Abstract
Unmodified gold nanoparticles (AuNPs)-based aptasensing (uGA) assay has been widely implemented in the determination of many different targets, but there are few reports on protein detection using uGA. Here, we designed a uGA assay for protein detection including the elimination of interfering proteins. Positively charged protein can be absorbed directly on the surface of AuNPs to form “protein corona”, which results in the aggregation of AuNPs even without salt addition, thereby preventing target protein detection. To overcome this problem, we systematically investigated the effect of modifying the pH of the solution during the uGA assay. A probe solution with a pH slightly higher than the isoelectric points (pI) of the target protein was optimal for protein detection in the uGA assay, allowing the aptamer to selectively detect the target protein. Three proteins (beta-lactoglobulin, lactoferrin, and lysozyme) with different pI were chosen as model proteins to validate our method. Positively charged interfering proteins (with pIs higher than the optimal pH) were removed by centrifugation of protein corona/AuNPs aggregates before the implementation of actual sample detection. Most importantly, the limit of detection (LOD) for all three model proteins was comparable to that of other methods, indicating the significance of modulating the pH. Moreover, choosing a suitable pH for a particular target protein was validated as a universal method, which is significant for developing a novel, simple, cost-effective uGA assay for protein detection.
1. Introduction
Gold nanoparticle (AuNP)-based colorimetric biosensors have been widely used in diagnostics [1,2], environmental monitoring [3,4], and food analysis [5,6]. Among these, an unmodified AuNPs-based aptasensing (uGA) assay [7,8], which incorporated DNA aptamer (in vitro selected oligonucleotides with specific target binding abilities) [9,10] and unmodified AuNPs in the design of colorimetric biosensors, arouse considerable attention due to their high specificity, ease-of-use, and label-free properties. These biosensors have been implemented in the determination of many targets with high sensitivity, ranging from metal ions, and toxins to pesticide residues [11,12,13,14,15,16,17]. The mechanism of the uGA assay relies on controlling the equilibrium between the attractive and repulsive forces. Briefly, the citrate-stabilized AuNPs in solution avoid aggregation due to the strong van der Waals attraction between AuNPs, but upon adding salt form aggregates when the surface charges are neutralized [18]. The aptamer, a single-strand oligonucleotide, is flexible to partially uncoil its bases so that the attractive van der Waals forces between the bases and AuNPs are sufficient to cause the aptamer to stick on the surface of AuNPs, as well asprotect the AuNPs against salt-induced aggregation [19,20,21]. Subsequently, the aptamer binds to its target and is removed from the surface of AuNPs, inducing the aggregation of AuNPs under high salt. However, these biosensors have rarely been reported for protein detection [22]. The reason for this may be the fact that the protein can bind to the surface of the nanomaterials in suitable conditions, leading to the formation of the so-called “protein corona” [23,24]. Under these conditions, the aptamer is prevented from being absorbed on the surface of the AuNPs and cannot protect them from aggregation under high salt solution. The lack of effective binding sites will then prevent protein detection. Therefore, preventing the formation of the protein corona is a prerequisite to the use of AuNPs for protein sensing.
Electrostatic interactions are the dominant factor for protein adsorption on the surface of nanoparticles [25]. Therefore, adjustment of the total charge of proteins is one approach to control their release and adsorption. Fortunately, proteins are polyelectrolytes and the total charge of a protein is usually expressed with respect to its isoelectric point (pI) when the total charge of the protein is neutral.In a typical aptasensing mechanism the aptamers can protect AuNPs from aggregation in a high salt solution [26,27]. Meanwhile, the AuNPs solution only is negligible changes of the color and the absorbance under the pH value from 4.1 to 11.2 (Figure S1), which allows the survey of protein-dependent stability of the AuNPs. In view of this, we established the hypothesis of a systemic assay for protein sensing using uGA. When the pH of the solution is lower than the pI of protein, the protein will be positively charged and surround the surface of AuNPs to form the protein corona, which causes the citrate-stabilized AuNPs to aggregate, even in the presence of an aptamer in the absence of salt (Figure 1a, top). Conversely, when the pH is much above the pI of the protein, the protein corona is destroyed and the protein is released into the solution due to its negative charge. The dispersed proteins can be identified by aptamers, but the effect of pH-induced carboxylic acid deprotonation for the AuNPs can sufficient against the total electrostatic attraction (for example, salt-induced ionic force and van der Waals interactions between AuNPs). Therefore, the charged nanoparticles are stable under high salt conditions (Figure 1a, bottom) [28]. The UV-vis spectra of AuNPs in the presence of 20.00 mM KCl at different pH confirm this effect (Figure S2). Only the pH near the pI a target protein is expected to be chosen for application in the uGA assay (Figure 1b). In order to confirm our hypothesis, three proteins with different pI values (beta-lactoglobulin (β-LG, pI = 5.2), lactoferrin (LF, pI = 8.7), and lysozyme (LZM, pI = 10.7)) were chosen as model proteins [29,30,31].Furthermore, the uGA assay, which relies on regulating pH to detect proteins, has been proposed for the first time and may be universally applied, whether for basic proteins or acidic proteins.
