Extracting Information and Enhancing the Quality of Separation Data: A Review on Chemometrics-Assisted Analysis of Volatile, Soluble and Colloidal Samples
Abstract
1. Introduction
2. Overview of the Main Chemometric Techniques and Their Advances
2.1. Principal Component Analysis (PCA)
2.2. Clusters Analysis (CA)
2.3. Design of Experiments (DoE)
2.4. Linear Discriminant Analysis (LDA)
2.5. Partial Least Square (PLS)
3. Gas Chromatography (GC) and Chemometrics
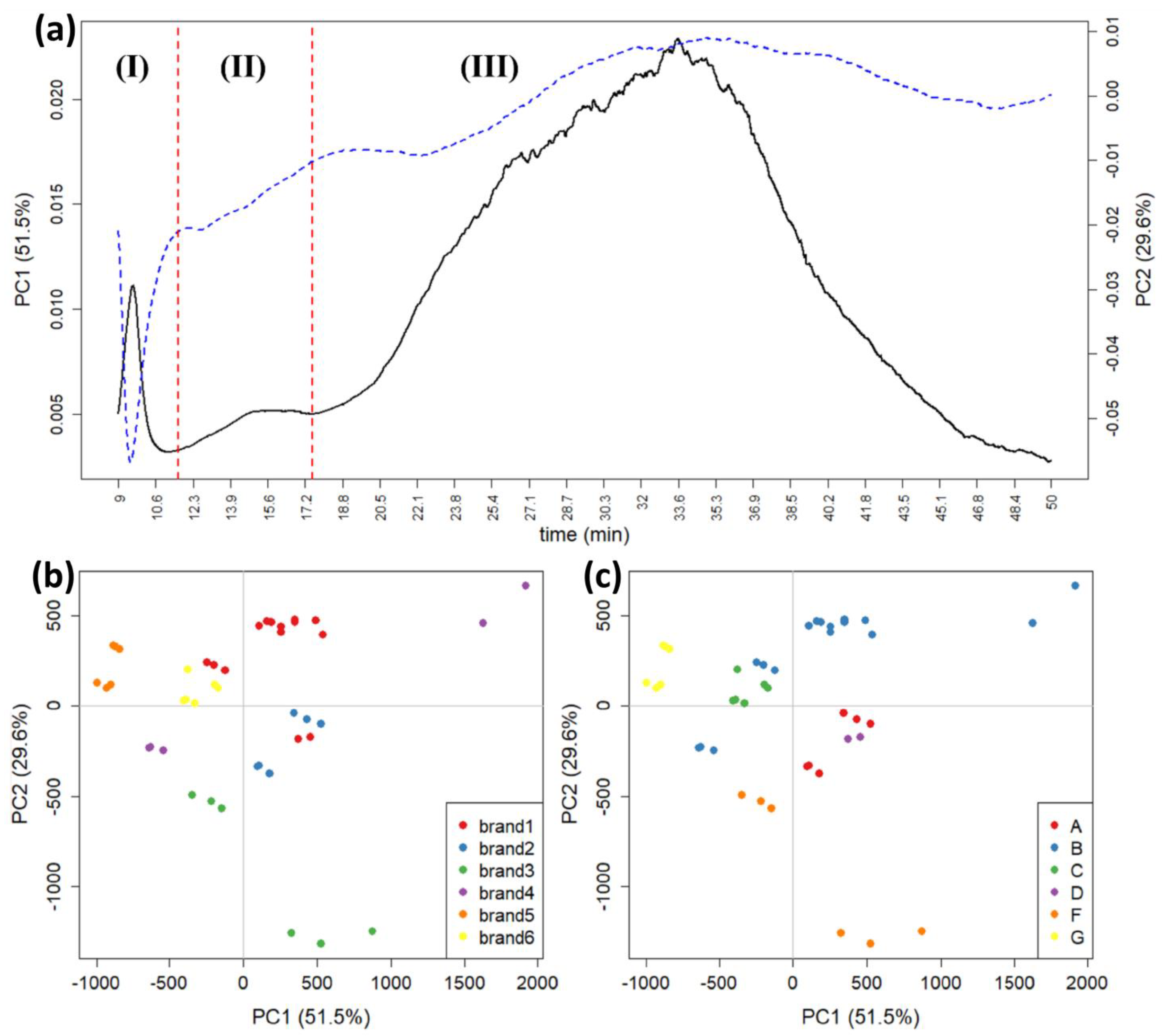
4. High-Performance Liquid Chromatography (HPLC) and Chemometrics
5. Colloidal Analysis and Chemometrics
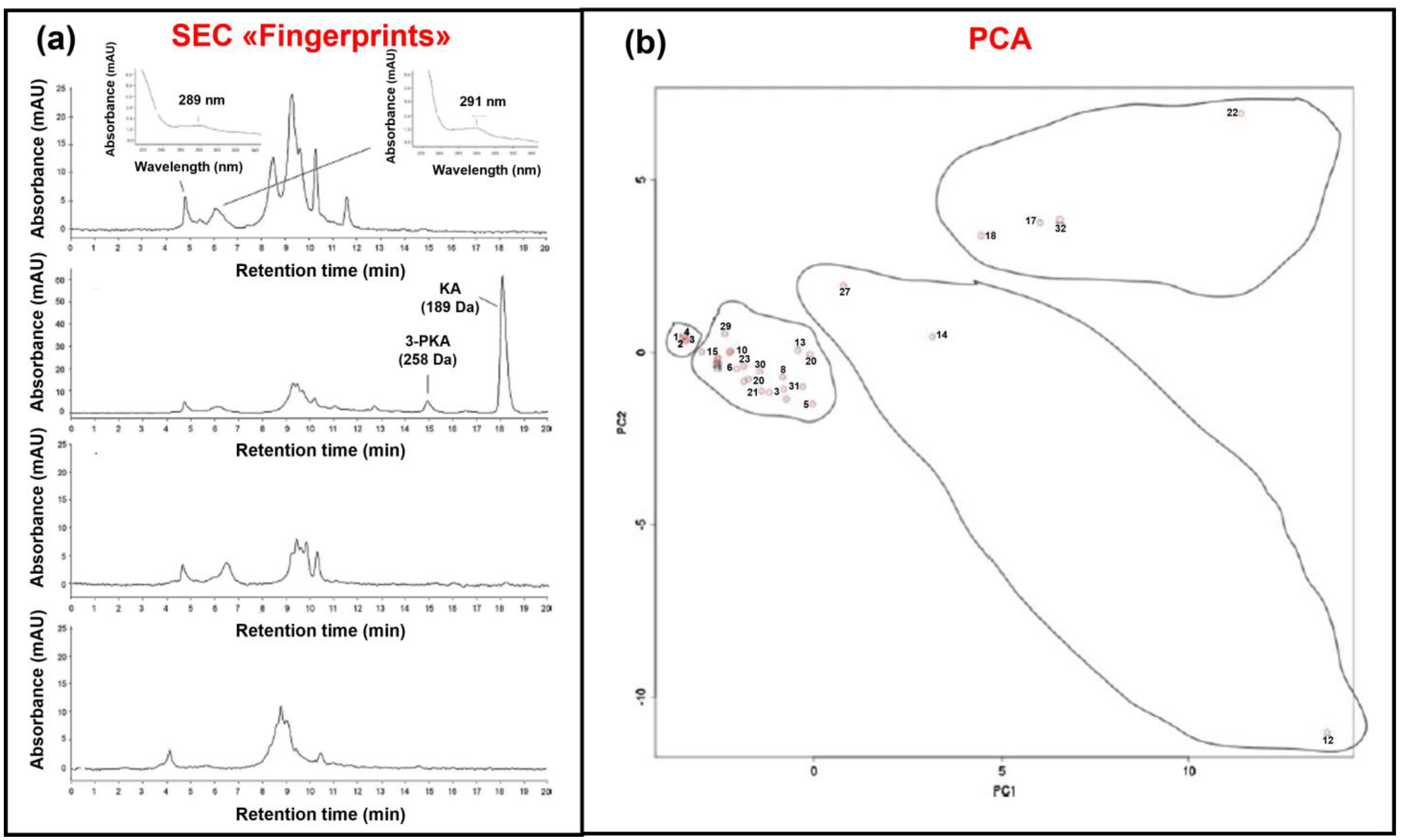
5.1. SEC
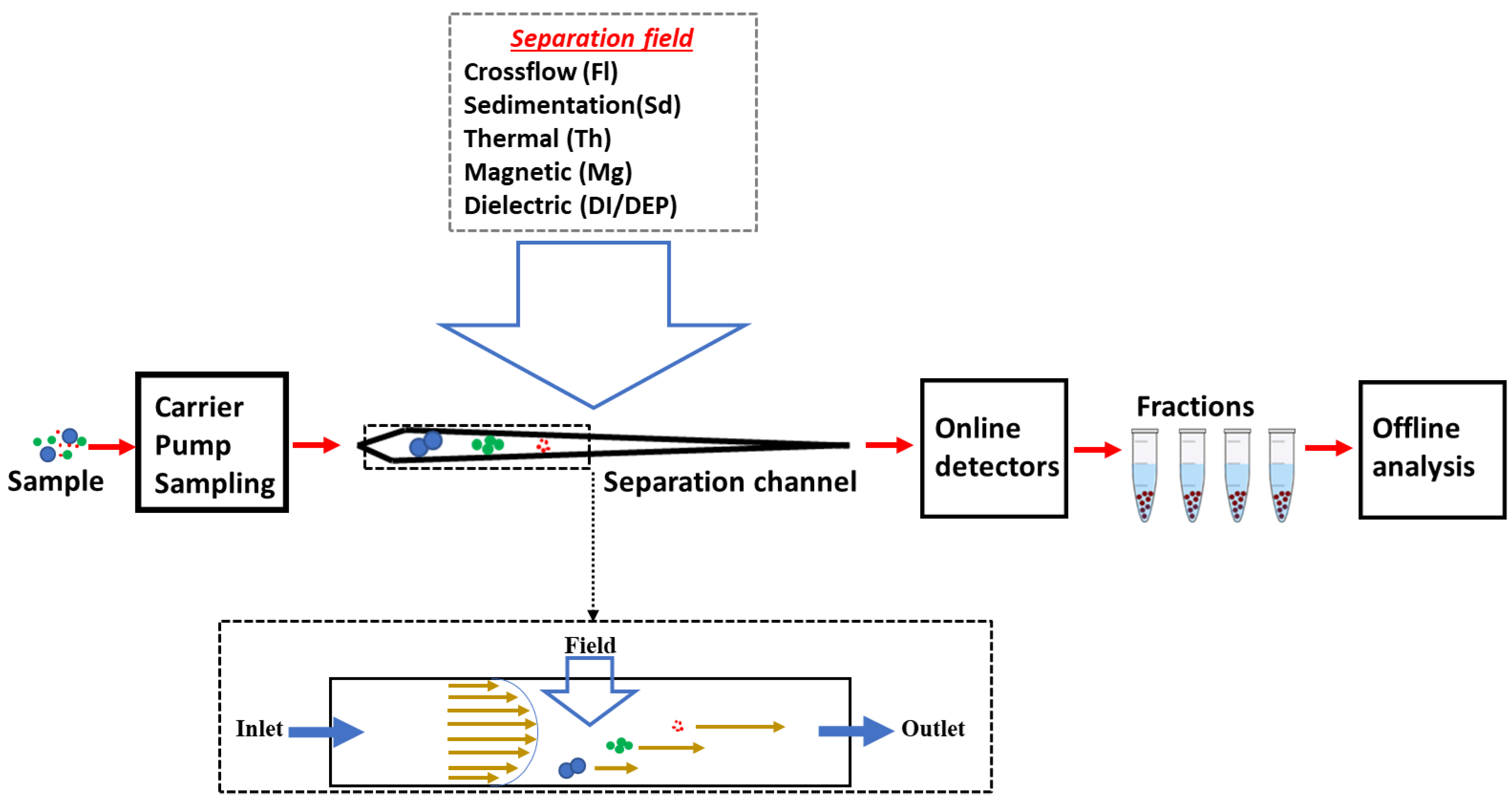
5.2. FFF
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef]
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef] [PubMed]
- Melucci, D.; Bendini, A.; Tesini, F.; Barbieri, S.; Zappi, A.; Vichi, S.; Conte, L.; Gallina Toschi, T. Rapid direct analysis to discriminate geographic origin of extra virgin olive oils by flash gas chromatography electronic nose and chemometrics. Food Chem. 2016, 204, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Marassi, V.; Roda, B.; Casolari, S.; Ortelli, S.; Blosi, M.; Zattoni, A.; Costa, A.L.; Reschiglian, P. Hollow-fiber flow field-flow fractionation and multi-angle light scattering as a new analytical solution for quality control in pharmaceutical nanotechnology. Microchem. J. 2018, 136, 149–156. [Google Scholar] [CrossRef]
- Marassi, V.; Macis, M.; Giordani, S.; Ferrazzano, L.; Tolomelli, A.; Roda, B.; Zattoni, A.; Ricci, A.; Reschiglian, P.; Cabri, W. Application of Af4-Multidetection to Liraglutide in Its Formulation: Preserving and Representing Native Aggregation. Molecules 2022, 27, 5485. [Google Scholar] [CrossRef] [PubMed]
- Reschiglian, P.; Melucci, D.; Zattoni, A.; Torsi, G. A new, low-cost separation technique for the characterization of particulate matter of environmental relevance: The Gravitational Field-Flow Fractionation (GrFFF). Ann. Chim. 1997, 87, 677–686. [Google Scholar]
- Zia, S.; Roda, B.; Maggio, A.; Marrazzo, P.; Pizzuti, V.; Alviano, F.; Bonsi, L.; Marassi, V.; Zattoni, A.; Reschiglian, P. Celector®: An Innovative Technology for Quality Control of Living Cells. Appl. Sci. 2022, 12, 9967. [Google Scholar] [CrossRef]
