Design of Peptide Ligand for Lactoferrin and Study of Its Binding Specificity
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Modelling
- (1)
- Supramolecular biostructures are built on the basis of the principle of continuity of type systems:where: R—Zi—simple and Qi–R = Xi—resonance groups, containing instead of R, Z, Q, X, atoms of organogenic elements (C, N, O, P, S).R R
| |
…HQ1–R = X1 … Z1 … HQ2–R = X2 … HQ2–R = X2 … …HQn–R = Xn … Zm…. - (2)
- These systems are utilized in biostructures as channels for energy and charge transfer. Formation of CIHBS in the area of subunit contact considerably promotes it stabilization.
2.2. Peptide Synthesis
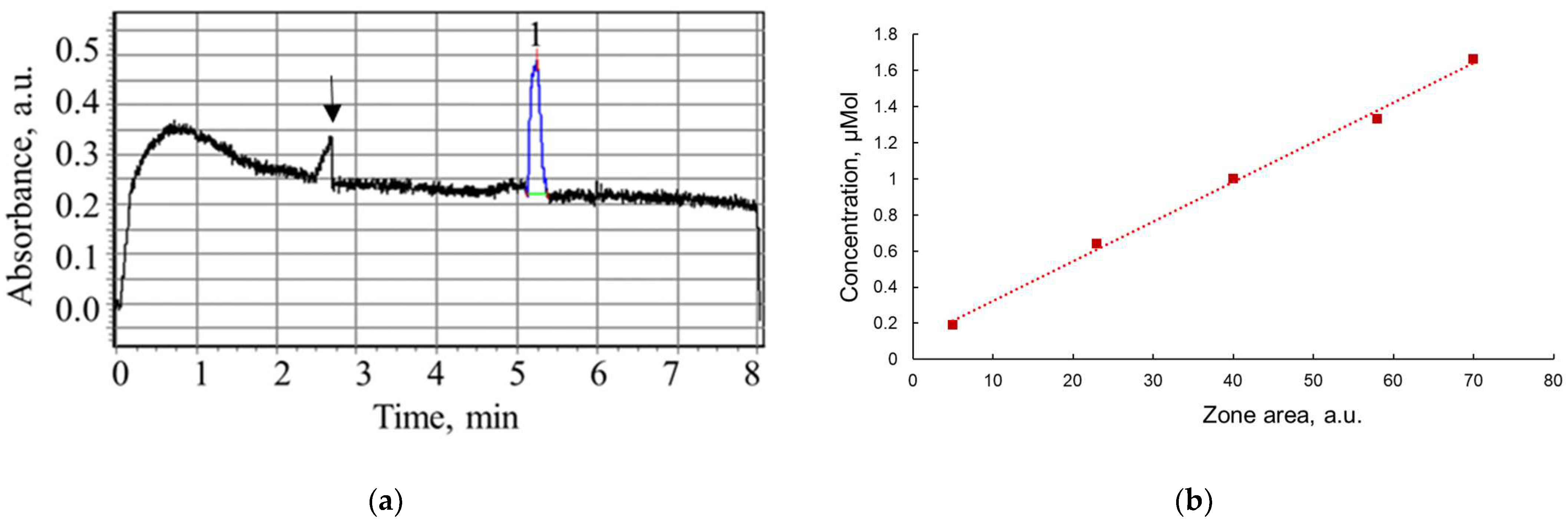
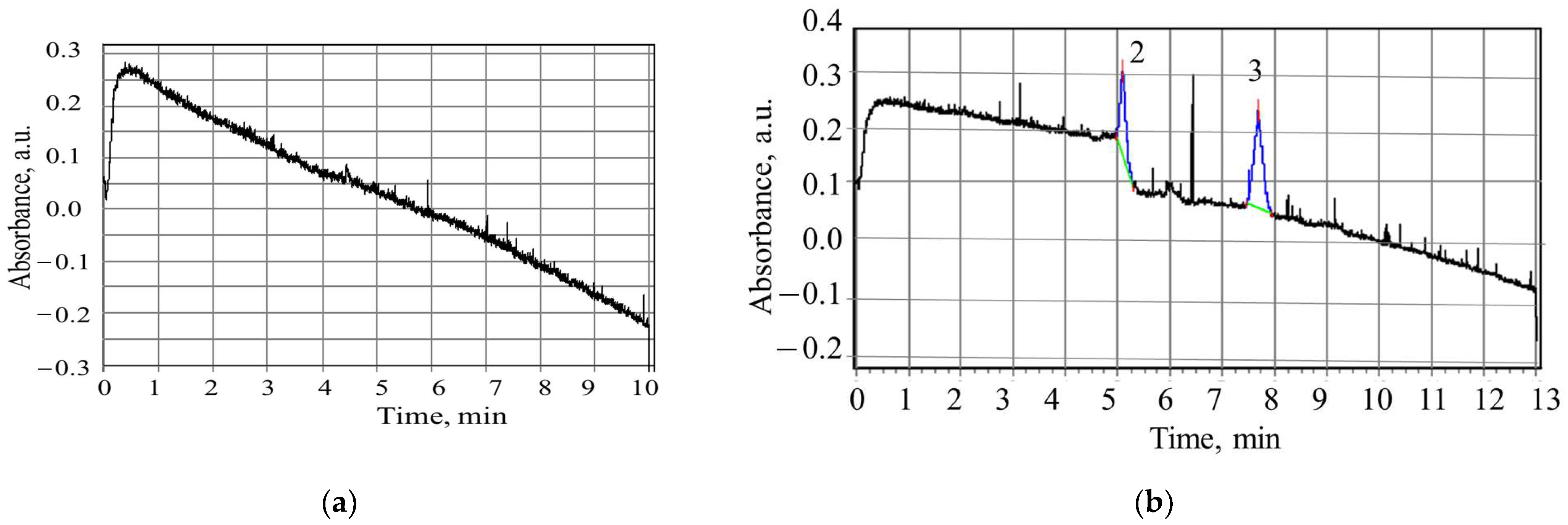
2.3. Capillary Electrophoresis
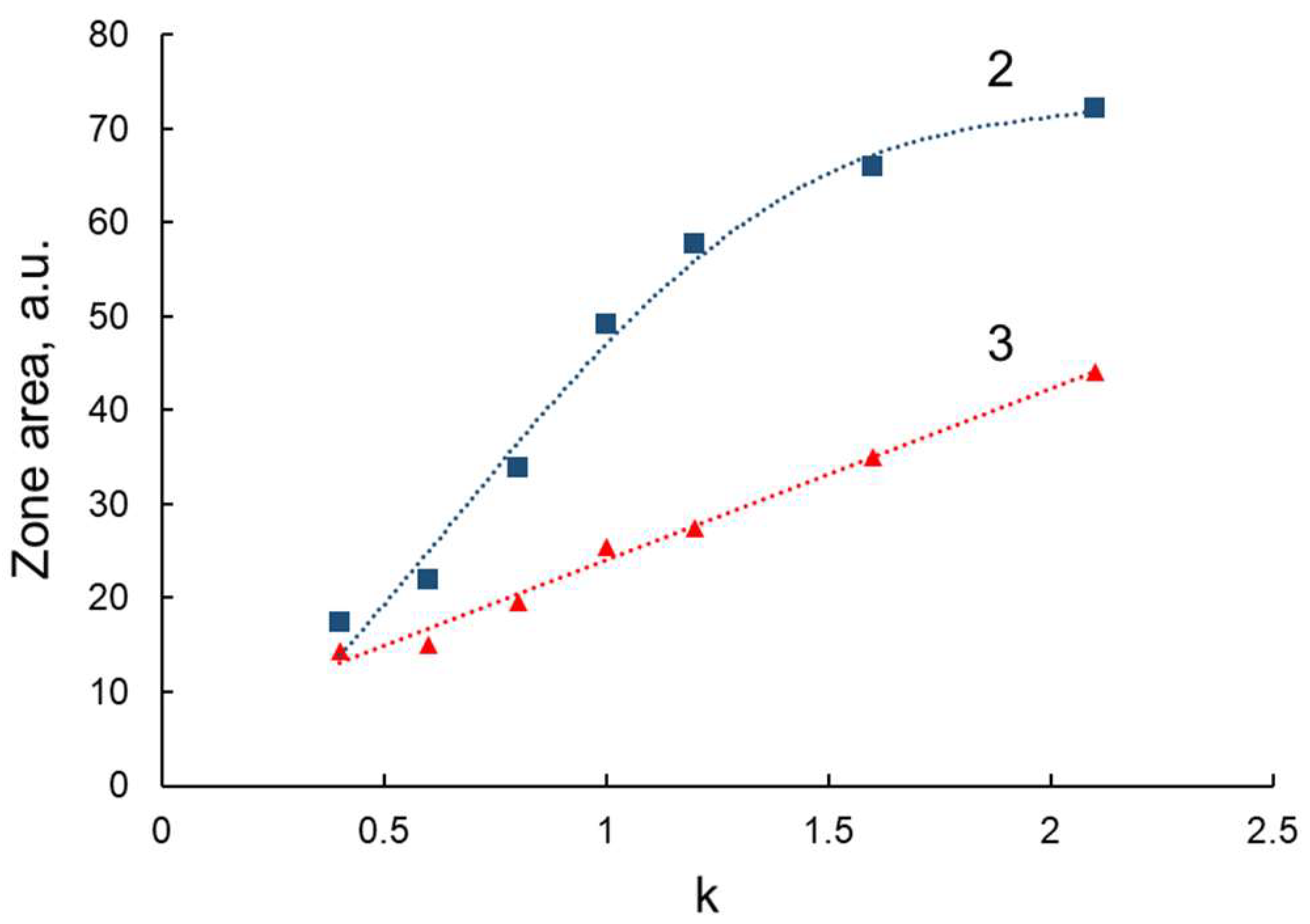
