosphatase conjugate. DPV detection of enzyme activity via redox current of 1-naphotl formed from 1-naphtylphosphate as enzyme substrate. | Maize flour | LOD 0.086 ng/mL, 0.1–10 ng/mL | [162] |
| Au electrode modified with thiolated stem-loop aptamer bearing methylene blue at internal thymine fragment | Reaction with mycotoxin makes methylene blue available for electron transfer measured in SWV mode. | Wine, milk, and corn flour | LOD 6 pM | [163] |
| Aflatoxin M1 | ||||
| Poly(neutral red) with carboxylated pillar[5]arene bearing monomeric dye and aminated aptamer | EIS with [Fe(CN)6]3−/4− redox indicator | Milk and milk products | LOD 0.5 ng/mL, 5–120 ng/L | [164] |
| Au modified with self-assembled layer of n-doped graphene nanosheets and carboxylated polystyrene nanospheres followed by carbodiimide binding of aminated aptamer | EIS with [Fe(CN)6]3−/4− redox indicator | Oil, soy sauce | LOD 2 pg/mL, 0.01–10 ng/mL | [165] |
| Hairpin shaped aptamer with Au nanoparticles and complementary strand immobilized on golden screen-printed electrode | DPV of diffusionally free methylene blue added after the analyte incubation | Human blood serum, milk | LOD 0.9 ng/L, 2–600 ng/L | [166] |
| Deoxynivalenol | ||||
| Iron nanoflorets graphene nickel foam as electrode, aptamer covalently attached via glutaraldehyde linking | Changes in the electric conductivity monitored by polarization curves | Plant extracts | LOD 2.11 pg/mL, 1 fg/mL–1 ng/mL | [167] |
| Ochratoxin A | ||||
| GCE modified with Au nanoparticles with attached aptamer, sandwich protocol with Cd containing MOF particles as labels | DPV signal of Cd (II) ions in the structure of the label measured without its dissolution | Red wine | LOD 10 pg/mL, 0.05–100 ng/mL | [168] |
| Screen-printed carbon electrode covered with polythiophene-carboxylic acid with covalently attached aptamer | EIS with [Fe(CN)6]3−/4− redox indicator | Coffee | LOD 0.125 ng/mL, 0.125–20.0 ng/mL | [169] |
| Au electrode covered with β-cyclodextrin onto MoS2.nanoAu layer; aptamer is attached to the surface via supramolecular interaction with terminal Methylene blue group | Target interaction removes aptamer from the cyclodextrin moiety. Instead, ferrocene carboxylic acid is captured. The DPV signals of both methylene blue and ferrocene change synchronously. | Wine | LOD 0.06 nM, 0.1–50 nM | [170] |
| Au electrode covered with thiolated aptamer | Target interaction prevents aptamer cleavage caused by exonuclease enzyme; signal is enhanced by silver metallization of aptamer molecule. Ag oxidation DPV signal. | Beer | LOD 0.7 pg/mL, 1 pg/mL–0.1 μg/mL | [171] |
| Pencil graphite electrode electrografted with 4-amionobenzoic acid followed by covalent immobilization of aminated aptamer | EIS with [Fe(CN)6]3−/4− redox indicator | Beer | LOD 0.1 ng/mL, 0.1–2.0 ng/mL | [172] |
| GCE covered with nitrogen doped graphene and saturated with Methylene blue | Aptamer hybridized with complementary DNA strand reacts with OTA, released DNA is adsorbed on the electrode and increases redox signal of methylene blue measured by SWV. | - | LOD 0.71 fg/mL, 1 fg/mL–1 μg/mL | [173] |
| Au electrode with covalently attached thiolated auxiliary DNA sequence complementary to aptamer | Signaling DNA probe bears Au nanoparticles and ferrocene label. Displacement protocol with DPV detection of ferrocene signal. | Wine | LOD 0.001 ppb, 0.001–500 ppb | [174] |
| Dual mode paper-based sensor. Aptamers were immobilized on chitosan functionalized MoS2–Au@Pt. | CV, EIS. Catalyzed reduction of H2O2. | Corn | LOD 0.025 pg/mL, 0.0001–200 ng/mL | [175] |