- Zattoni, A.; Rambaldi, D.C.; Reschiglian, P.; Melucci, M.; Krol, S.; Garcia, A.M.C.; Sanz-Medel, A.; Roessner, D.; Johann, C. Asymmetrical flow field-flow fractionation with multi-angle light scattering detection for the analysis of structured nanoparticles. J. Chromatogr. A 2009, 1216, 9106–9112. [Google Scholar] [CrossRef] [PubMed]
- Bro, R.; Smilde, A.K. Principal component analysis. Analytical Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Christophe, B.Y.C. PCA: The Basic Building Block of Chemometrics. In Analytical chemistry; Ira, S.K., Ed.; IntechOpen: Rijeka, Croatia, 2012; Chapter 1. [Google Scholar]
- Leardi, R. Experimental design in chemistry: A tutorial. Anal. Chim. Acta 2009, 652, 161–172. [Google Scholar] [CrossRef]
- Cela, R.; Claeys-Bruno, M.; Phan-Tan-Luu, R. Screening Strategies; Elsevier: Amsterdam, The Netherlands, 2010; Volume 1, pp. 251–300. [Google Scholar]
- Hibbert, D.B. Experimental design in chromatography: A tutorial review. J. Chromatogr. B 2012, 910, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Murty, M.N.; Flynn, P.J. Data clustering: A review. ACM Comput. Surv. 1999, 31, 264–323. [Google Scholar] [CrossRef]
- Tharwat, A.; Gaber, T.; Ibrahim, A.; Hassanien, A.E. Linear discriminant analysis: A detailed tutorial. AI Commun. 2017, 30, 169–190. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Smith, R.; Ventura, D.; Prince, J.T. LC-MS alignment in theory and practice: A comprehensive algorithmic review. Brief. Bioinform. 2015, 16, 104–117. [Google Scholar] [CrossRef]
- Matthiesen, R. Methods, algorithms and tools in computational proteomics: A practical point of view. Proteomics 2007, 7, 2815–2832. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Wiley Online Library: Hoboken, NJ, USA, 2002. [Google Scholar]
- Kumar, K. Principal component analysis: Most favourite tool in chemometrics. Resonance 2017, 22, 747–759. [Google Scholar] [CrossRef]
- Todeschini, R.; Ballabio, D.; Consonni, V.; Grisoni, F. A new concept of higher-order similarity and the role of distance/similarity measures in local classification methods. Chemom. Intell. Lab. Syst. 2016, 157, 50–57. [Google Scholar] [CrossRef]
