Aptamer-Conjugated Quantum Dot Optical Biosensors: Strategies and Applications
Abstract
1. Introduction
2. Aptamer-Based QD Biosensors for Molecule Detection and Monitoring
2.1. QD as Sensing Material
2.2. Strategy for the Change in the Optical Signal Emission of the QDs for Biosensing Application
3. Current Aptamer-Based QD Biosensors for Chemical and Biomolecule Detection
3.1. Biosensor Based on Change in Signal Emission of QD by Aptamer-Analyte Binding
3.2. Biosensor Using Change in Signal Emission of QD by Materials Directly Attaching to the Aptamer
3.3. Biosensor Using Change in Signal Emission of QD by Materials Indirectly Linking with the Aptamer
3.4. Biosensor Using Change in Signal Emission of QD by DNA-Modifying Enzyme-Assisted Reaction
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mehrotra, P. Biosensors and their applications—A review. J. Oral Biol. Craniofac. Res. 2016, 6, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Alhadrami, H.A. Biosensors: Classifications, medical applications, and future prospective. Biotechnol. Appl. Biochem. 2018, 65, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.M.; Lee, S.Y. Optical Biosensors for the Detection of Pathogenic Microorganisms. Trends Biotechnol. 2016, 34, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Khalid, K.; Tan, X.; Zaid, H.F.M.; Tao, Y.; Chew, C.L.; Chu, D.-T.; Lam, M.K.; Ho, Y.-C.; Lim, J.W.; Wei, L.C. Advanced in developmental organic and inorganic nanomaterial: A review. Bioengineered 2020, 11, 328–355. [Google Scholar] [CrossRef]
- Hubbe, H.; Mendes, E.; Boukany, P.E. Polymeric Nanowires for Diagnostic Applications. Micromachines 2019, 10, 225. [Google Scholar] [CrossRef]
- Rahong, S.; Yasui, T.; Kaji, N.; Baba, Y. Recent developments in nanowires for bio-applications from molecular to cellular levels. Lab Chip 2016, 16, 1126–1138. [Google Scholar] [CrossRef][Green Version]
- Syedmoradi, L.; Ahmadi, A.; Norton, M.L.; Omidfar, K. A review on nanomaterial-based field effect transistor technology for biomarker detection. Microchim. Acta 2019, 186, 739. [Google Scholar] [CrossRef]
- Saliev, T. The Advances in Biomedical Applications of Carbon Nanotubes. C 2019, 5, 29. [Google Scholar] [CrossRef]
- Ansari, L.; Hallaj, S.; Hallaj, T.; Amjadi, M. Doped-carbon dots: Recent advances in their biosensing, bioimaging and therapy applications. Colloids Surf. B Biointerfaces 2021, 203, 111743. [Google Scholar] [CrossRef]
- Ji, C.; Zhou, Y.; Leblanc, R.M.; Peng, Z. Recent Developments of Carbon Dots in Biosensing: A Review. ACS Sens. 2020, 5, 2724–2741. [Google Scholar] [CrossRef]
- Yu, S.; Chen, T.; Zhang, Q.; Zhou, M.; Zhu, X. Application of DNA nanodevices for biosensing. Analyst 2020, 145, 3481–3489. [Google Scholar] [CrossRef]
- Ayodele, O.; Adesina, A.; Pourianejad, S.; Averitt, J.; Ignatova, T. Recent Advances in Nanomaterial-Based Aptasensors in Medical Diagnosis and Therapy. Nanomaterials 2021, 11, 932. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Li, C.-C.; Zhang, C.-Y. Development of quantum dot-based biosensors: Principles and applications. J. Mater. Chem. B 2018, 6, 6173–6190. [Google Scholar] [CrossRef]
- Kargozar, S.; Hoseini, S.J.; Milan, P.B.; Hooshmand, S.; Kim, H.; Mozafari, M. Quantum Dots: A Review from Concept to Clinic. Biotechnol. J. 2020, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-J.; Xia, L.; Xie, H.-Y.; Zhang, Z.-L.; Pang, D.-W. Quantum Dot Based Biotracking and Biodetection. Anal. Chem. 2019, 91, 532–547. [Google Scholar] [CrossRef]
- Wen, L.; Qiu, L.; Wu, Y.; Hu, X.; Zhang, X. Aptamer-Modified Semiconductor Quantum Dots for Biosensing Applications. Sensors 2017, 17, 1736. [Google Scholar] [CrossRef] [PubMed]
- Foubert, A.; Beloglazova, N.V.; Rajkovic, A.; Sas, B.; Madder, A.; Goryacheva, I.Y.; De Saeger, S. Bioconjugation of quantum dots: Review & impact on future application. TrAC Trends Anal. Chem. 2016, 83, 31–48. [Google Scholar] [CrossRef]
- Wagner, A.M.; Knipe, J.M.; Orive, G.; Peppas, N.A. Quantum dots in biomedical applications. Acta Biomater. 2019, 94, 44–63. [Google Scholar] [CrossRef]
- Díaz-González, M.; De La Escosura-Muñiz, A.; Fernandez-Argüelles, M.T.; Alonso, F.J.G.; Costa-Fernandez, J.M. Quantum Dot Bioconjugates for Diagnostic Applications. Top. Curr. Chem. 2020, 378, 35. [Google Scholar] [CrossRef]
