A Label-Free Fluorescent Sensor Based on the Formation of Poly(thymine)-Templated Copper Nanoparticles for the Sensitive and Selective Detection of MicroRNA from Cancer Cells
Abstract
1. Introduction
2. Experimental
2.1. Reagents and Materials
2.2. Amplification Reaction
2.3. Formation of PolyT-CuNPs
2.4. Cell Culture, RNA Extraction, and qPCR
2.5. Apparatus
3. Results and Discussion
3.1. Principle of MiRNA Detection
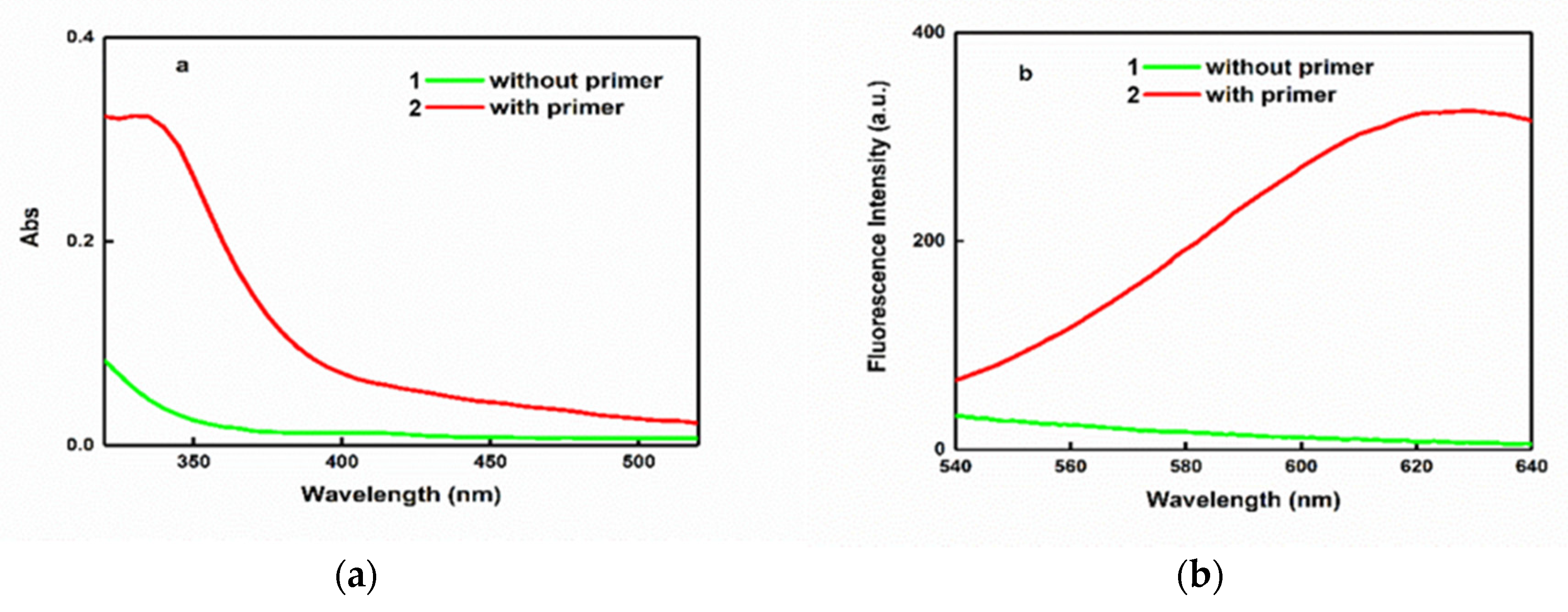
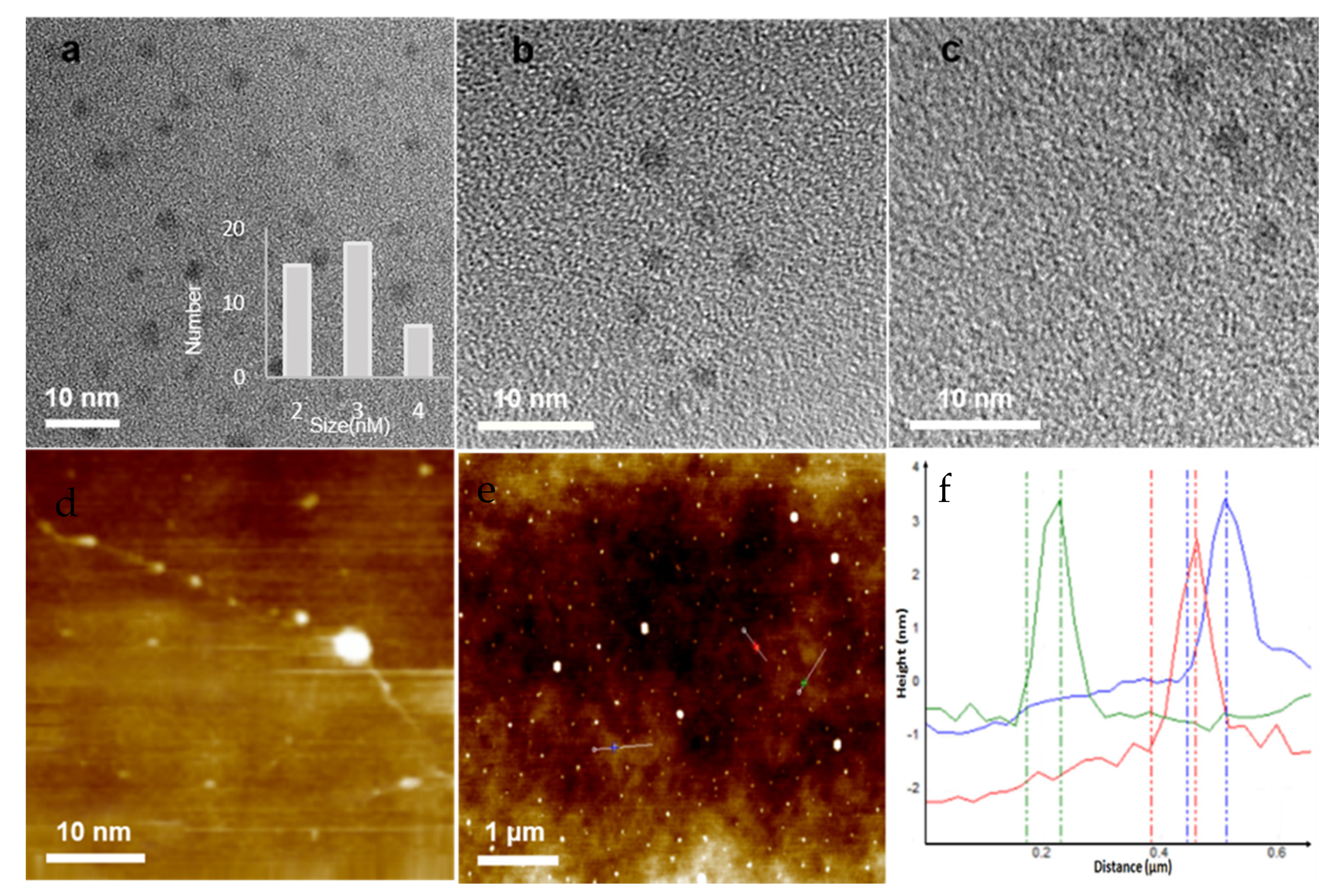
3.2. Characterization of CuNPs
3.3. Optimization of Sensor Conditions
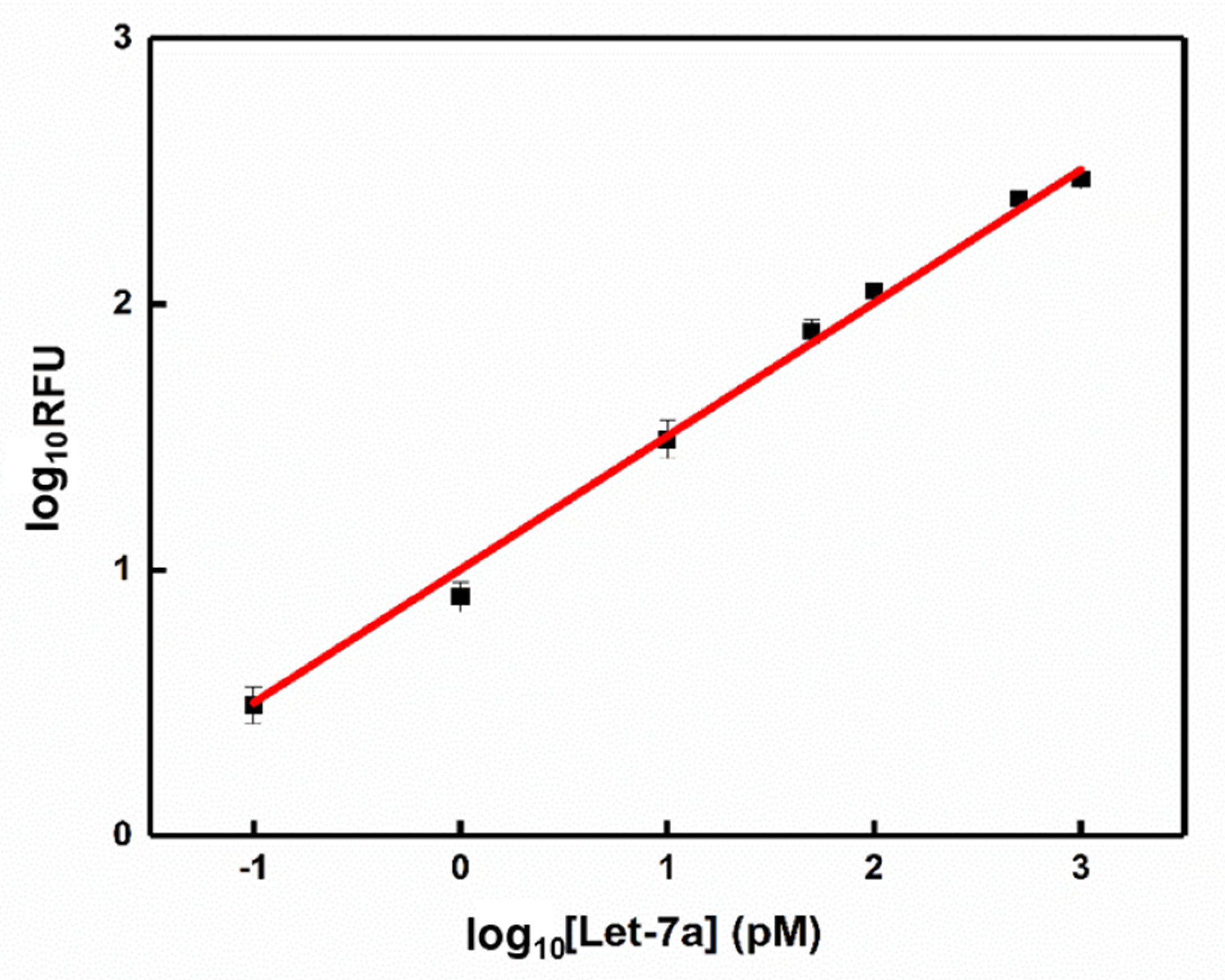
3.4. Detection of MiRNA
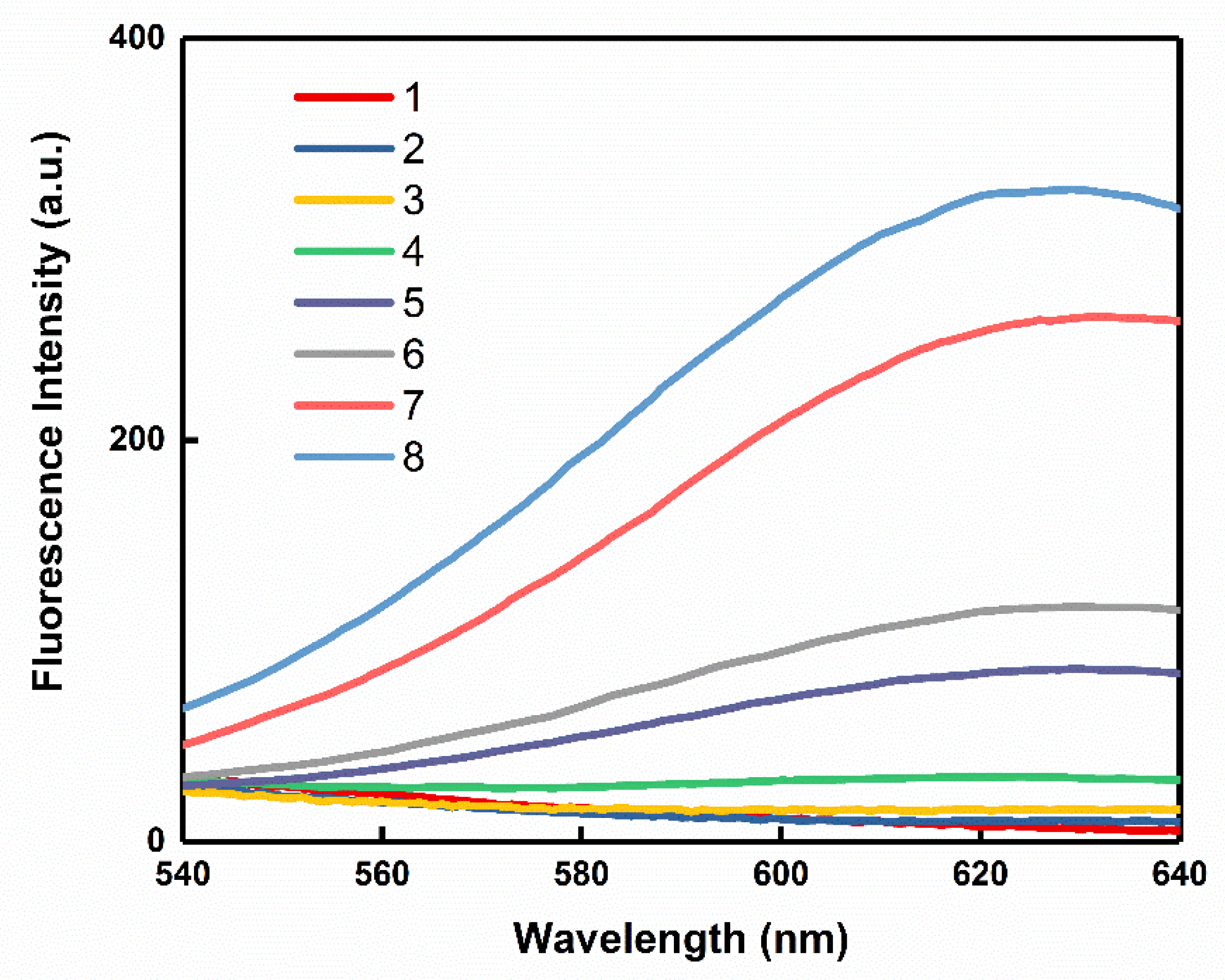
3.5. Specificity Study
3.6. Detection of MiRNA in Real Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Bartel, B. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-Y.; Ye, B.-C.; Yin, B.-C. A universal real-time PCR assay for rapid quantification of microRNAs via the enhancement of base-stacking hybridization. Chem. Commun. 2013, 49, 8247. [Google Scholar] [CrossRef]
- Zhang, J.; Li, C.; Zhi, X.; Ramón, G.A.; Liu, Y.; Zhang, C.; Pan, F.; Cui, D. Hairpin DNA-Templated silver nanoclusters as novel beacons in strand displacement amplification for MicroRNA detection. Anal. Chem. 2015, 88, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Tang, L.; Tian, Q.; Wang, Y.; Lin, L.; Li, J. Toehold initiated rolling circle amplification for visualizing individual MicroRNAs in situ in single cells. Angew. Chem. Int. Ed. 2014, 53, 2389–2393. [Google Scholar] [CrossRef]
- Wang, X.-P.; Yin, B.-C.; Ye, B.-C. A novel fluorescence probe of dsDNA-Templated copper nanoclusters for quantitative detection of microRNAs. RSC Adv. 2013, 3, 8633. [Google Scholar] [CrossRef]