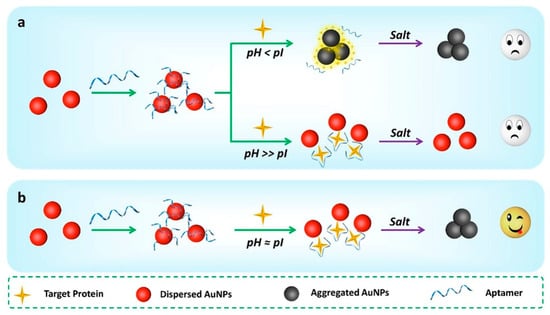
Figure 1.
Overview of the pH-dependent unmodified gold nanoparticles-based aptasensing (uGA) assay for protein detection. (a) At pH < isoelectric points (pI), the proteins directly induce the aggregation of unmodified gold nanoparticles (AuNPs) in the absence of salt; at pH >> pI, the proteins do not induce the aggregation of AuNPs even in the presence of salt. (b) The pH near the pI of protein can be used to implement the uGA assay because the aggregation of AuNPs is not affected by the charge of the protein but can be triggered by adding salt in this situation.
2. Materials and Methods
2.1. Reagents and Chemicals
All oligonucleotides were synthesized and purified by Sangon Biotechnology Co. Ltd. (Shanghai, China), and their sequences are shown in Table 1. Chloroauric acid (HAuCl4·4H2O) was purchased from Aladdin (Shanghai, China). Beta-lactoglobulin (β-LG), bovine serum albumin (BSA), γ-globulin, casein, and protamine were purchased from Shanghai Yuanye Biotechnology Co. Ltd. (Shanghai, China). Lactoferrin (LF) and lysozyme (LZM) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Physical properties of the three proteins are shown in Table 2. Trisodium citrate and all other chemical reagents were purchased from Sinopharm Chemical Reagent Co. Ltd. (Beijing, China). All reagents were of at least analytical grade and were used without any other further purification. All oligonucleotide solutions in this work were obtained by diluting the stock solution with 20 mM Tris-HCl buffer (pH 7.4).

Table 1.
List of aptamer DNA sequences used in the study.

Table 2.
Physical properties of proteins used in this study.
2.2. Apparatus
All absorbance spectra were recorded using a Shimadzu 2600 UV-vis spectrophotometer (Shimadzu, Japan) at room temperature. Transmission electron microscopy (TEM) images were recorded with a JEM-2100 (JEOL, Tokyo, Japan). Ultrapure water (18.2 MΩ·cm) was prepared using a milliQ purification-system (Millipore, Burlington, MA, USA). The AuNPs solution was adjusted with a pH meter (Fe-20K plus, Mettler Toledo instruments Co. Ltd., Shanghai, China). All reactions are performed in centrifuge tubes (1.5 mL). Images were collected with a smartphone (Honor 20, Huawei Technologies Co., Ltd., Guangzhou, China), and then the typical chromogenic images were extracted from the original photos by ppt software for showing in the schematic diagram.
2.3. Synthesis of AuNPs
We prepared the AuNPs using a previously published method [32]. In brief, 0.6 mL trisodium citrate (0.2 mM) solution was added to 100 mL of a boiling HAuCl4 (2.428 mM) solution and consistently stirred for an additional 20 min until the color of the mixture turned from light yellow to wine red. The diameter of the resultant AuNPs was determined by TEM and found to be 16.4 ± 1.93 nm. The concentration of the AuNPs (1.86 nM) was calculated by the absorbance measurements based on an extinction coefficient of 2.67 × 108 M‒1 cm−1 at a wavelength of 450 nm [33]. Finally, the AuNP solution was stored at 4 °C in the dark and was used in the subsequent experiments.
2.4. Adjusting of pH in the uGA Sensing System
Adjusting the pH of the sensing system solution is a key step to measure the target protein by the uGA assay. Proteins (β-LG, LF, and LZM) at a final concentration of 10‒8 M were incubated with 500 μL AuNPs at different pH values (β-LG: 4.1, 5.3, 6.5, 7.5, 9.0, 11.1; LF: 5.9, 7.0, 8.0, 9.5, 10.0, 11.1; LZM: 5.9, 7.0, 8.0, 9.0, 10.0, 11.1, 11.5). The tolerance of AuNPs solutions to salt solutions (20 mM) was investigated at different pH values. The absorbance was recorded by a UV-vis spectrophotometer in the range of 400 ~ 800 nm, and the corresponding images were obtained by a mobile camera. In order to screen out the optimal pH, the range of pH values was further reduced to be suitable for the determination of three target proteins (β-LG: 3.8, 4.3, 4.8, 5.3, 5.5, 5.7, 6.2, 6.7, 7.2; LF: 8, 8.5, 9.0, 9.5, 10, 10.5; LZM: 9.2, 9.7, 10.2, 10.7, 11.2,12.7). The color changes of the protein/AuNPs solutions with or without salt at the above-mentioned pH values were recorded. All the measurements were repeated 3 times for each sample.