3. Results and Discussion
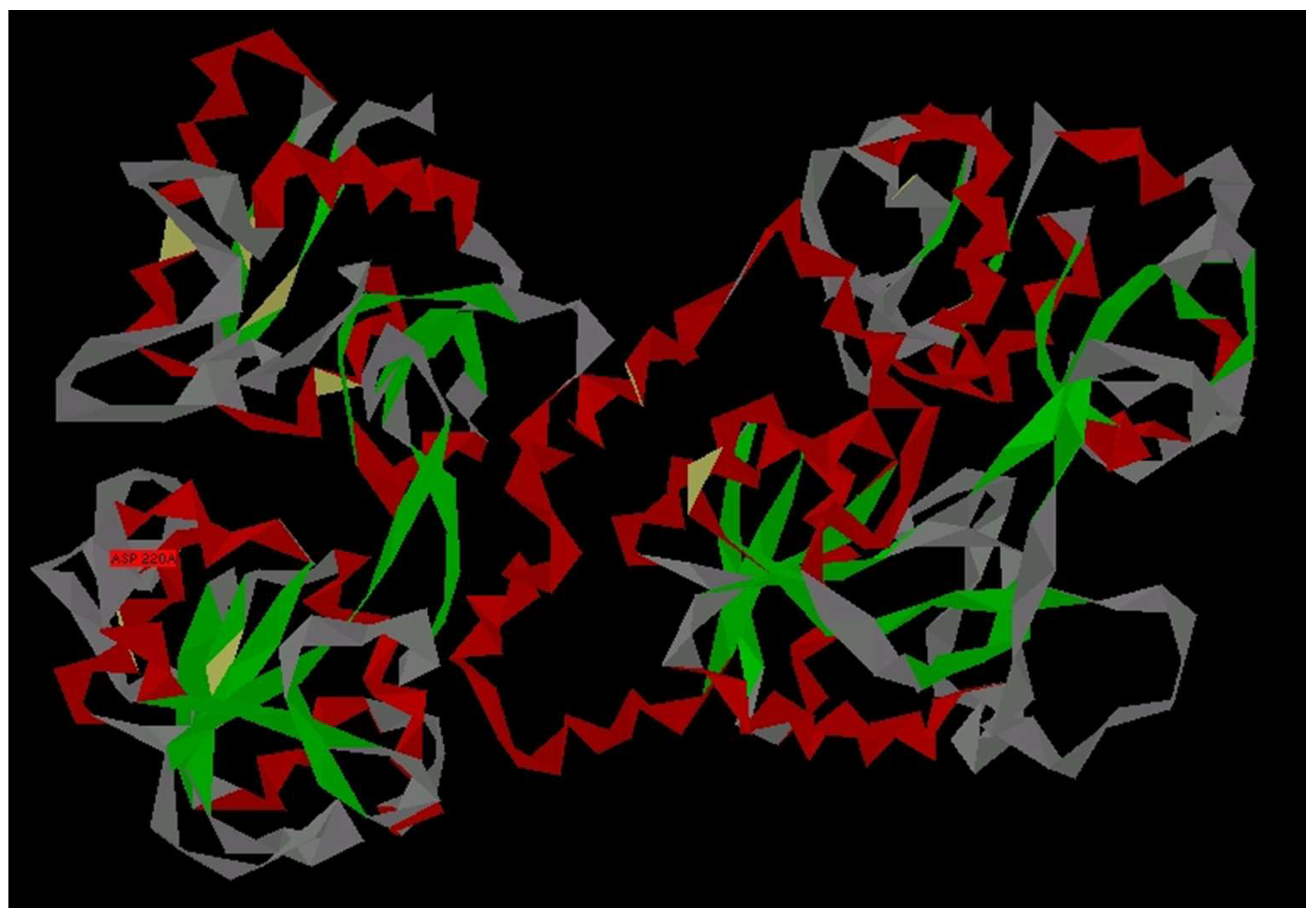
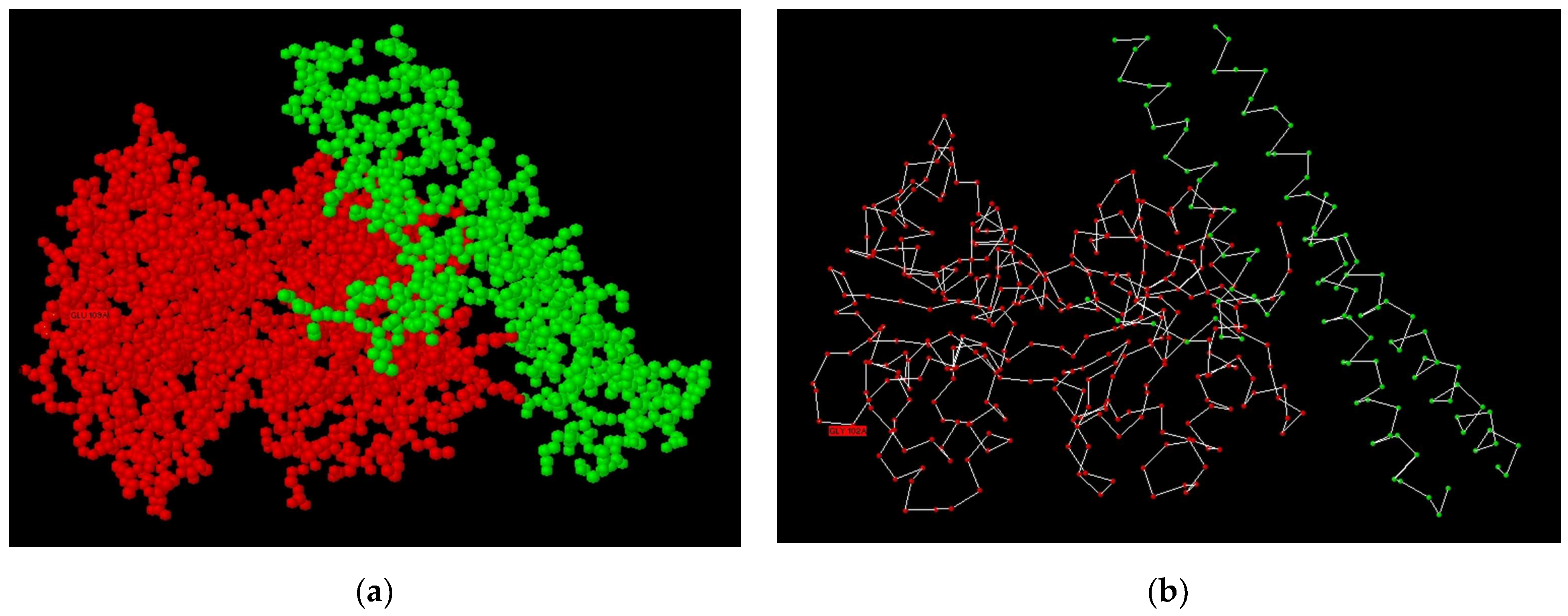
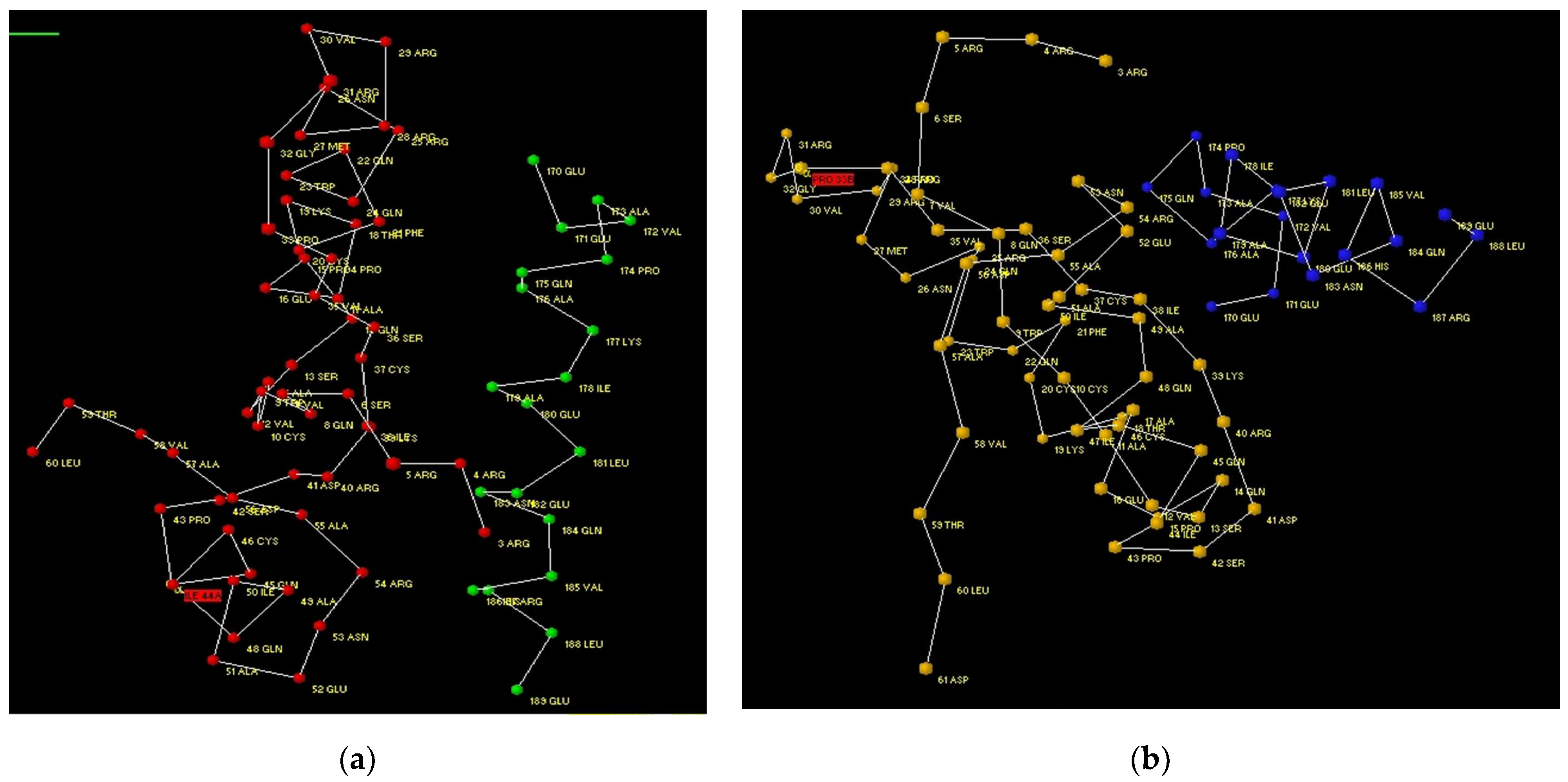
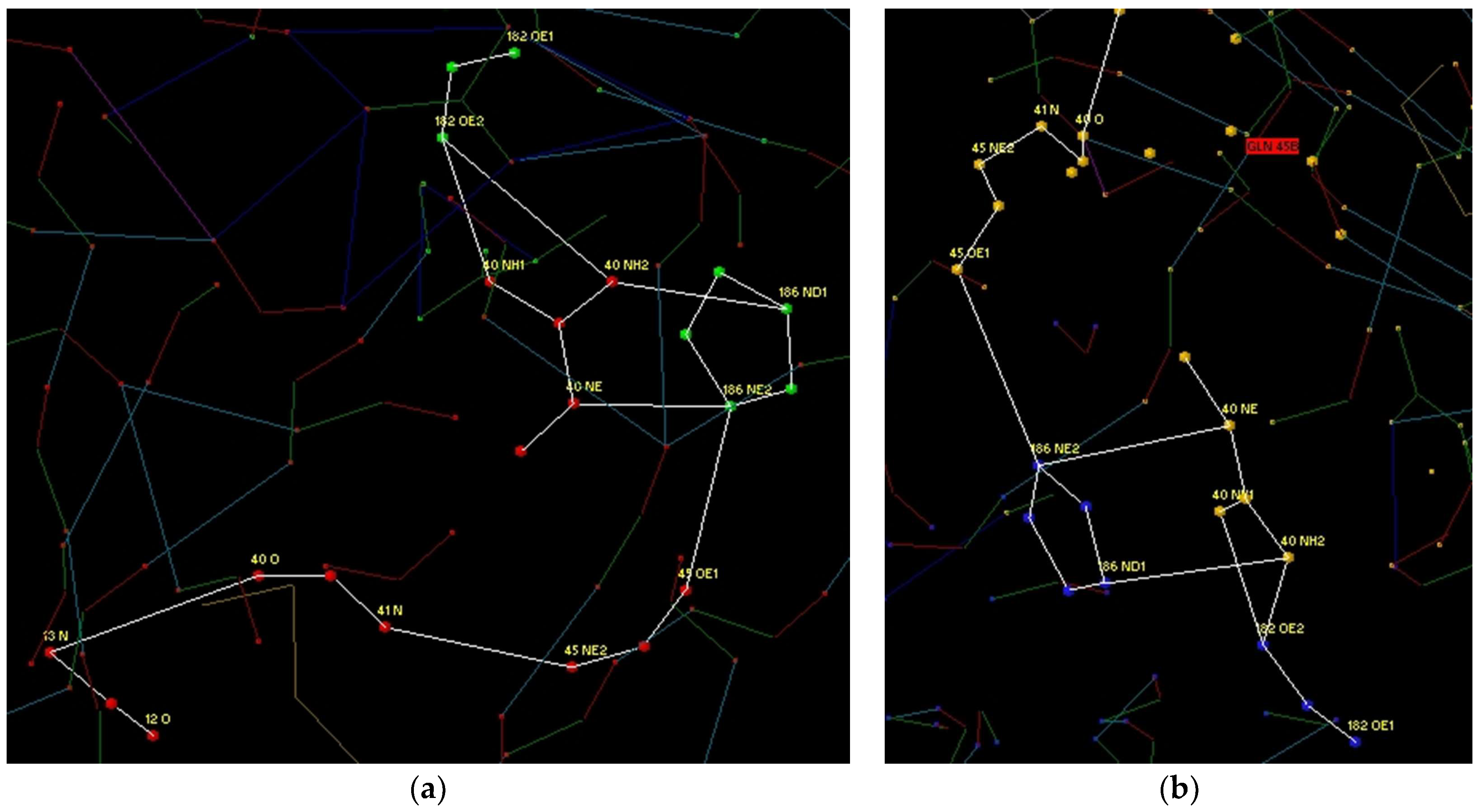
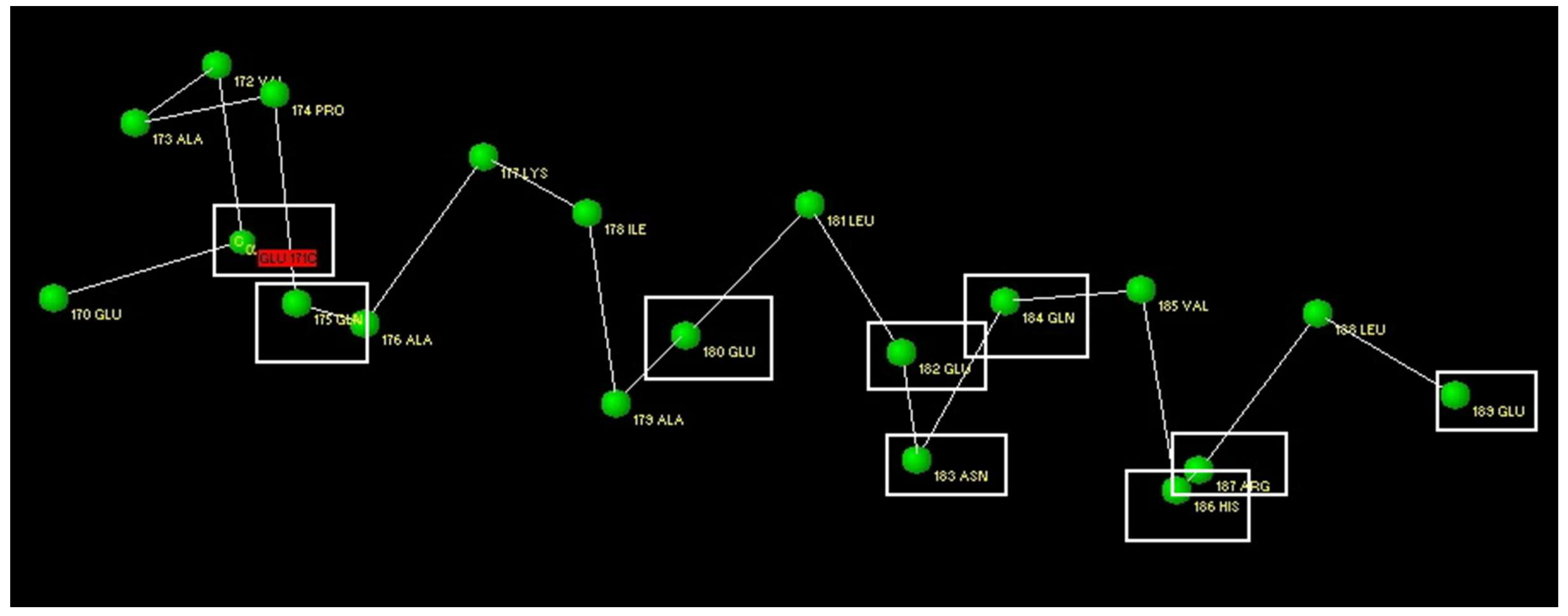
3.1. Modelling of Peptide Ligand for Lactoferrin
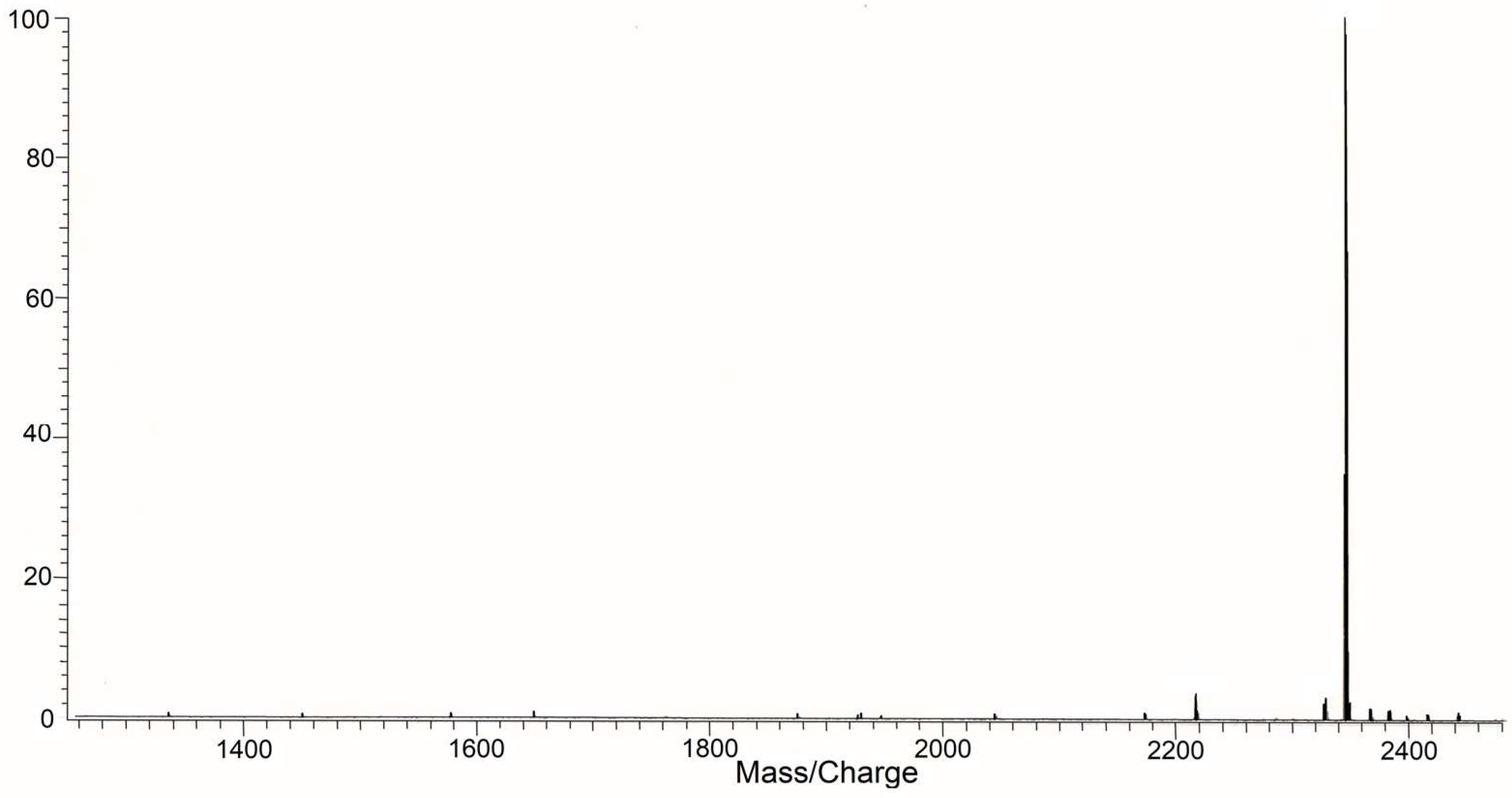
3.2. Study of Peptide LETI-11 and Its Selective Binding of Lactoferrin
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhosale, S.D.; Moulder, R.; Venäläinen, M.S.; Koskinen, J.S.; Pitkänen, N.; Juonala, M.T.; Kähönen, M.A.P.; Lehtimäki, T.J.; Viikari, J.S.A.; Elo, L.L.; et al. Serum Proteomic Profiling to Identify Biomarkers of Premature Carotid Atherosclerosis. Sci. Rep. 2018, 8, 9209. [Google Scholar] [CrossRef] [PubMed]
- Emilsson, V.; Gudmundsdottir, V.; Gudjonsson, A.; Jonmundsson, T.; Jonsson, B.G.; Karim, M.A.; Ilkov, M.; Staley, J.R.; Gudmundsson, E.F.; Launer, L.J.; et al. Coding and Regulatory Variants Are Associated with Serum Protein Levels and Disease. Nat. Commun. 2022, 13, 481. [Google Scholar] [CrossRef] [PubMed]
- WHO. Noncommunicable Diseases. Key Facts. Available online: https://www.who.int/news-room/fact-sheets/detail/noncommunicable-diseases (accessed on 30 November 2022).