- Stobiecka, M.; Chalupa, A. DNA strand replacement mechanism in molecular beacons encoded for the detection of cancer biomarkers. J. Phys. Chem. B 2016, 120, 4782–4790. [Google Scholar] [CrossRef]
- Stobiecka, M.; Dworakowska, B.; Jakiela, S.; Lukasiak, A.; Chalupa, A.; Zembrzycki, K. Sensing of survivin mRNA in malignant astrocytes using graphene oxide nanocarrier-supported oligonucleotide molecular beacons. Sens. Actuators B Chem. 2016, 235, 136–145. [Google Scholar] [CrossRef]
- Ratajczak, K.; Krazinski, B.E.; Kowalczyk, A.; Dworakowska, B.; Jakiela, S.; Stobiecka, M. Optical biosensing system for the detection of survivin mRNA in colorectal cancer cells using a graphene oxide carrier-bound oligonucleotide molecular beacon. Nanomaterials 2018, 8, 510. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Deng, T.; Chu, X.; Tan, W.; Jiang, J.-H.; Shen, G.; Yu, R. Rolling circle amplification combined with gold nanoparticle aggregates for highly sensitive identification of single-nucleotide polymorphisms. Anal. Chem. 2010, 82, 2811–2816. [Google Scholar] [CrossRef] [PubMed]
- Stobiecka, M.; Molinero, A.A.; Chałupa, A.; Hepel, M. Mercury/Homocysteine ligation-induced ON/OFF-switching of a T–T mismatch-based oligonucleotide molecular beacon. Anal. Chem. 2012, 84, 4970–4978. [Google Scholar] [CrossRef] [PubMed]
- Becerril, H.A.; Woolley, A.T. DNA-Templated nanofabrication. Chem. Soc. Rev. 2009, 38, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Houlton, A.; Pike, A.R.; Galindo, M.A.; Horrocks, B.R. DNA-based routes to semiconducting nanomaterials. Chem. Commun. 2009, 1797–1806. [Google Scholar] [CrossRef]
- Thomas, A. Blue emitting gold nanoclusters templated by poly-cytosine DNA at low pH and poly-adenine DNA at neutral pH. Chem. Commun. 2012, 48, 6845–6847. [Google Scholar]