- Taylor, A.I.; Holliger, P. Selecting Fully-Modified XNA Aptamers Using Synthetic Genetics. Curr. Protoc. Chem. Biol. 2018, 10, e44. [Google Scholar] [CrossRef]
- Gatto, B.; Palumbo, M.; Sissi, C. Nucleic Acid Aptamers Based on the G-Quadruplex Structure: Therapeutic and Diagnostic Potential. Curr. Med. Chem. 2009, 16, 1248–1265. [Google Scholar] [CrossRef]
- Kim, D.-M.; Go, M.-J.; Lee, J.; Na, D.; Yoo, S.-M. Recent Advances in Micro/Nanomaterial-Based Aptamer Selection Strategies. Molecules 2021, 26, 5187. [Google Scholar] [CrossRef]
- Dunn, M.R.; Jimenez, R.M.; Chaput, J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017, 1, 0076. [Google Scholar] [CrossRef]
- Cole, K.H.; Lupták, A. High-throughput methods in aptamer discovery and analysis. Methods Enzymol. 2019, 621, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Zhuo, Z.; Pan, Y.; Yu, Y.; Li, F.; Liu, J.; Wang, L.; Wu, X.; Li, D.; Wan, Y.; et al. Recent Progress in Aptamer Discoveries and Modifications for Therapeutic Applications. ACS Appl. Mater. Interfaces 2021, 13, 9500–9519. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.; Khan, I.M.; Wang, Z. Capture-SELEX for aptamer selection: A short review. Talanta 2021, 229, 122274. [Google Scholar] [CrossRef] [PubMed]
- Stanciu, L.A.; Wei, Q.; Barui, A.K.; Mohammad, N. Recent Advances in Aptamer-Based Biosensors for Global Health Applications. Annu. Rev. Biomed. Eng. 2021, 23, 433–459. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Bayat, P.; Nosrati, R.; Alibolandi, M.; Rafatpanah, H.; Abnous, K.; Khedri, M.; Ramezani, M. SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie 2018, 154, 132–155. [Google Scholar] [CrossRef]
- Matea, C.T.; Mocan, T.; Tabaran, F.; Pop, T.; Mosteanu, O.; Puia, C.; Iancu, C.; Mocan, L. Quantum dots in imaging, drug delivery and sensor applications. Int. J. Nanomed. 2017, 12, 5421–5431. [Google Scholar] [CrossRef] [PubMed]
- Alivisatos, A.P. Semiconductor clusters, nanocrystals, and quantum dots. Science 1996, 271, 933–937. [Google Scholar] [CrossRef]
- Michalet, X.; Pinaud, F.F.; Bentolila, L.A.; Tsay, J.M.; Doose, S.; Li, J.J.; Sundaresan, G.; Wu, A.M.; Gambhir, S.S.; Weiss, S. Quantum Dots for Live Cells, in Vivo Imaging, and Diagnostics. Science 2005, 307, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Datta, D.; Chaudhry, S.; Dutta, M.; Stroscio, M.A. Rapid Detection of Tumor Necrosis Factor-Alpha Using Quantum Dot-Based Optical Aptasensor. IEEE Trans. NanoBioscience 2018, 17, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Chen, Y.; Sebastian, J.; George, A.; Dutta, M.; Stroscio, M.A. A study on the response of FRET based DNA aptasensors in intracellular environment. Sci. Rep. 2020, 10, 13250. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Chen, Y.; George, A.; Dutta, M.; Stroscio, M.A. Fluorescence Resonant Energy Transfer-Based Quantum Dot Sensor for the Detection of Calcium Ions. Front. Chem. 2020, 8, 594. [Google Scholar] [CrossRef]
- Medintz, I.L.; Mattoussi, H. Quantum dot-based resonance energy transfer and its growing application in biology. Phys. Chem. Chem. Phys. 2009, 11, 17–45. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Pálmai, M.; Saed, B.; George, A.; Snee, P.T.; Hu, Y.S. Cytosolic delivery of membrane-penetrating QDs into T cell lymphocytes: Implications in immunotherapy and drug delivery. Nanoscale 2021, 13, 5519–5529. [Google Scholar] [CrossRef]
- Kovalenko, M.V.; Manna, L.; Cabot, A.; Hens, Z.; Talapin, D.V.; Kagan, C.R.; Klimov, V.I.; Rogach, A.L.; Reiss, P.; Milliron, D.J.; et al. Prospects of Nanoscience with Nanocrystals. ACS Nano 2015, 9, 1012–1057. [Google Scholar] [CrossRef]
- Zhao, P.; Xu, Q.; Tao, J.; Jin, Z.; Pan, Y.; Yu, C.; Yu, Z. Near infrared quantum dots in biomedical applications: Current status and future perspective: Near infrared quantum dots in biomedical applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2018, 10, e1483. [Google Scholar] [CrossRef]
- De, C.K.; Routh, T.; Roy, D.; Mandal, S.; Mandal, P.K. Highly Photoluminescent InP Based Core Alloy Shell QDs from Air-Stable Precursors: Excitation Wavelength Dependent Photoluminescence Quantum Yield, Photoluminescence Decay Dynamics, and Single Particle Blinking Dynamics. J. Phys. Chem. C 2018, 122, 964–973. [Google Scholar] [CrossRef]