- Nguyen, T.T.D.; Nguyen, L.T.T.; Nguyen, A.; Yun, U.; Vo, B. A method for efficient clustering of spatial data in network space. J. Intell. Fuzzy Syst. 2021, 40, 11653–11670. [Google Scholar] [CrossRef]
- Fisher, R.A. The design of experiments. Br. Med. J. 1936, 1, 554. [Google Scholar] [CrossRef]
- Biancolillo, A.; Næs, T. The Sequential and Orthogonalized PLS Regression for Multiblock Regression: Theory, Examples, and Extensions. Data Handl. Sci. Technol. 2019, 31, 157–177. [Google Scholar] [CrossRef]
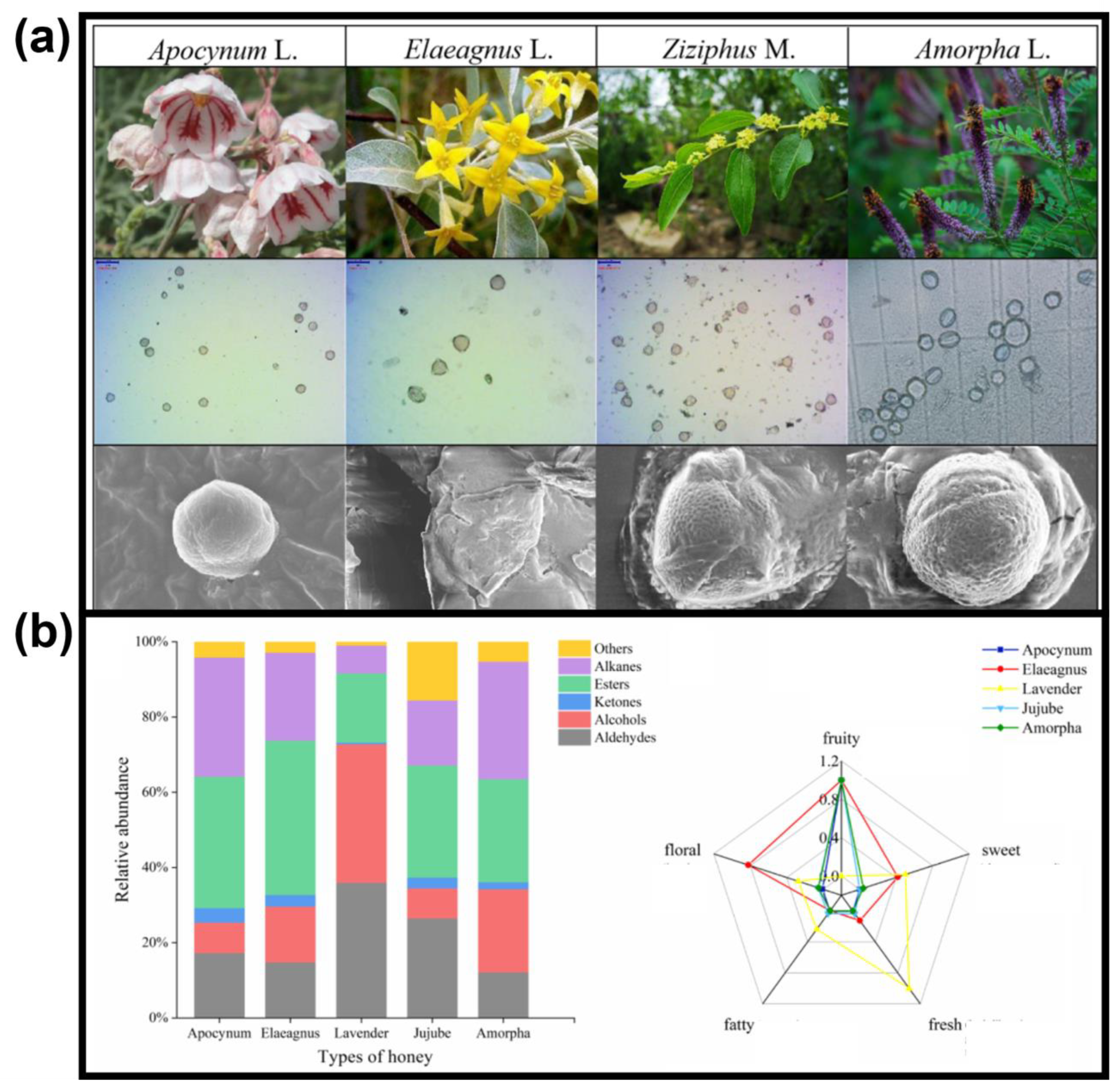
- Zappi, A.; Melucci, D.; Scaramagli, S.; Zelano, A.; Marcazzan, G.L. Botanical traceability of unifloral honeys by chemometrics based on head-space gas chromatography. Eur. Food Res. Technol. 2018, 244, 2149–2157. [Google Scholar] [CrossRef]
- Forleo, T.; Zappi, A.; Gottardi, F.; Melucci, D. Rapid discrimination of Italian Prosecco wines by head-space gas-chromatography basing on the volatile profile as a chemometric fingerprint. Eur. Food Res. Technol. 2020, 246, 1805–1816. [Google Scholar] [CrossRef]
- Morozzi, P.; Zappi, A.; Gottardi, F.; Locatelli, M.; Melucci, D. A quick and efficient non-targeted screening test for saffron authentication: Application of chemometrics to gas-chromatographic data. Molecules 2019, 24, 2602. [Google Scholar] [CrossRef] [PubMed]
- Zappi, A.; Marassi, V.; Kassouf, N.; Giordani, S.; Pasqualucci, G.; Garbini, D.; Roda, B.; Zattoni, A.; Reschiglian, P.; Melucci, D. A Green Analytical Method Combined with Chemometrics for Traceability of Tomato Sauce Based on Colloidal and Volatile Fingerprinting. Molecules 2022, 27, 5507. [Google Scholar] [CrossRef] [PubMed]
- Abdelwareth, A.; Zayed, A.; Farag, M.A. Chemometrics-based aroma profiling for revealing origin, roasting indices, and brewing method in coffee seeds and its commercial blends in the Middle East. Food Chem. 2021, 349, 129162. [Google Scholar] [CrossRef]
- Zakidou, P.; Plati, F.; Matsakidou, A.; Varka, E.M.; Blekas, G.; Paraskevopoulou, A. Single Origin Coffee Aroma: From Optimized Flavor Protocols and Coffee Customization to Instrumental Volatile Characterization and Chemometrics. Molecules 2021, 26, 4609. [Google Scholar] [CrossRef]