- Petty, J.T.; Zheng, J.; Hud, N.V.; Dickson, R.M. DNA-Templated Ag Nanocluster formation. J. Am. Chem. Soc. 2004, 126, 5207–5212. [Google Scholar] [CrossRef]
- Richards, C.I.; Choi, S.; Hsiang, J.-C.; Antoku, Y.; Vosch, T.; Bongiorno, A.; Tzeng, Y.-L.; Dickson, R.M. Oligonucleotide-stabilized Ag Nanocluster fluorophores. J. Am. Chem. Soc. 2008, 130, 5038–5039. [Google Scholar] [CrossRef]
- Jia, X.; Li, J.; Han, L.; Ren, J.; Yang, X.; Wang, E. DNA-hosted copper Nanoclusters for fluorescent identification of single nucleotide polymorphisms. ACS Nano 2012, 6, 3311–3317. [Google Scholar] [CrossRef]
- Rotaru, A.; Dutta, S.; Jentzsch, E.; Gothelf, K.; Mokhir, A. Selective dsDNA-Templated formation of copper nanoparticles in solution. Angew. Chem. Int. Ed. 2010, 49, 5665–5667. [Google Scholar] [CrossRef]
- Song, Q.; Shi, Y.; He, D.-C.; Xu, S.; Ouyang, J. Sequence-dependent dsDNA-Templated formation of fluorescent copper nanoparticles. Chem. A Eur. J. 2014, 21, 2417–2422. [Google Scholar] [CrossRef]
- Qing, Z.; He, X.; He, D.; Wang, K.; Xu, F.; Qing, T.; Yang, X. Poly (thymine) Templated selective formation of fluorescent copper nanoparticles. Angew. Chem. Int. Ed. 2013, 52, 9719–9722. [Google Scholar] [CrossRef]
- Liu, G.; Shao, Y.; Peng, J.; Dai, W.; Liu, L.; Xu, S.; Wu, F.; Wu, X. Highly thymine-dependent formation of fluorescent copper nanoparticles templated by ss-DNA. Nanotechnology 2013, 24, 345502. [Google Scholar] [CrossRef]
- Chen, W.-Y.; Lan, G.-Y.; Chang, H.-T. Use of fluorescent DNA-Templated Gold/Silver Nanoclusters for the detection of sulfide ions. Anal. Chem. 2011, 83, 9450–9455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, C.-F.; Zhang, L.; Tan, C.; Yang, J.; Chen, B.; Wang, L.; Zhang, H. DNA-Templated silver Nanoclusters for multiplexed fluorescent DNA detection. Small 2014, 11, 1385–1389. [Google Scholar] [CrossRef]