- Ji, B.; Koley, S.; Slobodkin, I.; Remennik, S.; Banin, U. ZnSe/ZnS Core/Shell Quantum Dots with Superior Optical Properties through Thermodynamic Shell Growth. Nano Lett. 2020, 20, 2387–2395. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Kim, K.-H.; Kim, S.; Choi, S.-M.; Jang, H.; Seo, H.-K.; Lee, H.; Chung, D.-Y.; Jang, E. Efficient and stable blue quantum dot light-emitting diode. Nature 2020, 586, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Guo, M.; Tan, L.; Geng, Y.; Huang, S.; Tang, Y.; Su, C.; Lin, C.C.; Liang, Y. Highly efficient fluorescent QDs sensor for specific detection of protein through double recognition of hybrid aptamer-molecular imprinted polymers. Sens. Actuators B Chem. 2018, 274, 627–635. [Google Scholar] [CrossRef]
- Li, D.; Guo, J.; Zhao, L.; Zhang, G.; Yan, G. A label-free RTP sensor based on aptamer/quantum dot nanocomposites for cytochrome c detection. RSC Adv. 2019, 9, 31953–31959. [Google Scholar] [CrossRef]
- Ren, J.; Liang, G.; Man, Y.; Li, A.; Jin, X.; Liu, Q.; Pan, L. Aptamer-based fluorometric determination of Salmonella Typhimurium using Fe3O4 magnetic separation and CdTe quantum dots. PLoS ONE 2019, 14, e0218325. [Google Scholar] [CrossRef]
- He, Z.-J.; Kang, T.-F.; Lu, L.-P.; Cheng, S.-Y. An electrochemiluminescence aptamer sensor for chloramphenicol based on GO-QDs nanocomposites and enzyme-linked aptamers. J. Electroanal. Chem. 2020, 860, 113870. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, X.; Han, H. Turn-on near-infrared electrochemiluminescence sensing of thrombin based on resonance energy transfer between CdTe/CdS coresmall/shellthick quantum dots and gold nanorods. Biosens. Bioelectron. 2016, 82, 26–31. [Google Scholar] [CrossRef]
- Zhao, M.; Chen, A.-Y.; Huang, D.; Chai, Y.-Q.; Zhuo, Y.; Yuan, R. MoS2 Quantum Dots as New Electrochemiluminescence Emitters for Ultrasensitive Bioanalysis of Lipopolysaccharide. Anal. Chem. 2017, 89, 8335–8342. [Google Scholar] [CrossRef]
- Fan, Z.; Yao, B.; Ding, Y.; Xie, M.; Zhao, J.; Zhang, K.; Huang, W. Electrochemiluminescence aptasensor for Siglec-5 detection based on MoS2@Au nanocomposites emitter and exonuclease III-powered DNA walker. Sens. Actuators B Chem. 2021, 334, 129592. [Google Scholar] [CrossRef]
- Wu, C.; Gan, N.; Ou, C.; Tang, H.; Zhou, Y.; Cao, J. A homogenous “signal-on” aptasensor for antibiotics based on a single stranded DNA binding protein-quantum dot aptamer probe coupling exonuclease-assisted target recycling for signal amplification. RSC Adv. 2017, 7, 8381–8387. [Google Scholar] [CrossRef]
- Tang, J.; Huang, N.; Zhang, X.; Zhou, T.; Tan, Y.; Pi, J.; Pi, L.; Cheng, S.; Zheng, H.; Cheng, Y. Aptamer-conjugated PEGylated quantum dots targeting epidermal growth factor receptor variant III for fluorescence imaging of glioma. Int. J. Nanomed. 2017, 12, 3899–3911. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Sivils, J.C. Further characterization and independent validation of a DNA aptamer-quantum dot-based magnetic sandwich assay for Campylobacter. Folia Microbiol. 2017, 62, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Q.; Liao, Z.; Sun, Y.; Cheng, Q.; Song, Y.; Song, E.; Tan, W. Magnetism-Resolved Separation and Fluorescence Quantification for Near-Simultaneous Detection of Multiple Pathogens. Anal. Chem. 2018, 90, 9621–9628. [Google Scholar] [CrossRef]
- Kurt, H.; Yüce, M.; Hussain, B.; Budak, H. Dual-excitation upconverting nanoparticle and quantum dot aptasensor for multiplexed food pathogen detection. Biosens. Bioelectron. 2016, 81, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Isildak, I.; Navaeipour, F.; Afsharan, H.; Kanberoglu, G.S.; Agir, I.; Ozer, T.; Annabi, N.; Totu, E.E.; Khalilzadeh, B. Electrochemiluminescence methods using CdS quantum dots in aptamer-based thrombin biosensors: A comparative study. Microchim. Acta 2019, 187, 25. [Google Scholar] [CrossRef]
- Wang, Q.; Cheng, X.; Li, H.; Yu, F.; Wang, Q.; Yu, M.; Liu, D.; Xia, J. A novel DNA quantum dots/aptamer-modified gold nanoparticles probe for detection of Salmonella typhimurium by fluorescent immunoassay. Mater. Today Commun. 2020, 25, 101428. [Google Scholar] [CrossRef]