2.5. Detection of Proteins in Aqueous Solutions
The pH of the three protein sensing systems was adjusted to their respective optimum values (LZM: 11.2, LF: 9.0, β-LG: 5.3). Ten microliters of aptamer at the optimal concentration (β-LG: 0.007 µM, LF: 0.498 µM, LZM: 0.016 µM) was added to 500 μL AuNPs in solution. Subsequently, 10 μL protein with different final concentrations (β-LG: 0, 0.001, 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 5 nM; LF: 0.01, 0.02, 0.1, 0.2, 0.5, 1, 2, 5, 10 nM; LZM: 0.82, 1.64, 8.2, 16.4, 82, 164 nM) was added. Finally, 100 μL salt with different final concentrations (20 mM, 57.14mM, and 50 mM for β-LG, LF, and LZM detection, respectively) was mixed with the above solution. The absorbance value at 522 nm, 630 nm and 700 nm after reaction were recorded.
The reaction temperature and time were evaluated under various operating temperatures (4, 12, 24, and 37 °C) and time (5, 10, 15, 20, 25, 30, 35, and 40 min) through the detection of three proteins (0.1 nM β-LG, 0.5 nM LF, and 100 nM LZM) as examples.
2.6. Removal of the Interfering Proteins from the Assay
For β-LG detection, 10 μL β-LG at various concentrations (1.5, 1.2, 0.6, and 0.3 μM) and six kinds of interfering proteins (10 μL, 30 nM) were added into 500 μL of a solution of AuNPs. The sample was mixed thoroughly and centrifuged at 12,000 r/min for 15 min. Then, 10 μL supernatant was added to another 500 μL AuNPs. Thus, 10 μL of 0.007 µM aptamer was mixed with solution. After the addition of 100 μL 20 mM salt, the color change and absorbance value at 522 nm and 700 nm were recorded. Under those conditions, the final concentrations of the four β-LG detection samples were 0.5, 0.4, 0.2, and 0.1 nM, respectively. The detected concentration was compared with the spiked value, and the recovery was calculated. For the detection of LF and LZM, the experimental processes were the same as for the β-LG assay, except that the spiked target proteins were different. For the detection of LF, different concentrations (1.5, 1.2, 0.6, 0.3 μM) of LF (10 μL) were added, and the final concentrations of the four LF detection samples were 0.5, 0.4, 0.2, and 0.1 nM, respectively. For LZM detection, different concentrations (240, 30, 15, 3 μM) of 10 μL LZM were added, and the final concentrations of the four LZM detection samples were 80, 10, 5, and 1 nM, respectively.
3. Results
3.1. Feasibility and Analytical Performance of the uGA Assay
The effect of pH on the stability of the β-LG (acidic protein)/AuNPs system is shown in Figure 2a. Citrate-stabilized AuNPs remained dispersed at a pH above the pI (>5.2) and the color remained. At a pH lower than the pI, the β-LG was positively charged causing the aggregation of the AuNPs, even though no salt was added [34,35,36,37], implying that a pH < 5.2 is not suitable for the colorimetric detection of β-LG. Furthermore, the optimum pH for β-LG detection was explored and found to lie in a narrow pH range between 3.8 and 7.2 under salt-free conditions and upon the addition of salt to the β-LG/AuNPs solution (Figure S3a). The results show that β-LG could directly induce aggregation of AuNPs along with color changes without or with the addition of salt at pH < 5.2. While the color of the β-LG/AuNPs solution without salt remained constant at pH ≥ 5.3, it significantly changed upon the addition of salt at pH 5.3–5.7. Therefore, the detection of β-LG could be accomplished at pH 5.3. As shown in Figure 2b, a series of UV-vis spectra of AuNPs confirmed the feasibility of uGA for the detection of β-LG at pH 5.3. Followed by the typical mechanism of uGA, the presence of KCl can trigger the aggregation of AuNPs (curve 4), while the protection by aptamer suppresses the aggregation of Au NPs in the presence of salt (curve 2). The formation of a β-LG/aptamer complex in the presence of salt caused the color of the AuNPs to change from red to gray (curve 5). The transmission electron microscopy (TEM) images further revealed the dispersion/aggregation of AuNPs under different conditions (Figure S3b). These results indicated that the specific interaction between aptamer and protein was sufficient for the absorption of the aptamer on the surface of AuNPs at the appropriate pH, which was in good agreement with our hypothesis. As critical factors, we explored the effect of varying the concentration of KCl and the aptamer in the uGA assay. Our results showed that the salt-induced aggregation reaction is saturated by triggering with 20 mM KCl (Figure 2c). As the pH of the solution decreased, repulsion between negatively charged aptamers and the AuNPs surface decreased, which facilitated the attachment of aptamers on AuNPs surface. On the contrary, with increased the pH, a decrease in the number of aptamers grafted to the AuNPs. Fortunately, with the change of pH, the secondary structure of the aptamer on AuNPs does not change [38]. Based on these factors, it is particularly important to optimize the concentration of Aptamer in the uGA assay. Figure 2d shows the degree of aggregation of AuNPs triggered by 20 mM KCl in the presence of increasing concentrations of aptamer in the presence or absence of β-LG. The ΔA700/A522 value was calculated to evaluate the performance of aptamer (ΔA700/A522 = A1 − A2, A1 and A2 represent the A700/A522 of AuNPs/KCl/aptamer solution in presence and absence of β-LG, respectively.), and the ΔF value reached the highest value when the concentration of aptamer was 7 nM with a satisfactory colorimetric signal. Subsequently, we optimized the reaction time of three protein detection systems (β-LG, LF and LZM) ranging from 5 to 40 min. There is a slight increase in the absorbance ratio at beginning of reaction, and nearly reached a plateau after a 30-min incubation (Figure S4a). To ensure adequate reaction, 40 min was chosen as the reaction time. Then, Figure S4b shows the change of absorbance ratio of three protein detection systems with increasing temperature. There is no obvious change in the absorbance ratio with increasing temperature. Considering the simplicity of the experiment, the room temperature was employed as the reaction temperature of protein detection systems.