- Gancarz, M.; Dobrzański, B.; Malaga-Toboła, U.; Tabor, S.; Combrzyński, M.; Ćwikła, D.; Strobel, W.R.; Oniszczuk, A.; Karami, H.; Darvishi, Y.; et al. Impact of Coffee Bean Roasting on the Content of Pyridines Determined by Analysis of Volatile Organic Compounds. Molecules 2022, 27, 1559. [Google Scholar] [CrossRef]
- Bressanello, D.; Marengo, A.; Cordero, C.; Strocchi, G.; Rubiolo, P.; Pellegrino, G.; Ruosi, M.R.; Bicchi, C.; Liberto, E. Chromatographic Fingerprinting Strategy to Delineate Chemical Patterns Correlated to Coffee Odor and Taste Attributes. J. Agric. Food Chem. 2021, 69, 4550–4560. [Google Scholar] [CrossRef]
- Sotiropoulou, N.S.; Xagoraris, M.; Revelou, P.K.; Kaparakou, E.; Kanakis, C.; Pappas, C.; Tarantilis, P. The Use of SPME-GC-MS IR and Raman Techniques for Botanical and Geographical Authentication and Detection of Adulteration of Honey. Foods 2021, 10, 1671. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Sun, J.; Zhao, H.; Wu, F.; Xue, X.; Wu, L.; Cao, W. Volatile compounds of five types of unifloral honey in Northwest China: Correlation with aroma and floral origin based on HS-SPME/GC–MS combined with chemometrics. Food Chem. 2022, 384, 132461. [Google Scholar] [CrossRef] [PubMed]
- Karabagias, I.K.; Karabagias, V.K.; Nayik, G.A.; Gatzias, I.; Badeka, A.V. A targeted chemometric evaluation of the volatile compounds of Quercus ilex honey in relation to its provenance. LWT 2022, 154, 112588. [Google Scholar] [CrossRef]
- Duru, M.E.; Taş, M.; Çayan, F.; Küçükaydın, S.; Tel-Çayan, G. Characterization of volatile compounds of Turkish pine honeys from different regions and classification with chemometric studies. Eur. Food Res. Technol. 2021, 247, 2533–2544. [Google Scholar] [CrossRef]
- Karabagias, I.K. Headspace volatile compounds fluctuations in honeydew honey during storage at in-house conditions. Eur. Food Res. Technol. 2022, 248, 715–726. [Google Scholar] [CrossRef]
- Karabagias, I.K. HS-SPME/GC-MS metabolomic analysis for the identification of exogenous volatile metabolites of monofloral honey and quality control suggestions. Eur. Food Res. Technol. 2022, 248, 1815–1821. [Google Scholar] [CrossRef]