- Sitkov, N.O.; Zimina, T.M.; Soloviev, A.V.; Lemozerskii, V.E.; Karasev, V.A. Development of Peptide Aptamers—3d Complementary Ligands for Selective Binding of Target Biomarkers of Diseases in Multiparametric Sensor Systems. In Proceedings of the 2019 IEEE Conference of Russian Young Researchers in Electrical and Electronic Engineering (EIConRus), St. Petersburg, Russia, 28–30 January 2019. [Google Scholar] [CrossRef]
- Molvin, J.; Pareek, M.; Jujic, A.; Melander, O.; Råstam, L.; Lindblad, U.; Daka, B.; Leósdóttir, M.; Nilsson, P.M.; Olsen, M.H.; et al. Using a Targeted Proteomics Chip to Explore Pathophysiological Pathways for Incident Diabetes—The Malmö Preventive Project. Sci. Rep. 2019, 9, 272. [Google Scholar] [CrossRef] [PubMed]
- Sitkov, N.; Zimina, T.; Kolobov, A.; Karasev, V.; Romanov, A.; Luchinin, V.; Kaplun, D. Toward Development of a Label-Free Detection Technique for Microfluidic Fluorometric Peptide-Based Biosensor Systems. Micromachines 2021, 12, 691. [Google Scholar] [CrossRef]
- Neeli, I.; Radic, M. Current Challenges and Limitations in Antibody-Based Detection of Citrullinated Histones. Front. Immunol. 2016, 7, 528. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lai, B.; Juhas, M. Recent Advances in Aptamer Discovery and Applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Fathil, M.F.M.; Md Arshad, M.K.; Gopinath, S.C.B.; Hashim, U.; Adzhri, R.; Ayub, R.M.; Ruslinda, A.R.; Nuzaihan, M.N.M.; Azman, A.H.; Zaki, M.; et al. Diagnostics on Acute Myocardial Infarction: Cardiac Troponin Biomarkers. Biosens. Bioelectron. 2015, 70, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on Aptamer Research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef]
- Li, J.; Tan, S.; Chen, X.; Zhang, C.-Y.; Zhang, Y. Peptide Aptamers with Biological and Therapeutic Applications. Curr. Med. Chem. 2011, 18, 4215–4222. [Google Scholar] [CrossRef]
- Gold, L. SELEX: How It Happened and Where It Will Go. J. Mol. Evol. 2015, 81, 140–143. [Google Scholar] [CrossRef]
- Dausse, E.; Barré, A.; Aimé, A.; Groppi, A.; Rico, A.; Ainali, C.; Salgado, G.; Palau, W.; Daguerre, E.; Nikolski, M.; et al. Aptamer Selection by Direct Microfluidic Recovery and Surface Plasmon Resonance Evaluation. Biosens. Bioelectron. 2016, 80, 418–425. [Google Scholar] [CrossRef]
- Morris, G.M.; Lim-Wilby, M. Molecular Docking. Mol. Modeling Proteins 2008, 443, 365–382. [Google Scholar] [CrossRef]
- Lensink, M.F.; Velankar, S.; Wodak, S.J. Modeling Protein-Protein and Protein-Peptide Complexes: CAPRI 6th Edition. Proteins Struct. Funct. Bioinform. 2016, 85, 359–377. [Google Scholar] [CrossRef]
- Agrawal, P.; Singh, H.; Srivastava, H.K.; Singh, S.; Kishore, G.; Raghava, G.P.S. Benchmarking of Different Molecular Docking Methods for Protein-Peptide Docking. BMC Bioinform. 2019, 19, 105–124. [Google Scholar] [CrossRef]
- Rhinehardt, K.L.; Mohan, R.V.; Srinivas, G. Computational Modeling of Peptide–Aptamer Binding. Comput. Pept. 2014, 313–333. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Senn, H.M.; Thiel, W. QM/MM Methods for Biomolecular Systems. Angew. Chem. Int. Ed. 2009, 48, 1198–1229. [Google Scholar] [CrossRef] [PubMed]
- Karasev, V. A Model of Molecular Vector Machine of Proteins. Biosystems 2019, 180, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Visualizer of Supramolecular Biostructures «Protein 3d». Available online: http://protein-3d.ru/index_e.html (accessed on 30 November 2022).
- Karasev, V.A.; Stefanov, V.E. Topological Nature of the Genetic Code. J. Theor. Biol. 2001, 209, 303–317. [Google Scholar] [CrossRef]
- Pokrovskii, V.N. The Mesoscopic Theory of Polymer Dynamics; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar] [CrossRef]
- Karasev, V.A.; Luchinin, V.V.; Stefanov, V.E. Topological Coding: Towards New Materials for Molecular Electronics. Adv. Funct. Mater. 2002, 12, 461–469. [Google Scholar] [CrossRef]
- Sitkov, N.O.; Zimina, T.M.; Karasev, V.A.; Lemozerskii, V.E.; Kolobov, A.A. Design of Peptide Ligands (Aptamers) for Determination of Myeloperoxidase Level in Blood Using Biochips. In Proceedings of the 2020 IEEE Conference of Russian Young Researchers in Electrical and Electronic Engineering (EIConRus), St. Petersburg, Russia, 27–30 January 2020. [Google Scholar] [CrossRef]
- RCSB Protein Data. Available online: https://www.rcsb.org/ (accessed on 30 November 2022).