- Kitte, S.A.; Tafese, T.; Xu, C.; Saqib, M.; Li, H.; Jin, Y. Plasmon-enhanced quantum dots electrochemiluminescence aptasensor for selective and sensitive detection of cardiac troponin I. Talanta 2021, 221, 121674. [Google Scholar] [CrossRef]
- Babamiri, B.; Salimi, A.; Hallaj, R. Switchable electrochemiluminescence aptasensor coupled with resonance energy transfer for selective attomolar detection of Hg2+ via CdTe@CdS/dendrimer probe and Au nanoparticle quencher. Biosens. Bioelectron. 2018, 102, 328–335. [Google Scholar] [CrossRef]
- Wang, Y.; Gan, N.; Li, T.; Cao, Y.; Hu, F.; Chen, Y. A novel aptamer–quantum dot fluorescence probe for specific detection of antibiotic residues in milk. Anal. Methods 2016, 8, 3006–3013. [Google Scholar] [CrossRef]
- Tan, L.; Kang, C.; Xu, S.; Tang, Y. Selective room temperature phosphorescence sensing of target protein using Mn-doped ZnS QDs-embedded molecularly imprinted polymer. Biosens. Bioelectron. 2013, 48, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Aboulaich, A.; Geszke, M.; Balan, L.; Ghanbaja, J.; Medjahdi, G.; Schneider, R. Water-Based Route to Colloidal Mn-Doped ZnSe and Core/Shell ZnSe/ZnS Quantum Dots. Inorg. Chem. 2010, 49, 10940–10948. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.A. Molecular imprinting polymers and their composites: A promising material for diverse applications. Biomater. Sci. 2017, 5, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.; Tamboli, V.; Harniman, R.L.; Estrela, P.; Allender, C.J.; Bowen, J.L. Aptamer–MIP hybrid receptor for highly sensitive electrochemical detection of prostate specific antigen. Biosens. Bioelectron. 2016, 75, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, J. Molecularly Imprinted Polymers with DNA Aptamer Fragments as Macromonomers. ACS Appl. Mater. Interfaces 2016, 8, 6371–6378. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Spivak, D.A. A Double-Imprinted Diffraction-Grating Sensor Based on a Virus-Responsive Super-Aptamer Hydrogel Derived from an Impure Extract. Angew. Chem. Int. Ed. 2014, 53, 2095–2098. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, S.; Snee, P.T.; Ramachandran, A.; George, A. Acidic domain in dentin phosphophoryn facilitates cellular uptake: Implications in targeted protein delivery. J. Biol. Chem. 2013, 288, 16098–16109. [Google Scholar] [CrossRef] [PubMed]
- Koshman, Y.E.; Waters, S.B.; Walker, L.A.; Los, T.; de Tombe, P.; Goldspink, P.H.; Russell, B. Delivery and visualization of proteins conjugated to quantum dots in cardiac myocytes. J. Mol. Cell. Cardiol. 2008, 45, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Xiong, E.; Zhang, X.; Liu, Y.; Zhou, J.; Yu, P.; Li, X.; Chen, J. Ultrasensitive Electrochemical Detection of Nucleic Acids Based on the Dual-Signaling Electrochemical Ratiometric Method and Exonuclease III-Assisted Target Recycling Amplification Strategy. Anal. Chem. 2015, 87, 7291–7296. [Google Scholar] [CrossRef]
- Fu, X.; Tan, X.; Yuan, R.; Chen, S. A dual-potential electrochemiluminescence ratiometric sensor for sensitive detection of dopamine based on graphene-CdTe quantum dots and self-enhanced Ru(II) complex. Biosens. Bioelectron. 2017, 90, 61–68. [Google Scholar] [CrossRef]
- Spring, S.; Goggins, S.; Frost, C. Ratiometric Electrochemistry: Improving the Robustness, Reproducibility and Reliability of Biosensors. Molecules 2021, 26, 2130. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wang, J.; Luo, F.; Guo, L.; Qiu, B.; Lin, Z. Highly reproducible ratiometric aptasensor based on the ratio of amplified electrochemiluminescence signal and stable internal reference electrochemical signal. Electrochim. Acta 2018, 283, 798–805. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Zhou, H.; Wu, P.; Zhang, H.R.; Xu, J.J.; Chen, H.Y. In situ activation of CdS electrochemiluminescence film and its application in H2S detection. Anal. Chem. 2014, 86, 8657–8664. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.; Zhu, Q.; Zhao, K.; Deng, A.; Li, J. Multiple Signal Amplified Electrochemiluminescent Immunoassay for Hg2+ Using Graphene-Coupled Quantum Dots and Gold Nanoparticles-Labeled Horseradish Peroxidase. Environ. Sci. Technol. 2015, 49, 5013–5020. [Google Scholar] [CrossRef] [PubMed]