- Passarella, S.; Guerriero, E.; Quici, L.; Ianiri, G.; Cerasa, M.; Notardonato, I.; Protano, C.; Vitali, M.; Russo, M.V.; De Cristofaro, A.; et al. Dataset of PAHs determined in home-made honey samples collected in Central Italy by means of DLLME-GC-MS and cluster analysis for studying the source apportionment. Data Brief 2022, 42, 108136. [Google Scholar] [CrossRef]
- Cecchi, L.; Migliorini, M.; Giambanelli, E.; Cane, A.; Zanoni, B.; Canuti, V.; Mulinacci, N.; Melani, F. Is the volatile compounds profile a suitable tool for authentication of virgin olive oils (Olea europaea L.) according to cultivars? A study by using HS-SPME-GC-MS and chemometrics. Food Control 2022, 139, 109092. [Google Scholar] [CrossRef]
- Lioupi, A.; Sampsonidis, I.; Virgiliou, C.; Papoti, V.T.; Zinoviadou, K.G.; Spyros, A.; Theodoridis, G. Optimisation of the HS-SPME/GC-MS Approach by Design of Experiments Combined with Chemometrics for the Classification of Cretan Virgin Olive Oils. Metabolites 2022, 12, 114. [Google Scholar] [CrossRef]
- Drira, M.; Guclu, G.; Portolés, T.; Jabeur, H.; Kelebek, H.; Selli, S.; Bouaziz, M. Safe and Fast Fingerprint Aroma Detection in Adulterated Extra Virgin Olive Oil Using Gas Chromatography–Olfactometry-Mass Spectrometry Combined with Chemometrics. Food Anal. Methods 2021, 14, 2121–2135. [Google Scholar] [CrossRef]
- Shi, T.; Wu, G.; Jin, Q.; Wang, X. Detection of camellia oil adulteration using chemometrics based on fatty acids GC fingerprints and phytosterols GC-MS fingerprints. Food Chem. 2021, 352, 129422. [Google Scholar] [CrossRef] [PubMed]
- De Flaviis, R.; Mutarutwa, D.; Sacchetti, G.; Mastrocola, D. Could environmental effect overcome genetic? A chemometric study on wheat volatiles fingerprint. Food Chem. 2022, 372, 131236. [Google Scholar] [CrossRef] [PubMed]
- Cervellieri, S.; Lippolis, V.; Mancini, E.; Pascale, M.; Logrieco, A.F.; De Girolamo, A. Mass spectrometry-based electronic nose to authenticate 100% Italian durum wheat pasta and characterization of volatile compounds. Food Chem. 2022, 383, 132548. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.F.C.; Tega, D.U.; Duarte, G.H.B.; Barbosa, L.D.; Ribeiro, H.C.; Castello, A.C.D.; Sawaya, A.C.H.F.; Sussulini, A. Foodomics for agroecology: Differentiation of volatile profile in mint (Mentha × gracilis Sole) from permaculture, organic and conventional agricultural systems using HS-SPME/GC–MS. Food Res. Int. 2022, 155, 111107. [Google Scholar] [CrossRef] [PubMed]