- Misra, S.; Kumar, A.; Kumar, P.; Yadav, A.K.; Mohania, D.; Pandit, A.K.; Prasad, K.; Vibha, D. Blood-Based Protein Biomarkers for Stroke Differentiation: A Systematic Review. PROTEOMICS—Clin. Appl. 2017, 11, 1700007. [Google Scholar] [CrossRef] [PubMed]
- Barro, C.; Zetterberg, H. The Blood Biomarkers Puzzle—A Review of Protein Biomarkers in Neurodegenerative Diseases. J. Neurosci. Methods 2021, 361, 109281. [Google Scholar] [CrossRef]
- Ananthan, K.; Lyon, A.R. The Role of Biomarkers in Cardio-Oncology. J. Cardiovasc. Transl. Res. 2020, 13, 431–450. [Google Scholar] [CrossRef]
- Sokolov, A.V.; Zakahrova, E.T.; Kostevich, V.A.; Samygina, V.R.; Vasilyev, V.B. Lactoferrin, Myeloperoxidase, and Ceruloplasmin: Complementary Gearwheels Cranking Physiological and Pathological Processes. BioMetals 2014, 27, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Lepanto, M.S.; Rosa, L.; Paesano, R.; Valenti, P.; Cutone, A. Lactoferrin in Aseptic and Septic Inflammation. Molecules 2019, 24, 1323. [Google Scholar] [CrossRef]
- Kruzel, M.L.; Zimecki, M.; Actor, J.K. Lactoferrin in a Context of Inflammation-Induced Pathology. Front. Immunol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Sokolov, A.V.; Dubrovskaya, N.M.; Kostevich, V.A.; Vasilev, D.S.; Voynova, I.V.; Zakharova, E.T.; Runova, O.L.; Semak, I.V.; Budevich, A.I.; Nalivaeva, N.N.; et al. Lactoferrin Induces Erythropoietin Synthesis and Rescues Cognitive Functions in the Offspring of Rats Subjected to Prenatal Hypoxia. Nutrients 2022, 14, 1399. [Google Scholar] [CrossRef] [PubMed]
- Skarżyńska, E.; Żytyńska-Daniluk, J.; Lisowska-Myjak, B. Correlations between Ceruloplasmin, Lactoferrin and Myeloperoxidase in Meconium. J. Trace Elem. Med. Biol. 2017, 43, 58–62. [Google Scholar] [CrossRef]
- Grudinin, N.V.; Karev, V.E.; Bunenkov, N.S.; Komok, V.V.; Kostevitch, V.V.; Gorbunov, N.P.; Sokolov, A.V.; Karpov, A.A.; Lepik, K.V.; Shvetsov, A.N.; et al. Histological features and blood plasma changes after heterotopic heart transplantation. Cell. Ther. Transplant. 2018, 7, 59–60. [Google Scholar]
- Zhang, Y.; Lu, C.; Zhang, J. Lactoferrin and Its Detection Methods: A Review. Nutrients 2021, 13, 2492. [Google Scholar] [CrossRef] [PubMed]
- Monnard, C.; Vernet, M. Détermination de la lactoferrine plasmatique par une méthode immunoenzymatique “double sandwich” en phase hétérogène [Determination of plasma lactoferrin by the “double sandwich” immunoenzymatic method in heterogeneous phase]. Pathol. -Biol. 1988, 36, 941–944. [Google Scholar] [PubMed]
- Mantel, C.; Miyazawa, K.; Broxmeyer, H.E. Physical Characteristics and Polymerization During Iron Saturation of Lactoferrin, A Myelopoietic Regulatory Molecule With Suppressor Activity. Adv. Exp. Med. Biol. 1994, 121–132. [Google Scholar] [CrossRef]
- Bagby, G.J.; Bennett, R. Feedback Regulation of Granulopoiesis: Polymerization of Lactoferrin Abrogates Its Ability to Inhibit CSA Production. Blood 1982, 60, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Sitkov, N.; Zimina, T.; Kolobov, A.; Sevostyanov, E.; Trushlyakova, V.; Luchinin, V.; Krasichkov, A.; Markelov, O.; Galagudza, M.; Kaplun, D. Study of the Fabrication Technology of Hybrid Microfluidic Biochips for Label-Free Detection of Proteins. Micromachines 2021, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, D.M. Thirteen Decades of Peptide Synthesis: Key Developments in Solid Phase Peptide Synthesis and Amide Bond Formation Utilized in Peptide Ligation. Amino Acids 2017, 50, 39–68. [Google Scholar] [CrossRef] [PubMed]
- Zaia, J.; Annan, R.S.; Biemann, K. The Correct Molecular Weight of Myoglobin, a Common Calibrant for Mass Spectrometry. Rapid Commun. Mass Spectrom. 1992, 6, 32–36. [Google Scholar] [CrossRef]
- Myoglobin from Equine Heart. Available online: https://www.sigmaaldrich.com/RU/en/product/sigma/m9267 (accessed on 30 November 2022).
- Davies, M.J. Myeloperoxidase-Derived Oxidation: Mechanisms of Biological Damage and Its Prevention. J. Clin. Biochem. Nutr. 2010, 48, 8–19. [Google Scholar] [CrossRef]
- Malle, E.; Furtmüller, P.G.; Sattler, W.; Obinger, C. Myeloperoxidase: A Target for New Drug Development? Br. J. Pharmacol. 2007, 152, 838–854. [Google Scholar] [CrossRef]
- Mathy-Hartert, M.; Bourgeois, E.; Grülke, S.; Deby-Dupont, G.; Caudron, I.; Deby, C.; Lamy, M.; Serteyn, D. Purification of myeloperoxidase from equine polymorphonuclear leucocytes. Can. J. Vet. Res. 1998, 62, 127. [Google Scholar] [PubMed]
- Li, J.; Ding, X.; Chen, Y.; Song, B.; Zhao, S.; Wang, Z. Determination of Bovine Lactoferrin in Infant Formula by Capillary Electrophoresis with Ultraviolet Detection. J. Chromatogr. A 2012, 1244, 178–183. [Google Scholar] [CrossRef]
- Shimazaki, K.-I.; Kawaguchi, A.; Sato, T.; Ueda, Y.; Tomimura, T.; Shimamura, S. Analysis of Human and Bovine Milk Lactoferrins by Rotofor and Chromatofocusing. Int. J. Biochem. 1993, 25, 1653–1658. [Google Scholar] [CrossRef]
- Senkovich, O.; Cook, W.J.; Mirza, S.; Hollingshead, S.K.; Protasevich, I.I.; Briles, D.E.; Chattopadhyay, D. Structure of a Complex of Human Lactoferrin N-Lobe with Pneumococcal Surface Protein A Provides Insight into Microbial Defense Mechanism. J. Mol. Biol. 2007, 370, 701–713. [Google Scholar] [CrossRef]
- Isoelectric Point Calculator. Available online: https://www.bachem.com/knowledge-center/peptide-calculator/ (accessed on 30 November 2022).