- Delices, A.; Moodelly, D.; Hurot, C.; Hou, Y.; Ling, W.L.; Saint-Pierre, C.; Gasparutto, D.; Nogues, G.; Reiss, P.; Kheng, K. Aqueous synthesis of DNA-functionalized near-infrared AgInS2/ZnS core/shell quantum dots. ACS Appl. Mater. Interfaces 2020, 12, 44026–44038. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Kuang, H.; Wang, L.; Xu, C. Gold nanorod ensembles as artificial molecules for applications in sensors. J. Mater. Chem. 2011, 21, 16759–16782. [Google Scholar] [CrossRef]
- Wang, J.; Han, H.-Y. Near-infrared electrogenerated chemiluminescence from quantum dots. Rev. Anal. Chem. 2013, 32, 43. [Google Scholar] [CrossRef]
- Stobiecka, M. Novel plasmonic field-enhanced nanoassay for trace detection of proteins. Biosens. Bioelectron. 2014, 55, 379–385. [Google Scholar] [CrossRef]
- Stobiecka, M.; Chalupa, A. Modulation of Plasmon-Enhanced Resonance Energy Transfer to Gold Nanoparticles by Protein Survivin Channeled-Shell Gating. J. Phys. Chem. B 2015, 119, 13227–13235. [Google Scholar] [CrossRef]
- Kim, D.M.; Yoo, S.M. DNA-modifying enzyme reaction-based biosensors for disease diagnostics: Recent biotechnological advances and future perspectives. Crit. Rev. Biotechnol. 2020, 40, 787–803. [Google Scholar] [CrossRef]
- Gopalakrishnan, D.; Damien, D.; Shaijumon, M.M. MoS2 Quantum Dot-Interspersed Exfoliated MoS2 Nanosheets. ACS Nano 2014, 8, 5297–5303. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tang, M. The cytotoxicity of core-shell or non-shell structure quantum dots and reflection on environmental friendly: A review. Environ. Res. 2020, 194, 110593. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhu, L.; Yan, M.; Zhu, Q.; Lu, Q.; Huang, J.; Cui, H.; Yang, X. Dual-Wavelength Ratiometric Electrochemiluminescence Immunosensor for Cardiac Troponin I Detection. Anal. Chem. 2019, 91, 1524–1531. [Google Scholar] [CrossRef]
- Du, Y.; Zhong, Y.; Dong, J.; Qian, C.; Sun, S.; Gao, L.; Yang, D. The effect of PEG functionalization on the in vivo behavior and toxicity of CdTe quantum dots. RSC Adv. 2019, 9, 12218–12225. [Google Scholar] [CrossRef]
- Zhang, L.; Dou, X.; Zhang, C.; Ying, G.; Liu, C.; Luo, J.; Li, Q.; Li, P.; Wang, Y.; Yang, M. Facile preparation of stable PEG-functionalized quantum dots with glycine-enhanced photoluminescence and their application for screening of aflatoxin B1 in herbs. Sens. Actuators B Chem. 2018, 261, 188–195. [Google Scholar] [CrossRef]
- Lemon, C.M.; Karnas, E.; Han, X.; Bruns, O.T.; Kempa, T.J.; Fukumura, D.; Bawendi, M.G.; Jain, R.K.; Duda, D.G.; Nocera, D.G. Micelle-Encapsulated Quantum Dot-Porphyrin Assemblies as in Vivo Two-Photon Oxygen Sensors. J. Am. Chem. Soc. 2015, 137, 9832–9842. [Google Scholar] [CrossRef] [PubMed]
- Chinnathambi, S.; Abu, N.; Hanagata, N. Biocompatible CdSe/ZnS quantum dot micelles for long-term cell imaging without alteration to the native structure of the blood plasma protein human serum albumin. RSC Adv. 2017, 7, 2392–2402. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Chen, G.; Li, Y.; Xu, W.; Gong, S. Quantum-Dot-Based Theranostic Micelles Conjugated with an Anti-EGFR Nanobody for Triple-Negative Breast Cancer Therapy. ACS Appl. Mater. Interfaces 2017, 9, 30297–30305. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, H.; Kang, S.-M.; Bae, B.-S. Two-Step-Enhanced Stability of Quantum Dots via Silica and Siloxane Encapsulation for the Long-Term Operation of Light-Emitting Diodes. ACS Appl. Mater. Interfaces 2019, 11, 22801–22808. [Google Scholar] [CrossRef]
- Kim, Y.H.; Koh, S.; Lee, H.; Kang, S.-M.; Lee, D.C.; Bae, B.-S. Photo-Patternable Quantum Dots/Siloxane Composite with Long-Term Stability for Quantum Dot Color Filters. ACS Appl. Mater. Interfaces 2020, 12, 3961–3968. [Google Scholar] [CrossRef]
- Choi, Y.; Lee, S.Y. Biosynthesis of inorganic nanomaterials using microbial cells and bacteriophages. Nat. Rev. Chem. 2020, 4, 638–656. [Google Scholar] [CrossRef]