- Xu, F.; Shi, H.; He, X.; Wang, K.; He, D.; Guo, Q.; Qing, Z.; Yan, L.; Ye, X.; Li, D.; et al. Concatemeric dsDNA-Templated copper nanoparticles strategy with improved sensitivity and stability based on rolling circle replication and its application in MicroRNA detection. Anal. Chem. 2014, 86, 6976–6982. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Sargent, E.H.; Kelley, S.O. One-step DNA-programmed growth of luminescent and biofunctionalized nanocrystals. Nat. Nanotechnol. 2008, 4, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, J.; Fang, Z.; Zeng, L. Random dsDNA-templated formation of copper nanoparticles as novel fluorescence probes for label-free lead ions detection. Chem. Commun. 2012, 48, 1057–1059. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Duan, M.; Zhang, H.; Jiang, J.-H.; Yu, R. Inhibition of dsDNA-Templated copper nanoparticles by pyrophosphate as a label-free fluorescent strategy for alkaline phosphatase assay. Anal. Chem. 2013, 85, 3797–3801. [Google Scholar] [CrossRef]
- Qing, Z.; Zhu, L.; Yang, S.; Cao, Z.; He, X.; Wang, K.; Tan, W. In situ formation of fluorescent copper nanoparticles for ultrafast zero-background Cu2+ detection and its toxicides screening. Biosens. Bioelectron. 2016, 78, 471–476. [Google Scholar] [CrossRef]
- Yin, J.; He, X.; Wang, K.; Qing, Z.; Wu, X.; Shi, H.; Yang, X. One-step engineering of silver nanoclusters–aptamer assemblies as luminescent labels to target tumor cells. Nanoscale 2012, 4, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Monson, C.; Woolley, A.T. DNA-Templated construction of copper nanowires. Nano Lett. 2003, 3, 359–363. [Google Scholar] [CrossRef]