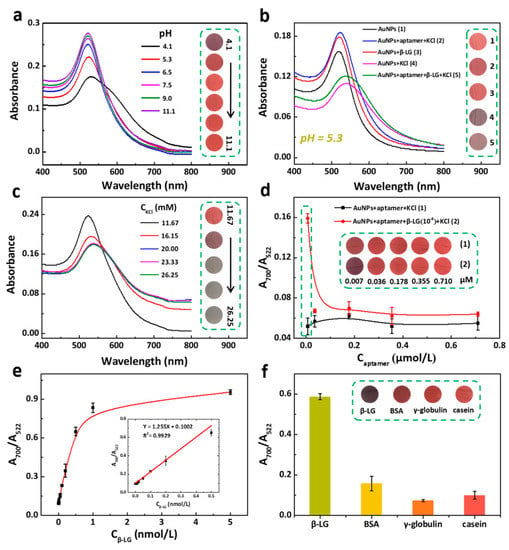
Figure 2.
Determination of beta-lactoglobulin (β-LG) using the uGA assay. (a) UV-vis spectra of AuNPs with β-LG at different pH values (4.1, 5.3, 6.5, 7.5, 9.0, and 11.1). Concentrations: AuNPs, 1.86 nM; β-LG, 10‒8 M. (b) UV-vis spectra of (1) AuNPs, (2) AuNPs + aptamer + KCl, (3) AuNPs + β-LG, (4) AuNPs + KCl, (5) AuNPs + aptamer + β-LG + KCl. Concentrations: AuNPs, 1.86 nM; β-LG, 10‒8 M; aptamer, 7 nM; KCl, 20 mM.(c) The UV-vis spectra of AuNPs/β-LG solution with different concentrations (11.67, 16.15, 20.00, 23.33, and 26.25 mM) of KCl. (d) plots of A700/A522 ratio of AuNPs/KCl and β-LG/KCl/AuNPs with the concentration of aptamer from 0.007 to 0.710 µM. (e) Plot of the absorption ratio (A700/A522) versus β-LG concentration from 0.01 to 0.5 nM. (f) Selectivity assay of the sensing method toward 10‒8 M β-LG, 10‒7 M casein, 10‒7 M bovine serum albumin (BSA), and 10‒7 M γ-globulin. (All insets are the corresponding photographs.).
The sensitivity of the uGA for β-LG detection was investigated in the optimized reaction at 20.00 mM KCl and an aptamer probe concentration of 7 nM at pH 5.3. Figure 2e displayed a good linear relationship between A700/A522 versus the β-LG concentration from 1 to 500 pM. The linear equation was A700/A522 = 1.255 [β-LG] + 0.1002 with R2 = 0.9929. Outstandingly, the limit of detection (LOD) was determined to be 0.9 pM based on the 3σ/S calculation (σ, the standard deviation for a blank solution; S, the slope of the calibration curve).
The pI of the protein plays a decisive role in the specificity tests. For the interfering proteins with pI < 5.3, aptamers can dominate the selectivity. We evaluated the detection specificity of the uGA assay for β-LG by introducing bovine serum albumin (BSA), γ-globulin, and casein as the control groups whose pIs < 5.3 (Table 2). Figure 2f showed that the A700/A522 value of β-LG was much higher than that of the other three proteins, although the concentration of the other three proteins was 10 times higher. This result further indicated that the uGA assay showcased outstanding specificity for discriminating the target, β-LG, at pH 5.3.
3.2. Anti-Interference Performance of the uGA Assay
The coexistence of several proteins with different pIs in one sample is an inevitable key issue during the detection. Proteins whose pI are greater than the chosen pH (5.3) will be positively charged and will directly induce aggregation of the AuNPs (see Figure 1a), thereby interfering the detection of the target protein. Therefore, the interference of these miscellaneous proteins should be eliminated in advance. We designed an interfering protein removal strategy. To this end, seven proteins (six interfering proteins, including BSA, γ-globulin, casein, LZM, LF and protamine, and β-LG as the target protein) with different pIs were added to well-dispersed red AuNPs and incubated at pH 5.3 (Figure 3a1–2). The proteins with a pI > 5.3 were positively charged and formed protein corona on the surface of AuNPs, thereby causing the AuNPs to aggregate. Following centrifugation, the AuNPs aggregates caused these proteins to precipitate. Next, the supernatant was resuspended in a solution of dispersed AuNPs, achieving the separation of interfering proteins with pIs greater than 5.3 (Figure 3a3–4). Subsequently, the introduction of the target protein aptamer can mediate the aggregation of AuNPs upon the addition of salt, performing a selective uGA assay for β-LG (Figure 3a5–6). Notably, the success of each separation and detection was confirmed by measuring the A700/A522 ratio of AuNPs (Figure 3b). To verify the effectiveness of the interfering proteins removal strategy, a mixture of six kinds of interfering proteins spiked with a β-LG sample was analyzed by implementing the proposed strategy. A series of samples with different concentrations of β-LG (0.5, 0.4, 0.2, 0.1 nM) was prepared by adding the six kinds of interfering proteins (30 nM) (Figure 3c). Importantly, the obtained results were in good concordance with the known spiked value. The average recovery of the β-LG assay was 103.6% (Figure 3d), indicating the feasibility and validity of this pretreatment process. The integration of the pretreatment process and the specificity of the aptamer for the selectivity satisfy the requirement of the uGA assay for β-LG.