- Mi, C.; Wang, Y.; Zhang, J.; Huang, H.; Xu, L.; Wang, S.; Fang, X.; Fang, J.; Mao, C.; Xu, S. Biosynthesis and characterization of CdS quantum dots in genetically engineered Escherichia coli. J. Biotechnol. 2011, 153, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, G.; Collao, B.; Araneda, M.; Escobar, B.; Álvarez, S.; Bravo, D.; Pérez-Donoso, J. Use of acidophilic bacteria of the genus Acidithiobacillus to biosynthesize CdS fluorescent nanoparticles (quantum dots) with high tolerance to acidic pH. Enzym. Microb. Technol. 2016, 95, 217–224. [Google Scholar] [CrossRef]
- Carrasco, V.; Amarelle, V.; Lagos-Moraga, S.; Quezada, C.P.; Espinoza-González, R.; Faccio, R.; Fabiano, E.; Pérez-Donoso, J.M. Production of cadmium sulfide quantum dots by the lithobiontic Antarctic strain Pedobacter sp. UYP1 and their application as photosensitizer in solar cells. Microb. Cell Factories 2021, 20, 41. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.-W.; Ivanov, I.N.; Duty, C.E.; Love, L.J.; Rondinone, A.J.; Wang, W.; Li, Y.-L.; Madden, A.S.; Mosher, J.J.; Hu, M.Z.; et al. Scalable economic extracellular synthesis of CdS nanostructured particles by a non-pathogenic thermophile. J. Ind. Microbiol. Biotechnol. 2013, 40, 1263–1271. [Google Scholar] [CrossRef] [PubMed]
- Órdenes-Aenishanslins, N.; Anziani-Ostuni, G.; Monrás, J.P.; Tello, A.; Bravo, D.; Toro-Ascuy, D.; Rifo, R.S.; Prasad, P.N.; Pérez-Donoso, J.M. Bacterial Synthesis of Ternary CdSAg Quantum Dots through Cation Exchange: Tuning the Composition and Properties of Biological Nanoparticles for Bioimaging and Photovoltaic Applications. Microorganisms 2020, 8, 631. [Google Scholar] [CrossRef] [PubMed]
- Monrás, J.P.; Diaz, V.; Bravo, D.; Montes, R.A.; Chasteen, T.G.; Osorio-Roman, I.O.; Vásquez, C.C.; Pérez-Donoso, J.M. Enhanced Glutathione Content Allows the In Vivo Synthesis of Fluorescent CdTe Nanoparticles by Escherichia coli. PLoS ONE 2012, 7, e48657. [Google Scholar] [CrossRef]
- Moon, J.W.; Ivanov, I.N.; Joshi, P.C.; Armstrong, B.L.; Wang, W.; Jung, H.; Rondinone, A.J.; Jellison, G.E.; Meyer, H.M., Jr.; Jang, G.G., 3rd; et al. Scalable production of microbially mediated zinc sulfide nanoparticles and application to functional thin films. Acta Biomater. 2014, 10, 4474–4483. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.-W.; Phelps, T.J.; Fitzgerald, C.L., Jr.; Lind, R.F.; Elkins, J.G.; Jang, G.G.; Joshi, P.C.; Kidder, M.; Armstrong, B.L.; Watkins, T.R.; et al. Manufacturing demonstration of microbially mediated zinc sulfide nanoparticles in pilot-plant scale reactors. Appl. Microbiol. Biotechnol. 2016, 100, 7921–7931. [Google Scholar] [CrossRef] [PubMed]
- Dameron, C.T.; Reese, R.N.; Mehra, R.K.; Kortan, A.R.; Carroll, P.J.; Steigerwald, M.L.; Brus, L.E.; Winge, D.R. Biosynthesis of cadmium sulphide quantum semiconductor crystallites. Nature 1989, 338, 596–597. [Google Scholar] [CrossRef]
- Al-Shalabi, Z.; Doran, P.M. Biosynthesis of fluorescent CdS nanocrystals with semiconductor properties: Comparison of microbial and plant production systems. J. Biotechnol. 2016, 223, 13–23. [Google Scholar] [CrossRef]
- Syed, A.; Ahmad, A. Extracellular biosynthesis of CdTe quantum dots by the fungus Fusarium oxysporum and their anti-bacterial activity. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2013, 106, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Hao, N.; Yang, Y.; Zhao, D. Biosynthesis of biocompatible cadmium telluride quantum dots using yeast cells. Nano Res. 2010, 3, 481–489. [Google Scholar] [CrossRef]
- Mala, J.G.S.; Rose, C. Facile production of ZnS quantum dot nanoparticles by Saccharomyces cerevisiae MTCC 2918. J. Biotechnol. 2014, 170, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.M.; Rajan, R.; Kurup, G.G. Biologically synthesized ZnS quantum dots as fluorescent probes for lead (II) sensing. Luminescence 2020, 35, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Gao, F.; Li, N. T4 Virus-Based Toolkit for the Direct Synthesis and 3D Organization of Metal Quantum Particles. Chemistry 2010, 16, 14397–14403. [Google Scholar] [CrossRef]
- Mao, C.; Flynn, C.E.; Hayhurst, A.; Sweeney, R.; Qi, J.; Georgiou, G.; Iverson, B.; Belcher, A.M. Viral assembly of oriented quantum dot nanowires. Proc. Natl. Acad. Sci. USA 2003, 100, 6946–6951. [Google Scholar] [CrossRef]