- Ford, P.W.; Berger, T.A.; Jackoway, G. Spice authentication by fully automated chemical analysis with integrated chemometrics. J. Chromatogr. A 2022, 1667, 462889. [Google Scholar] [CrossRef] [PubMed]
- Ghiasi, S.A.; Parastar, H. Chemometrics-assisted isotope ratio fingerprinting based on gas chromatography/combustion/isotope ratio mass spectrometry for saffron authentication. J. Chromatogr. A 2021, 1657, 462587. [Google Scholar] [CrossRef]
- Di Donato, F.; D’Archivio, A.A.; Maggi, M.A.; Rossi, L. Detection of Plant-Derived Adulterants in Saffron (Crocus sativus L.) by HS-SPME/GC-MS Profiling of Volatiles and Chemometrics. Food Anal. Methods 2021, 14, 784–796. [Google Scholar] [CrossRef]
- Farag, M.A.; Khaled, S.E.; El Gingeehy, Z.; Shamma, S.N.; Zayed, A. Comparative Metabolite Profiling and Fingerprinting of Medicinal Cinnamon Bark and Its Commercial Preparations via a Multiplex Approach of GC-MS, UV, and NMR Techniques. Metabolites 2022, 12, 614. [Google Scholar] [CrossRef]
- Farag, M.A.; Kabbash, E.M.; Mediani, A.; Döll, S.; Esatbeyoglu, T.; Afifi, S.M. Comparative Metabolite Fingerprinting of Four Different Cinnamon Species Analyzed via UPLC-MS and GC-MS and Chemometric Tools. Molecules 2022, 27, 2935. [Google Scholar] [CrossRef]
- Salem, M.A.; Zayed, A.; Alseekh, S.; Fernie, A.R.; Giavalisco, P. The integration of MS-based metabolomics and multivariate data analysis allows for improved quality assessment of Zingiber officinale Roscoe. Phytochemistry 2021, 190, 112843. [Google Scholar] [CrossRef]
- Yu, D.X.; Guo, S.; Wang, J.M.; Yan, H.; Zhang, Z.Y.; Yang, J.; Duan, J.A. Comparison of Different Drying Methods on the Volatile Components of Ginger ( Zingiber officinale Roscoe) by HS-GC-MS Coupled with Fast GC E-Nose. Foods 2022, 11, 1611. [Google Scholar] [CrossRef] [PubMed]