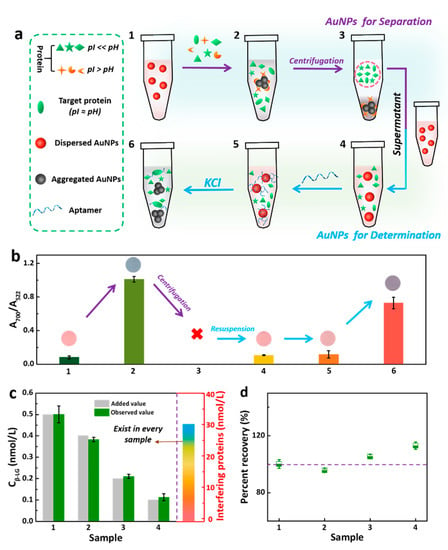
Figure 3.
(a) Schematic diagram of the separation of interfering proteins and detection of target proteins relying on the dual function of AuNPs. Ten microliters β-LG and six kinds of interfering proteins (10 μL, 30 nM) were added into 500 μL AuNPs. The sample was mixed thoroughly and centrifuged at 12,000 r/min for 15 min. Then, 10 μL supernatant was added to another 500 μL AuNPs. Thus, 10 μL of 0.007 µM aptamer was mixed. After the addition of 100 μL 20 mM salt, the color change and absorbance value at 522 nm and 700 nm were recorded. (b) Evolution of the A700/A522 ratio of AuNPs corresponding to processes in Figure 3a (Inset: the corresponding photographs). (c) Analysis of different concentrations β-LG in the mixture sample, which were prepared by mixing the various concentrations (0.5, 0.4, 0.2, 0.1 nM) β-LG and six kinds of interfering proteins (BSA, γ-globulin, casein, LZM, lactoferrin (LF) and protamine), the concentration of each interfering protein is 30 nM. (d) Recoveries of β-LG in the interfering proteins removal assay.
3.3. The Universality of the uGA Assay Confirmed by Using Two Different Target Proteins
The neutral protein LF, with a pI at 8.7, was successfully detected at pH 9. Although there was a negligible effect on AuNPs/LF system at pH values ranging from 9 to 10.5, high pH values triggered the denaturation of LF. Therefore, pH 9 was chosen as the optimal reaction condition and used in the subsequent assay (Figures S5a and S6a). The feasibility of the uGA assay for the detection of LF was confirmed by TEM (Figure S5b) and UV-vis absorption spectra (Figure S6b). KCl at 57.14 mM (Figure S6c) and LF aptamer at 0.498 μM (Figure S6d) were found to be suitable for the quantification of LF. Under these optimal conditions, the sensitivity of the LF detection assay was assessed by measuring the absorbance ratio at A700/A522 of LF solutions ranging in concentration from 0.01 to 10 nM (Figure S6e). A linear relation was obtained between the absorbance ratio and the concentration of LF in the 0.01–1 nM range (R2 = 0.9987) (Figure S6e, inset). The LOD measured 3.66 pM. Figure S5c,d showed the pretreatment of the LF solution that was analogous to that of β-LG solution. The interference proteins with pI > 9 were first removed by centrifugation of aggregated AuNPs/protein complexes formed by electrostatic adsorption at pH 9 (Figure S5c). Then, the aptamer performed was added for the detection of LF (Figure S6f). The high recovery rate of the spiked LF (Figure S5d) indicated the excellent reliability and reproducibility of the uGA assay.
For the basic protein, LZM with a pI of 10.7 was successfully detected at pH 11.2. Values of pH lower than its pI (<10.7) were confirmed unsuitable for detecting LZM by observing the color of AuNPs solution (Figure S7a) and measuring the UV-vis absorption spectra (Figure S8a). Although the pH values ranging from 10.7 to 11.2 could be used in the uGA assay (Figure S7a), a pH value of 11.2 was chosen and used in the subsequent assay. The feasibility and performance of uGA assay for the detection of LZM was confirmed by TEM (Figure S7b) and UV-vis absorption spectra (Figure S8b). The salt and aptamer concentrations were optimized to 58.33 mM (Figure S8c) and 0.016 μM (Figure S8d), respectively. Next, LZM detection was assessed under optimal conditions. A calibration curve was obtained for changes in the A630/A522 ratio versus the logarithmic value of the LZM concentration with R2 = 0.9816, (Figure S8e). The LOD was calculated to be about 0.71 nM. For the assay specificity, the interfering proteins with its pI > 11.2 were first removed by centrifugation and filtration of protein corona/AuNPs aggregates at pH 11.2 (Figure S7c). In the presence of interfering proteins, the average recovery of added LZM was 118.3% (Figure S7d). Thus, the aptamer was capable of the selective detection of LZM (Figure S8f). This section may be divided by subheadings. It should provide a concise and precise description of the experimental results and their interpretation, as well as the experimental conclusions that can be drawn.