| Au electrode modified with copolymer of pyrrole and pyrrole-3-qcetic acid followed by covalent binding of PAMAM G4 dendrimer and cross-lining of aptamer with glutaraldehyde | EIS with [Fe(CN)6]3−/4− redox indicator | Wine | LOD 2 ng/mL, 2–6000 ng/mL | [176] |
| Patulin | ||||
| Glassy carbon electrode modified with ZnO nanorods and Au nanoparticles | DPV signal of [Fe(CN)6]3−/4− redox indicator | Apple juice | LOD 0.25 pg/mL, 0.50 pg/mL–50 ng/mL | [125] |
| Screen-printed carbon electrode “activated” by chemical grafting with diazonium salt, aminated aptamer with long PEG linker | EIS with [Fe(CN)6]3−/4− redox indicator | Apple juice | LOD 1.25 ng/mL, 1–25 ng/mL | [177] |
| T2 toxin | ||||
| GCE covered with polyaniline-MoS2-chitosan-Au nanocomposite and thiolated aptamer. | GO-tetraethylene pentaamine–gold@platinum nanorods bearing auxiliary DNA complementary to aptamer are added together with analyte solution. Left free aptamer molecules form hybridization product amperometrically detected by electrocatalytic oxidation of hydrogen peroxide. | Canned beer | LOD 1.79 fg/mL, 10 fg/mL–100 ng/mL | [178] |
| Zearalenone | ||||
| GCE modified with chitosan, acetylene black and multiwalled carbon nanotubes followed by Au deposition and covalent attachment of thiolated DNA sequence complementary to aptamer | Aptamer is covalently attached to carboxylated rGO nanoflackes. Its reaction with mycotoxin prevents binding to the electrode. In the opposite way, hybridization results in a sharp decrease of the surface layer permeability detected with EIS by ferricyanide redox probe. | Corn oil and corn flour | LOD 3.64 fg/mL, 10 fg/mL–10 ng/mL | [179] |
| Au electrode with covalently attached zearalenone conjugate | Indirect competitive assay with SWV or EIS measurements of permeability of the surface layer in the presence of [Fe(CN)6]3−/4− redox indicator. | Maize grain | LOD 0.017 ng/mL, 0.01–1000 ng/mL | [180] |
| Fumonisin B1 and zearalenone | ||||
| GCE covered with co-reduced MoS2 and Au followed by covalent immobilization of thiolated aptamers against zearalenone and fumonisin B1 | Au nanoparticles modified with DNA sequences complementary to aptamers and saturated with thionine or 6-ferrocenelhexanthiol. Analyte binding resulted in release of the labels and changes in the signals of ferrocene and thionine recorded simultaneously in DPV mode. | Maize | Zearalenone: LOD 0.5 pg/mL, 0.001–10 ng/mL Fumonisin B1: LOD 0.5 pg/mL, 0.001–100 ng/mL | [181] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evtugyn, G.; Porfireva, A.; Kulikova, T.; Hianik, T. Recent Achievements in Electrochemical and Surface Plasmon Resonance Aptasensors for Mycotoxins Detection. Chemosensors 2021, 9, 180. https://doi.org/10.3390/chemosensors9070180
Evtugyn G, Porfireva A, Kulikova T, Hianik T. Recent Achievements in Electrochemical and Surface Plasmon Resonance Aptasensors for Mycotoxins Detection. Chemosensors. 2021; 9(7):180. https://doi.org/10.3390/chemosensors9070180
Chicago/Turabian StyleEvtugyn, Gennady, Anna Porfireva, Tatjana Kulikova, and Tibor Hianik. 2021. "Recent Achievements in Electrochemical and Surface Plasmon Resonance Aptasensors for Mycotoxins Detection" Chemosensors 9, no. 7: 180. https://doi.org/10.3390/chemosensors9070180
APA StyleEvtugyn, G., Porfireva, A., Kulikova, T., & Hianik, T. (2021). Recent Achievements in Electrochemical and Surface Plasmon Resonance Aptasensors for Mycotoxins Detection. Chemosensors, 9(7), 180. https://doi.org/10.3390/chemosensors9070180