- Perin, M.; Dallegrave, A.; Suchecki Barnet, L.; Zanchetti Meneghini, L.; de Araújo Gomes, A.; Pizzolato, T.M. Pharmaceuticals, pesticides and metals/metalloids in Lake Guaíba in Southern Brazil: Spatial and temporal evaluation and a chemometrics approach. Sci. Total Environ. 2021, 793, 148561. [Google Scholar] [CrossRef] [PubMed]
- Pourasil, R.S.M.; Cristale, J.; Lacorte, S.; Tauler, R. Non-targeted Gas Chromatography Orbitrap Mass Spectrometry qualitative and quantitative analysis of semi-volatile organic compounds in indoor dust using the Regions of Interest Multivariate Curve Resolution chemometrics procedure. J. Chromatogr. A 2022, 1668, 462907. [Google Scholar] [CrossRef]
- Mazur, D.M.; Detenchuk, E.A.; Sosnova, A.A.; Artaev, V.B.; Lebedev, A.T. GC-HRMS with Complementary Ionization Techniques for Target and Non-target Screening for Chemical Exposure: Expanding the Insights of the Air Pollution Markers in Moscow Snow. Sci. Total Environ. 2021, 761, 144506. [Google Scholar] [CrossRef] [PubMed]
- Press, I.; Press, I. The Proof and Measurement of Association between Two Things. Am. J. Psychol. 2013, 15, 72–101. [Google Scholar]
- Omokpariola, D.O.; Nduka, J.K.; Kelle, H.I.; Mgbemena, N.M.A.; Iduseri, E.O. Chemometrics, health risk assessment and probable sources of soluble total petroleum hydrocarbons in atmospheric rainwater, Rivers State, Nigeria. Sci. Rep. 2022, 12, 11829. [Google Scholar] [CrossRef]
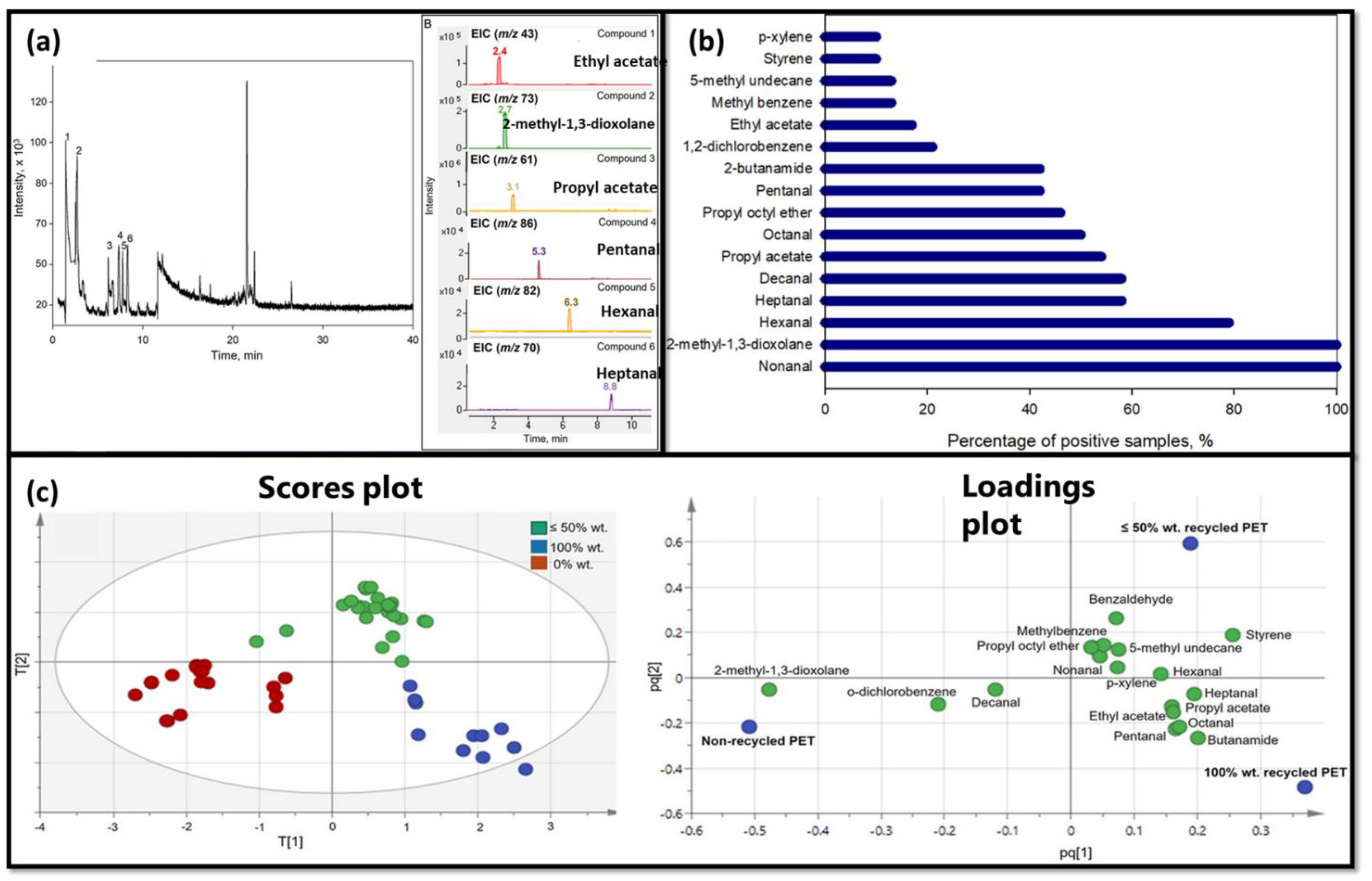
- Peñalver, R.; Marín, C.; Arroyo-Manzanares, N.; Campillo, N.; Viñas, P. Authentication of recycled plastic content in water bottles using volatile fingerprint and chemometrics. Chemosphere 2022, 297, 134156. [Google Scholar] [CrossRef]
- Hermelin, A.; Fabien, L.; Fischer, J.; Saric, N.; Massonnet, G.; Burnier, C. Analysis of condom evidence in forensic science: Background survey of the human vaginal matrix using DRIFTS and pyrolysis-GC/MS. Forensic Sci. Int. 2021, 321, 110724. [Google Scholar] [CrossRef]
- Podolskiy, I.I.; Mochalova, E.S.; Temerdashev, A.Z.; Gashimova, E.M. Application of Statistical Data Analysis Methods to Test the Degradation of Urine Samples for Doping Control Purposes. J. Anal. Chem. 2021, 76, 761–771. [Google Scholar] [CrossRef]