4. Conclusions
In conclusion, few reports on protein detection by using uGA assay exist, mainly because the proteins may be directly absorbed on the surface of AuNPs to form the protein corona. Therefore, disturbing the formation of protein corona is a prerequisite for the successful design of an uGA assay for protein detection. Adjustment of pH of the probe solution is one effective approach. In the current work, a probe solution with a pH slightly higher than the pI of the target protein was validated to be suitable for protein detection in a uGA assay. Three model proteins with different pIs (i.e., β-LG, LF, LZM) were utilized in a proof-of-principle concept of this essay. The uGA assay shows a better LOD for all three proteins compared with other methods (Table S1), suggesting the practical meaning of this problem-solving strategy. It is worth noting that the interfering proteins with pIs higher than the optimized pH of the assay form protein corona on the surface of AuNPs at optimized pH, which are removed by centrifugation of protein corona/AuNPs aggregation before the start of actual sample detection. Moreover, the successful performance of uGA assay for three model proteins shows that this strategy may be universally applied, whether for basic proteins or acidic proteins.
Supplementary Materials
The following are available online at https://www.mdpi.com/2227-9040/8/4/98/s1, Table S1: Comparison of different methods for the analysis of β-LG, LF, and LZM, Figure S1. UV-vis spectra and color change in the AuNPs solution with different pHs in the absence of protein. Figure S2: UV-vis spectra of AuNPs with KCl (20 mM) at different pH values, Inset: the corresponding photographs of the solution of AuNPs. Figure S3: (a) Illustration of AuNPs/β-LG solution with (bottom) and without (upper) salt at various pH. (b) TEM image of AuNPs. Figure S4: Optimization of reaction temperature and time. Figure S5: (a) Illustration of AuNPs/LF solution with (bottom) and without (upper) salt at different pH. (b) TEM images of AuNPs. (c,d) Analysis anti-interference performance of the uGA Assay for detection LF. Figure S6: Determination of LF using the uGA assay. Figure S7: (a) Illustration of AuNPs/LZM solution with (bottom) and without (upper) salt at different pH values. (b) TEM image of AuNPs. (c,d) Analysis of the anti-interference performance of the uGA Assay for detection LZM. Figure S8: Determination of LF using the uGA assay.
Author Contributions
This paper was prepared through the contributions of all authors. X.S. designed the experiment, secured the research funding and contributed to the final manuscript preparation. D.L. performed the experiment, wrote the initial manuscript. Q.Z. guided experiments and revised manuscript. D.Z. performed the experiment. X.L. guided experiments. All authors have read and agreed to the published version of the manuscript.
Funding
Authors deeply acknowledge the financial support for this work from the National Natural Science Foundation of China (NSFC, Project no.31972155, 31801473), Natural Science Foundation of Hunan Province (2019JJ40115, 2020JJ5249), China Postdoctoral Science Foundation (2018M642980), Changsha Municipal Natural Science Foundation (kq1905017), and Scientific Research Foundation of Hunan Provincial Education Department (18C0131).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; nor in the decision to publish the results.
References
- Lopa, N.S.; Rahman, M.M.; Ahmed, F.; Ryu, T.; Sutradhar, S.C.; Kim, J.; Kim, D.H.; Lee, Y.H.; Kim, W. Simple, Low-cost, Sensitive and Label-Free Aptasensor for the Detection of Cardiac Troponin I Based on a Gold Nanoparticles Modified Titanium Foil. Biosens. Bioelectron. 2019, 126, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Loiseau, A.; Zhang, L.; Hu, D.; Salmain, M.; Mazouzi, Y.; Flack, R.; Liedberg, B.; Boujday, S. Core-Shell Gold/Silver Nanoparticles for Localized Surface Plasmon Resonance-Based Naked-Eye Toxin Biosensing. ACS Appl. Mater. Interfaces 2019, 11, 46462–46471. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, X.; Liu, Q.; Chen, Z. Colorimetric Differentiation of Flavonoids Based on Effective Reactivation of Acetylcholinesterase Induced by Different Affnities between Flavonoids and Metal Ions. Anal. Chem. 2020, 92, 3361–3365. [Google Scholar] [CrossRef]