- Qing, Z.; He, X.; Qing, T.; Wang, K.; Shi, H.; He, D.; Zou, Z.; Yan, L.; Xu, F.; Ye, X.; et al. Poly(Thymine)-Templated fluorescent copper nanoparticles for ultrasensitive label-free nuclease assay and its inhibitors screening. Anal. Chem. 2013, 85, 12138–12143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, Q.; Li, J.; Ge, J.; Wang, J.-Y.; Dong, Z.-Z.; Li, Z. A label-free method for detecting biothiols based on poly(thymine)-templated copper nanoparticles. Biosens. Bioelectron. 2015, 69, 77–82. [Google Scholar] [CrossRef]
- Wang, H.-B.; Zhang, H.-D.; Chen, Y.; Liu, Y. A fluorescent biosensor for protein detection based on poly(thymine)-templated copper nanoparticles and terminal protection of small molecule-linked DNA. Biosens. Bioelectron. 2015, 74, 581–586. [Google Scholar] [CrossRef]
- Ou, L.; Li, X.; Liu, H.; Li, L.; Chu, X. Poly(thymine)-templated fluorescent copper nanoparticles for ultrasensitive label-free detection of Pb2+ ion. Anal. Sci. 2014, 30, 723–727. [Google Scholar] [CrossRef]
- Johnson, S.M.; Großhans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 MicroRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Pasquinelli, A.; Reinhart, B.; Slack, F.; Maller, B.; Ruvkun, G. Conservation across animal phylogeny of the sequence and temporal regulation of the 21 nucleotide C. elegans let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Takamizawa, J.; Chamoto, K.; Tsuji, T.; Funamoto, H.; Kosaka, A.; Matsuzaki, J.; Sato, T.; Konishi, H.; Fujio, K.; Yamamoto, K.; et al. Reduced expression of the let-7 MicroRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef]