- Huang, F.; Dang, Z.; Guo, C.-L.; Lu, G.-N.; Gu, R.R.; Liu, H.-J.; Zhang, H. Biosorption of Cd(II) by live and dead cells of Bacillus cereus RC-1 isolated from cadmium-contaminated soil. Colloids Surf. B Biointerfaces 2013, 107, 11–18. [Google Scholar] [CrossRef]
- He, X.; Ma, N. An overview of recent advances in quantum dots for biomedical applications. Colloids Surf. B Biointerfaces 2014, 124, 118–131. [Google Scholar] [CrossRef]
- Wang, F.-T.; Wang, L.-N.; Xu, J.; Huang, K.-J.; Wu, X. Synthesis and modification of carbon dots for advanced biosensing application. Analyst 2021, 146, 4418–4435. [Google Scholar] [CrossRef]
- Sivasankarapillai, V.S.; Jose, J.; Shanavas, M.S.; Marathakam, A.; Uddin, S.; Mathew, B. Silicon Quantum Dots: Promising Theranostic Probes for the Future. Curr. Drug Targets 2019, 20, 1255–1263. [Google Scholar] [CrossRef] [PubMed]
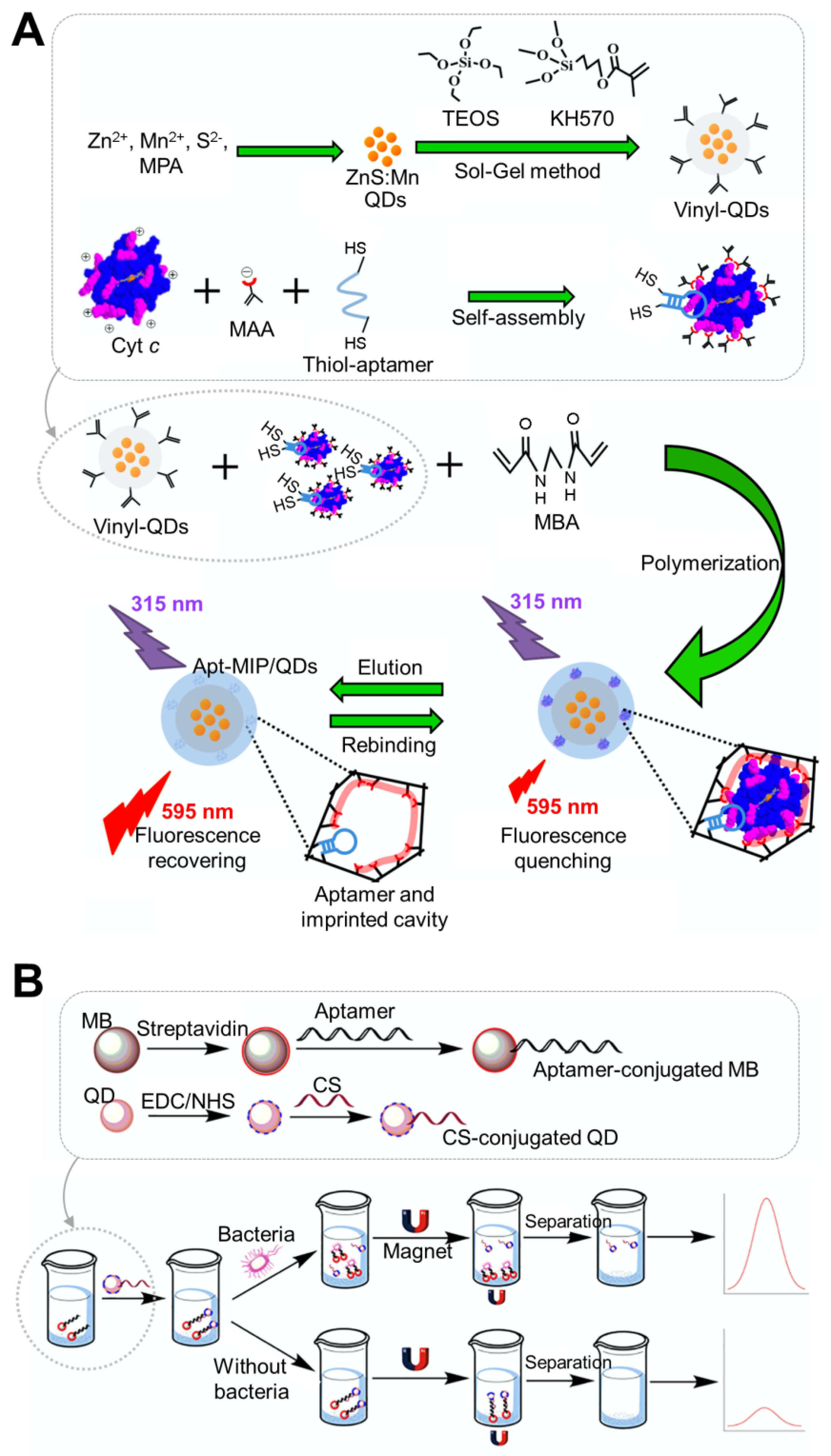


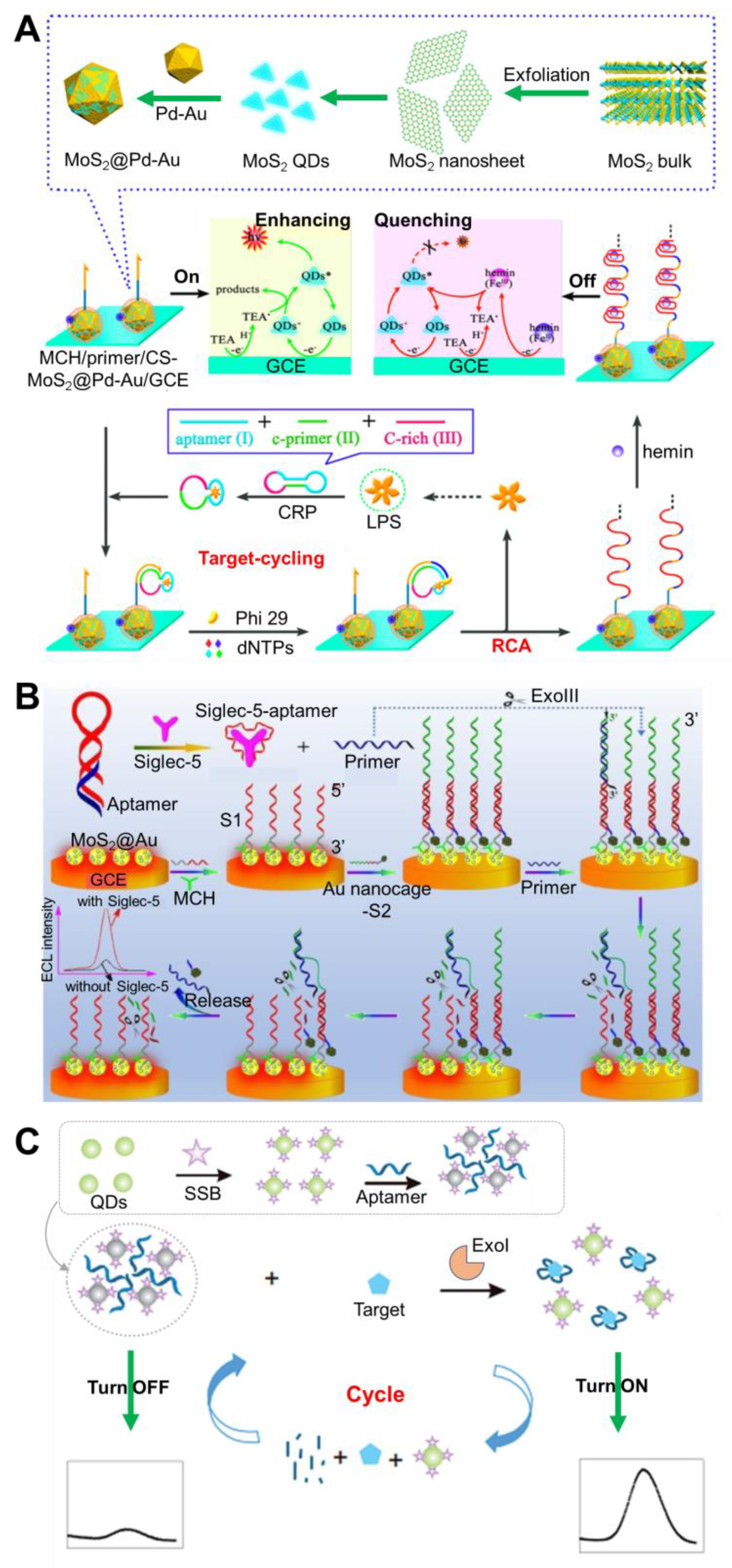
| Sensing Platform | QD Material | Aptamer Conjugation Method | Analyte | Sensing Type | Linear Range | LoD | Sample | Features | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Vinyl modified QD | Mn/ZnS | Conjugation of SH-aptamer to molecular imprinted polymer-QD | Cytochrome c (Cyt c) | Fluorescence | 0.20–2.00 μM | 54 nM | Urine, serum | Use of double recognition based on aptamer and imprinted cavity | [44]; Figure 1A |
| Polyethyleneimine (PEI)-capped core-shell QD | Mn/ZnS | Electrostatic attraction to (+) charged PEI-QD and (−) charged aptamer | Cyt c | PL | 0.166–9.96 μM | 84 nM | Human serum | Quenching effect by electron transfer between QD and Cyt c | [45] |
| Core/shell | PEG-CdSe/ZnS | Biotin aptamer-SA QD conjugation | EGFRvIII | Fluorescence | ND | ND | Mice bearing U87-EGFRvIII brain tumors | Small enough (20 nm) aptamer-QD conjugates to cross BBB | [52] |
| MNP-QD | CdTe | Hybridization of oligo with complementary strand of aptamer attached to QD | Salmonella typhimurium | Fluorescence | 10–1010 CFU/mL | 1 CFU/mL | Milk, water | Formation of MNP-QD composition by binding aptamer and complementary strand | [46]; Figure 1B |
| Core/shell | CdSe/ZnS | Covalent linking between COOH-QD and NH2-aptamer | Tumour necrosis factor-alpha (TNF-a) | PL | 0–22.3 nM | 97.2 pM | Human serum-based sample | Use of FRET. Sensing platform: QD (donor)-Aptamer-AuNP (acceptor) | [34] |
| Core/shell | CdSe/ZnS | Biotin aptamer-streptavidin QD conjugation | Campylobacter jejuni | Fluorescence | ND | 5 CFU/mL | Chicken rinsate | Sandwich assay with aptamer-conjugated MB and aptamer-attached QD | [53] |
| Core/shell | CdSe/ZnS | Biotin aptamer-avidin QD conjugation | Escherichia coli O157:H7, Salmonella typhimurium | Fluorescence | 63−108 CFU/mL for S. typhimurium; 40−108 CFU/mL for E. coli | 25 CFU/mL for S. typhimurium; 16 CFU/mL for E. coli | Milk, human serum, human urine | Use of aptamer-modified fluorescentmagnetic multifunctional nanoprobes consisting of (3-mercaptopropyl) trimethoxysilane, magnetic γ-Fe2O3, and fluorescent QDs | [54] |
| Single QD | CdTe | Covalent linking between COOH-QD and NH2-aptamer | S. typhimurium, Staphylococcus aureus | PL | 10–106 CFU/mL | 16 CFU/mL for S. aureus; 28 CFU/mL for S. typhimurium | None | Sensing platform; complementary strand-conjugated MB-aptamer-attached QD. Use of MB for simple separation | [55] |
| Core/shell | CdSe/ZnS | Covalent linking between COOH-QD and NH2-aptamer | TNF-a | PL | ND | ND | Mouse pre-osteocyte cells (MC3T3 E1), Mouse monocyte/macrophage cells (RAW264.7) | Use of FRET. Sensing platform: CPP-QD (donor)-Aptamer-AuNP (acceptor). Intracellular biomolecule detection | [35]; Figure 2 |
| Core/shell | CdSe/ZnS | Covalent linking between COOH-QD and NH2-aptamer | Ca2+ | PL | ND | 3.77 pM | Mouse pre-osteocyte cells (MC3T3 E1) | Use of FRET. Complex sensing platform: CPP-QD (donor)-Aptamer-AuNP (acceptor). Intracellular biomolecule detection | [36] |
| Single QD on GCE | CdS | Conjugation of SH-aptamer to QD-GCE | Thrombin | ECL | 500–5000 pg/mL in quenching method; 50–2000 pg/mL in amplification method; 5–500 pg/mL in ratiometric method | 92 pg/mL in quenching method; 6.5 pg/mL in amplification method; 500 fg/mL in ratiometric method | Human serum | Use of FRET. Enhancement of ECL emission of QD by treatment of H2O2 | [56] |