- Esmaelpourfarkhani, M.; Abnous, K.; Taghdisi, S.M.; Chamsaz, M. A Novel Turn-Off Fluorescent Aptasensor for Ampicillin Detection Based on Perylenetetracarboxylic Acid Diimide and Gold Nanoparticles. Biosens. Bioelectron. 2020, 164, 112329. [Google Scholar] [CrossRef] [PubMed]
- Zang, L.; Salmain, M.; Liedberg, B.; Boujday, S. Naked Eye Immunosensing of Food Biotoxins Using Gold Nanoparticle-Antibody Bioconjugates. ACS Appl. Nano Mater. 2019, 2, 4150–4158. [Google Scholar] [CrossRef]
- Luo, K.; Ryu, J.; Seol, I.H.; Jeong, K.B.; You, S.M.; Kim, Y.R. Paper-Based Radial Chromatographic Immunoassay for the Detection of Pathogenic Bacteria in Milk. ACS Appl. Mater. Interfaces 2019, 11, 46472–46478. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhou, X.; Liedberg, B.; Zhu, X.Y.; Memon, A.G.; Shi, H. Screening Criteria for Qualified Antibiotic Targets in Unmodified Gold Nanoparticles-Based Aptasensing. ACS Appl. Mater. Interfaces 2019, 9, 35492–35494. [Google Scholar] [CrossRef] [PubMed]
- Alsager, O.A.; Kumar, S.; Zhu, B.; Travas-Sejdic, J.; McNatty, K.P.; Hodgkiss, J.M. Label-Free Electrochemical Aptasensor for Femtomolar Detection of 17 Beta-Estradiol. Anal. Chem. 2015, 87, 4201–4209. [Google Scholar] [CrossRef]
- Eissa, S.; Siddiqua, A.; Chinnappan, R.; Zourob, M. Electrochemical SELEX Technique for the Selection of DNA Aptamers against the Small Molecule 11-Deoxycortisol. Anal. Chem. 2019, 2, 2624–2632. [Google Scholar] [CrossRef]
- Zhao, L.; Qi, X.; Yan, X.; Huang, Y.; Liang, X.; Zhang, L.; Wang, S.; Tan, W. Engineering Aptamer with Enhanced Affinity by Triple Helix-Based Terminal Fixation. J. Am. Chem. Soc. 2019, 141, 17493–17497. [Google Scholar] [CrossRef]
- Fan, D.; Zhai, Q.; Zhou, W.; Zhu, X.; Wang, E.; Dong, S. A Label-Free Colorimetric Aptasensor for Simple, Sensitive and Selective Detection of Pt (II) Based on Platinum (II)-Oligonucleotide Coordination Induced Gold Nanoparticles Aggregation. Biosens. Bioelectron. 2016, 85, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Huang, Y.; Ma, Y.; Jia, S.; Gao, M.; Li, J.; Zhang, H.; Xu, D.; Wu, M.; Chen, Y.; et al. Design and Synthesis of Target-Responsive Aptamer-Cross-linked Hydrogel for Visual Quantitative Detection of Ochratoxin A. ACS Appl. Mater. Interfaces 2015, 7, 6982–6990. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wang, S.; Liu, X.; He, C.; Tang, Y.; Li, Q.; Liu, J.; Su, H.; Tan, T.; Dong, Y. Aptamer-based Colorimetric Biosensing of Ochratoxin A in Fortified White Grape Wine Sample Using Unmodified Gold Nanoparticle. Aanl. Sci. 2017, 33, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Bhairab, M.; Shylaja, R.; Lavu, P.S.; Bhavanashri, N.; Kingston, J. Highly Sensitive Colorimetric Biosensor for Staphylococcal Enterotoxin B by a Label-Free Aptamer and Gold Nanoparticles. Front. Microbiol. 2018, 9, 179. [Google Scholar]
- Kasoju, A.; Shahdeo, D.; Khan, A.A.; Shrikrishna, N.S.; Mahari, S.; Alanazi, A.M.; Bhat, M.A.; Giri, J.; Gandhi, S. Fabrication of Microfluidic Device for Aflatoxin M1 Detection in Milk Samples with Specific Aptamers. Sci. Rep. 2020, 10, 4627. [Google Scholar] [CrossRef]
- Li, H.; Hu, W.; Hassan, M.M.; Zhang, Z.; Chen, Q. A Facile and Sensitive SERS-Based Biosensor for Colormetric Detection of Acetamiprid in Green Tea Based on Unmodified Gold Nanoparticles. J. Food Meas. Charact. 2019, 13, 259–268. [Google Scholar] [CrossRef]
- Bala, R.; Dhingra, S.; Kumar, M.; Bansal, K.; Mittal, S.; Sharma, R.K.; Wangoo, N. Detection of Organophosphorus Pesticide—Malathion in Environmental Samples Using Peptide and Aptamer Based Nanoprobes. Chem. Eng. J. 2016, 311, 111–116. [Google Scholar] [CrossRef]
- Li, H.; Rothberg, L. Colorimetric Detection of DNA Sequences Based on Electrostatic Interactions with Unmodified Gold Nanoparticles. Proc. Natl. Acad. Sci. USA 2004, 101, 14036–14039. [Google Scholar] [CrossRef]
- Lee, E.H.; Lee, S.; Kim, M.; Lee, S.W. Simple and Rapid Detection of Bisphenol A Using a Gold Nanoparticle-Based Colorimetric Aptasensor. Food Chem. 2019, 287, 205–213. [Google Scholar] [CrossRef]
- Gao, Z.; Qiu, Z.; Lu, M.; Shu, J.; Tang, D. Hybridization Chain Reaction-Based Colorimetric Aptasensor of Adenosine 5′-Triphosphate on Unmodified Gold Nanoparticles and Two Label-Free Hairpin Probes. Biosens. Bioelectron. 2017, 89, 1006–1012. [Google Scholar] [CrossRef]