- Broyde, S.; Wang, L.; Rechkoblit, O.; Geacintov, N.E.; Patel, D.J. Lesion processing: High-fidelity versus lesion-bypass DNA polymerases. Trends Biochem. Sci. 2008, 33, 209–219. [Google Scholar] [CrossRef][Green Version]
- Yeh, H.-C.; Sharma, J.; Han, J.J.; Martinez, J.S.; Werner, J.H. A DNA−silver nanocluster probe that fluoresces upon hybridization. Nano Lett. 2010, 10, 3106–3110. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Z.; Tao, Y.; Wang, Z.; Ren, J.; Qu, X. Hybridization chain reaction engineered dsDNA for Cu metallization: An enzyme-free platform for amplified detection of cancer cells and microRNAs. Chem. Commun. 2015, 51, 11496–11499. [Google Scholar] [CrossRef]
- Ye, T.; Chen, J.; Liu, Y.; Ji, X.; Zhou, G.; He, Z. Periodic fluorescent silver clusters assembled by rolling circle amplification and their sensor application. ACS Appl. Mater. Interfaces 2014, 6, 16091–16096. [Google Scholar] [CrossRef]
- Qiu, X.; Wang, P.; Cao, Z. Hybridization chain reaction modulated DNA-hosted silver nanoclusters for fluorescent identification of single nucleotide polymorphisms in the let-7 miRNA family. Biosens. Bioelectron. 2014, 60, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Chi, B.-Z.; Liang, R.-P.; Qiu, W.-B.; Yuan, Y.-H.; Qiu, J.-D. Direct fluorescence detection of microRNA based on enzymatically engineered primer extension poly-thymine (EPEPT) reaction using copper nanoparticles as nano-dye. Biosens. Bioelectron. 2017, 87, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Miao, P.; Zhang, T.; Xu, J.; Tang, Y. Electrochemical detection of miRNA combining T7 exonuclease-assisted cascade signal amplification and DNA-Templated copper nanoparticles. Anal. Chem. 2018, 90, 11154–11160. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, H.; Wang, L.; Zhu, J.; Jiang, W. Cascade signal amplification based on copper nanoparticle-reported rolling circle amplification for ultrasensitive electrochemical detection of the prostate cancer biomarker. ACS Appl. Mater. Interfaces 2016, 8, 2573–2581. [Google Scholar] [CrossRef]
- Park, K.W.; Batule, B.S.; Kang, K.S.; Park, K.S.; Park, H.G. Rapid and ultrasensitive detection of microRNA by target-assisted isothermal exponential amplification coupled with poly (thymine)-templated fluorescent copper nanoparticles. Nanotechnology 2016, 27, 425502. [Google Scholar] [CrossRef] [PubMed]



| Description | Linear Range | Detection Limit | Detection Method | Total Analysis Time | Ref. |
|---|---|---|---|---|---|
| DNA-templated silver nanoclusters/hairpin-DNA (AgNCs/HpDNA) and strand-displacement amplification | 0 to 0.05 µM | 0.05 µM | Fluorescence | ~6.0 h | [6] |
| ds-DNA-templated copper nanoparticles (CuNPs) and rolling circle amplification (RCA) | 10 to 400 pM | 10 pM | Fluorescence | ~16.5 h | [26] |
| Silver cluster and rolling circle amplification | 6 to 300 pM | 0.84 pM | Fluorescence | ~8.5 h | [45] |
| Hybridization chain reaction and DNA-hosted silver nanoclusters | 3.12 to 50 nM | 0.78 nM | Fluorescence | ~6.5 h | [46] |
| Poly(thymine)-templated CuNPs and enzymatically engineered primer extension | 1 pM to 1nM | 100 fM | Fluorescence | ~3.5 h | [47] |
| T7 exonuclease-assisted cascade signal amplification and DNA-templated copper nanoparticles | 0.1 fM to 0.1 pM | 0.045 fM | Electrochemical | ~5.0 h | [48] |
| Cascade signal amplification based on CuNPs and reported rolling circle amplification | Prostate cancer biomarker 0.05–500 fg/mL | 0.020 ± 0.001 fg/mL prostate specific antigen (PSA) | Electrochemical | ~16.0 h | [49] |