- Andreeva, A.; Budenkova, E.; Babich, O.; Sukhikh, S.; Ulrikh, E.; Ivanova, S.; Prosekov, A.; Dolganyuk, V. Production, Purification, and Study of the Amino Acid Composition of Microalgae Proteins. Molecules 2021, 26, 2767. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.C.; von Hippel, P.H. Calculation of Protein Extinction Coefficients from Amino Acid Sequence Data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.A. Interpretation of α-Synuclein UV Absorption Spectra in the Peptide Bond and the Aromatic Regions. J. Photochem. Photobiol. B Biol. 2020, 212, 112022. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-L.; Baker, H.M.; Shewry, S.C.; Jameson, G.B.; Baker, E.N. Structure of Recombinant Human Lactoferrin Expressed in Aspergillus Awamori. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Schindler, C.E.M.; de Vries, S.J.; Zacharias, M. Fully Blind Peptide-Protein Docking with PepATTRACT. Structure 2015, 23, 1507–1515. [Google Scholar] [CrossRef]
- De Vries, S.J.; Rey, J.; Schindler, C.E.M.; Zacharias, M.; Tuffery, P. The PepATTRACT Web Server for Blind, Large-Scale Peptide–Protein Docking. Nucleic Acids Res. 2017, 45, W361–W364. [Google Scholar] [CrossRef]

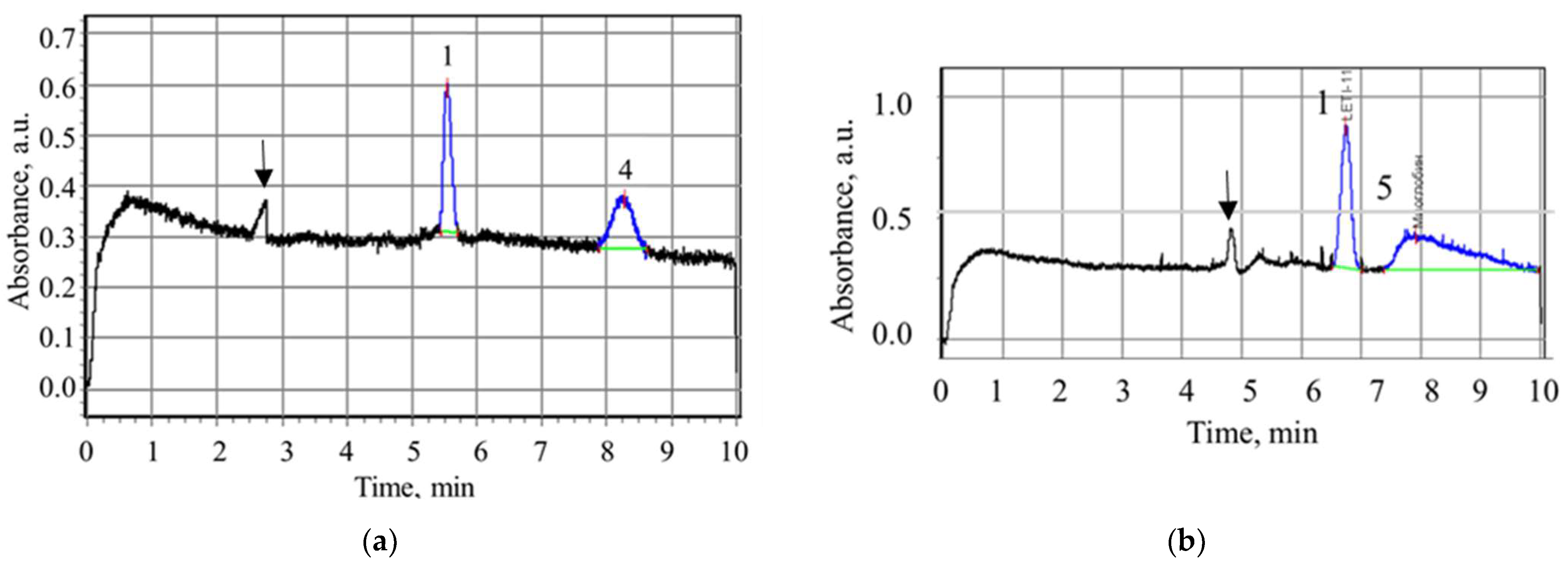
| No. | Sample | MW, Da | pI | Reference |
|---|---|---|---|---|
| 1 | Myoglobin from equine heart (MB); (Sigma-Aldrich, St. Louis, MO, USA) | 117,199 | 6.8 | [42,43] |
| 2 | Myeloperoxidase from human leucocytes in monomeric form (homoMPO); (IEM RAS, St. Petersburg, Russia) | 75,000 | 5.62 | [44,45,46] |
| 3 | Lactoferrin from human milk (LF) (IEM RAS, St. Petersburg) | 80,000 | 8–9 | [47,48,49] |
| 4 | Peptide (LETI-11): EEVAQAQAKIAELENQVHRLE (SPB ETU—IOPHS, St. Petersburg) | 2345 | 5.6 | [50] |
| 170 176 180 184 189 |
| GLU GLU VAL ALA PRO GLN ALA LYS ILE ALA GLU LEU GLU ASN GLN VAL HIS ARG LEU GLU |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zimina, T.; Sitkov, N.; Karasev, V.; Skorik, Y.; Kolobov, A.; Kolobov, A.; Bunenkov, N.; Luchinin, V. Design of Peptide Ligand for Lactoferrin and Study of Its Binding Specificity. Chemosensors 2023, 11, 162. https://doi.org/10.3390/chemosensors11030162
Zimina T, Sitkov N, Karasev V, Skorik Y, Kolobov A, Kolobov A, Bunenkov N, Luchinin V. Design of Peptide Ligand for Lactoferrin and Study of Its Binding Specificity. Chemosensors. 2023; 11(3):162. https://doi.org/10.3390/chemosensors11030162
Chicago/Turabian StyleZimina, Tatiana, Nikita Sitkov, Vladimir Karasev, Yury Skorik, Alexey Kolobov, Alexander Kolobov, Nikolay Bunenkov, and Viktor Luchinin. 2023. "Design of Peptide Ligand for Lactoferrin and Study of Its Binding Specificity" Chemosensors 11, no. 3: 162. https://doi.org/10.3390/chemosensors11030162
APA StyleZimina, T., Sitkov, N., Karasev, V., Skorik, Y., Kolobov, A., Kolobov, A., Bunenkov, N., & Luchinin, V. (2023). Design of Peptide Ligand for Lactoferrin and Study of Its Binding Specificity. Chemosensors, 11(3), 162. https://doi.org/10.3390/chemosensors11030162