- Lan, L.; Yao, Y.; Ping, J.; Ying, Y. Recent Progress in Nanomaterial-Based Optical Aptamer Assay for the Detection of Food Chemical Contaminants. ACS Appl. Mater. Interfaces 2017, 9, 23287–23301. [Google Scholar] [CrossRef]
- Xia, F.; Zuo, X.; Yang, R.; Xiao, Y.; Kang, D.; Valleebelisale, A.; Gong, X.; Yuen, J.; Hsu, B.B.; Heeger, A.J.; et al. Colorimetric Detection of DNA, Small Molecules, Proteins, and Ions using Unmodified Gold Nanoparticles and Conjugated Polyelectrolytes. Proc. Natl. Acad. Sci. USA 2010, 107, 10837–10841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hao, J.; Xu, X.; Chen, X.; Wang, J. Protein Corona-triggered Catalytic Inhibition of Insufficient POSS Polymer-caged Gold Nanoparticles for Sensitive Colorimetric Detection of Metallothioneins. Anal. Chem. 2020, 92, 2080–2087. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M.; Lohse, S.E.; Murphy, C.J.; Fathizadeh, A.; Montazeri, A.; Suslick, K.S. Variation of Protein Corona Composition of Gold Nanoparticles Following Plasmonic Heating. Nano Lett. 2014, 14, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Dewald, I.; Isakin, O.; Schubert, J.; Kraus, T.; Chanana, M. Protein Identity and Environmental Parameters Determine the Final Physicochemical Properties of Protein-Coated Metal Nanoparticles. J. Phys. Chem. C 2015, 119, 25482–25492. [Google Scholar] [CrossRef]
- Razmi, N.; Baradaran, B.; Hejazi, M.; Hasanzadeh, M.; Mosafer, J.; Mokhtarzadeh, A.; Guardia, M.D. Recent Advances on Aptamer-Based Biosensors to Detection of Platelet-Derived Growth Factor. Biosens. Bioelectron. 2018, 113, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Dai, P.; Xu, J.; Chen, H. A Sensitive Aptasensor for Colorimetric Detection of Adenosine Triphosphate Based on the Protective Effect of ATP-Aptamer Complexes on Unmodified Gold Nanoparticles. ACS Appl. Mater. Interfaces 2016, 8, 4434–4441. [Google Scholar] [CrossRef] [PubMed]
- Kairdolf, B.A.; Nie, S. Multidentate-Protected Colloidal Gold Nanocrystals: pH Control of Cooperative Precipitation and Surface Layer Shedding. J. Am. Chem. Soc. 2011, 133, 7268–7271. [Google Scholar] [CrossRef]
- Peng, D.; Yang, J.; Li, J.; Tang, C.; Li, B. Foams Stabilized by β-Lactoglobulin Amyloid Fibrils: Effect of pH. J. Agric. Food Chem. 2017, 65, 10658–10665. [Google Scholar] [CrossRef]
- Teepakorn, C.; Flaty, K.; Charcosset, C. Optimization of Lactoferrin and Bovine Serum Albumin Separation Using Ion-Exchange Membrane Chromatography. Sep. Purif. Technol. 2015, 151, 292–302. [Google Scholar]
- Santos, M.B.; Carvalho, C.W.; Garciarojas, E.E. Heteroprotein Complex Formation of Bovine Serum Albumin and Lysozyme: Structure and Thermal Stability. Food Hydrocolloid. 2018, 74, 267–274. [Google Scholar] [CrossRef]
- Shi, X.; Lu, D.; Wang, Z.; Zhang, D.; Gao, W.; Zhang, C.; Deng, J.; Guo, S. Colorimetric and visual determination of acrylamide via acrylamide-mediated polymerization of acrylamide-functionalized gold nanoparticles. Microchim. Acta 2018, 185, 1–9. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold Nanoparticles from UV−Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Aggarwal, P.; Hall, J.B.; McLeland, C.B.; Dobrovolskaia, M.A.; Mcneil, S.E. Nanoparticle Interaction with Plasma Proteins as It Relates to Particle Biodistribution, Biocompatibility and Therapeutic Efficacy. Adv. Drug. Deliver. Rev. 2009, 61, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Chen, Z.; Tan, L.; Lou, T.; Zhao, Y. DNA-Catalytically Active Gold Nanoparticle Conjugates-Based Colorimetric Multidimensional Sensor Array for Protein Discrimination. Anal. Chem. 2017, 89, 556–559. [Google Scholar] [CrossRef]
- Xi, H.; He, W.; Liu, Q.; Chen, Z. Protein Discrimination Using a Colorimetric Sensor Array Based on Gold Nanoparticle Aggregation Induced by Cationic Polymer. ACS Sustain. Chem. Eng. 2018, 6, 10751–10757. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Y.; Zhang, S.; Wang, S.; Zhang, S.; Zhang, X. Aptamer-Based Plasmonic Sensor Array for Discrimination of Proteins and Cells with the Naked Eye. Anal. Chem. 2013, 85, 6571–6574. [Google Scholar] [CrossRef]
- Dam, D.; Lee, H.; Lee, R.; Kim, K.; Kelleher, N.; Odom, T. Tunable Loading of Oligonucleotides with Secondary Structure on Gold Nanoparticles through a pH-Driven Method. Bioconjugate Chem. 2015, 26, 279–285. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).