| Target-assisted isothermal exponential amplification (TAIEA) and CuNPs | 70 fM to 700 nM | 0.27 fM | Fluorescence | ~1.0 h | [50] |
| ss-DNA-templated CuNP strand and rolling circle amplification | 100 fM to 1.0 nM | 70.6 fM | Fluorescence | ~5.5 h | This work |
| Lung Cancer Cells (106 copies μg−1) | HeLa Cells (106 copies μg−1) | Normal Cells (106 copies μg−1) | |
|---|---|---|---|
| This assay | 1.7 ± 0.32 | 3.7 ± 0.68 | 5.4 ± 0.91 |
| qPCR method | 1.8 ± 0.30 | 3.3 ± 0.80 | 5.6 ± 0.80 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Q.; Gao, Z.; Dayal, H.; Li, S.F.Y. A Label-Free Fluorescent Sensor Based on the Formation of Poly(thymine)-Templated Copper Nanoparticles for the Sensitive and Selective Detection of MicroRNA from Cancer Cells. Chemosensors 2020, 8, 52. https://doi.org/10.3390/chemosensors8030052
Ma Q, Gao Z, Dayal H, Li SFY. A Label-Free Fluorescent Sensor Based on the Formation of Poly(thymine)-Templated Copper Nanoparticles for the Sensitive and Selective Detection of MicroRNA from Cancer Cells. Chemosensors. 2020; 8(3):52. https://doi.org/10.3390/chemosensors8030052
Chicago/Turabian StyleMa, Qian, Zhiqiang Gao, Hiranya Dayal, and Sam Fong Yau Li. 2020. "A Label-Free Fluorescent Sensor Based on the Formation of Poly(thymine)-Templated Copper Nanoparticles for the Sensitive and Selective Detection of MicroRNA from Cancer Cells" Chemosensors 8, no. 3: 52. https://doi.org/10.3390/chemosensors8030052
APA StyleMa, Q., Gao, Z., Dayal, H., & Li, S. F. Y. (2020). A Label-Free Fluorescent Sensor Based on the Formation of Poly(thymine)-Templated Copper Nanoparticles for the Sensitive and Selective Detection of MicroRNA from Cancer Cells. Chemosensors, 8(3), 52. https://doi.org/10.3390/chemosensors8030052