| Single QD-GOx composite on GCE | CdS | Duplex formation with NH2-labelled complementary strand, which can bind to COOH-QD-GOx composite | Chloramphenicol | ECL | 100 nM–1 pM | 0.5 pM | Milk | Use of GOx-CdS QD-GCE as substrate. Use of HRP as quencher. Sensing platform: GCE-(GOx-QD)-Aptamer-HRP | [47]; Figure 3A |
| Core/shell on GCE | CdTe/CdS | Electrostatic attraction to NH2-QD and (-) charged aptamer | Thrombin | NECL | 100 aM–10 fM | 31 aM | Interfering agent (BSA, IgG, HSA)-containing samples | Coating of chitosan on GCE-QD for uniform surface for aptamer immobilization. Use of NIR QDs | [48]; Figure 3B |
| DNA-QD/AuNP | DNA | Conjugation of SH-aptamer to DNA-QD/AuNP conjugate | S. typhimurium | Fluorescence | 10–1.0 × 107 CFU/mL | 13.6 CFU/mL | Milk | Use of DNA-QD/AuNP for easy and simple aptamer conjugation | [57] |
| Single QD on GCE | CdS | Covalent linking between COOH-QD and NH2-aptamer | Cardiac troponin I | ECL | 1 fg/mL–10 ng/mL | 0.75 fg/mL | Human serum | Use of aptamer conjugated CdS QDs and AuNPs as ECL luminophores and plasmon sources, respectively. Sandwich assay with aptamer-conjugated QD and aptamer-attached AuNP. | [58] |
| Core/shell on ITO electrode and magentic | CdTe/CdS | Covalent linking between COOH-QD and NH2-aptamer | Hg2+ | ECL | 20 aM to 2 μM | 2 aM | Carp fish, saltwater fish, tap water | Sensing platform: Fe3O4@SiO2/dendrimers/QDs. Use of AuNP as a quencher. | [59] |
| MoS2@Pd-Au on GCE | MoS2 | Duplex formation with primer DNA crosslinked with chitosan-MoS2@Pd-Au | Lipopolysaccharide | ECL | 0.1 fg/mL–50 ng/mL | 0.07 fg/mL | None | Incorporation of target-cycling synchronized rolling circle amplification reaction | [49]; Figure 4A |
| MoS2@Au | MoS2 | Conjugation of SH-aptamer to QD | Siglec-5 | ECL | 10–500 pM | 8.9 pM | None | Use of ExoIII-assisted signal amplification of QD | [50]; Figure 4B |
| Single QD | CdSe | Covalent linking between COOH-QD and NH2-SSB, followed by binding with aptamer | Streptomycin | Fluorescence | 0.1–100 ng/mL | 0.03 ng/mL | Milk | Use of ExoI-assisted target recycling amplification | [51]; Figure 4C |
| Single QD | CdSe | Duplex formation, which can covalently bind to antibody-QD | Chloromycetin | Fluorescence | 0.05–100 ng/mL | 2 pg/mL | Milk | Reusable sensing platform | [60] |
| Purpose | Strategy | Feature | Reference(s) |
|---|---|---|---|
| Reduction of toxicity | Deposition of semiconductor shell layer on the core (production of core-shell composite) | Protection of the toxic component from degradation; Minimal surface defects of these QDs | [75] |
| Attachment of hydrophilic bifunctional molecule (e.g., PEG) | High stability under biologically relevant conditions; Less affected by size of QD | [52,84,85] | |
| Functionalization of amphiphilic polymer (micelle-forming) | Robust, commercially available, and cheap polymeric precursors; Easy control over number of functional units introduced into polymeric coating | [86,87,88] | |
| Coating surface of QD with silica | Non-toxic, biocompatible, chemically inert, optically transparent, protect from leaching of toxic QD components | [89,90] | |
| Use of eco-friendly synthetic method | Bacteria, fungi, virus-driven biosynthesis | Need for optimization of biosynthesis, recovery, and purification process | [91] |
|
| [92,93,94,95] | |
| [96] | ||
| [97] | ||
| [98,99] | ||
|
| [100,101] | |
| [102,103] | ||
| [104,105] | ||
|
| [106] | |
| [107] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Yoo, S. Aptamer-Conjugated Quantum Dot Optical Biosensors: Strategies and Applications. Chemosensors 2021, 9, 318. https://doi.org/10.3390/chemosensors9110318
Kim D, Yoo S. Aptamer-Conjugated Quantum Dot Optical Biosensors: Strategies and Applications. Chemosensors. 2021; 9(11):318. https://doi.org/10.3390/chemosensors9110318
Chicago/Turabian StyleKim, Dongmin, and Seungmin Yoo. 2021. "Aptamer-Conjugated Quantum Dot Optical Biosensors: Strategies and Applications" Chemosensors 9, no. 11: 318. https://doi.org/10.3390/chemosensors9110318
APA StyleKim, D., & Yoo, S. (2021). Aptamer-Conjugated Quantum Dot Optical Biosensors: Strategies and Applications. Chemosensors, 9(11), 318. https://doi.org/10.3390/chemosensors9110318