- Grocki, P.; Woollam, M.; Wang, L.; Liu, S.; Kalra, M.; Siegel, A.P.; Li, B.Y.; Yokota, H.; Agarwal, M. Chemometric Analysis of Urinary Volatile Organic Compounds to Monitor the Efficacy of Pitavastatin Treatments on Mammary Tumor Progression over Time. Molecules 2022, 27, 4277. [Google Scholar] [CrossRef]
- Yang, Q.; Shi, B.H.; Tian, G.L.; Niu, Q.Q.; Tang, J.; Linghu, D.D.; He, H.Q.; Wu, B.Q.; Yang, J.T.; Xu, L.; et al. GC–MS urinary metabolomics analysis of inherited metabolic diseases and stable metabolic biomarker screening by a comprehensive chemometric method. Microchem. J. 2021, 168, 106350. [Google Scholar] [CrossRef]
- Aliaño-González, M.J.; Montalvo, G.; García-Ruiz, C.; Ferreiro-González, M.; Palma, M. Assessment of Volatile Compound Transference through Firefighter Turnout Gear. Int. J. Environ. Res. Public Health 2022, 19, 3663. [Google Scholar] [CrossRef] [PubMed]
- Bogdal, C.; Schellenberg, R.; Höpli, O.; Bovens, M.; Lory, M. Recognition of gasoline in fire debris using machine learning: Part I, application of random forest, gradient boosting, support vector machine, and naïve bayes. Forensic Sci. Int. 2022, 331, 111146. [Google Scholar] [CrossRef]
- Bogdal, C.; Schellenberg, R.; Lory, M.; Bovens, M.; Höpli, O. Recognition of gasoline in fire debris using machine learning: Part II, application of a neural network. Forensic Sci. Int. 2022, 332, 111177. [Google Scholar] [CrossRef] [PubMed]
- Bro, R. PARAFAC. Tutorial and applications. Chemom. Intell. Lab. Syst. 1997, 38, 149–171. [Google Scholar] [CrossRef]
- Kruskal, J.B. More factors than subjects, tests and treatments: An indeterminacy theorem for canonical decomposition and individual differences scaling. Psychometrika 1976, 41, 281–293. [Google Scholar] [CrossRef]
- Giebelhaus, R.T.; Sorochan Armstrong, M.D.; de la Mata, A.P.; Harynuk, J.J. Untargeted region of interest selection for gas chromatography—Mass spectrometry data using a pseudo F-ratio moving window. J. Chromatogr. A 2022, 1682, 463499. [Google Scholar] [CrossRef]
- Ochoa, G.S.; Sudol, P.E.; Trinklein, T.J.; Synovec, R.E. Class comparison enabled mass spectrum purification for comprehensive two-dimensional gas chromatography with time-of-flight mass spectrometry. Talanta 2022, 236, 122844. [Google Scholar] [CrossRef]
- Biancolillo, A.; Boqué, R.; Cocchi, M.; Marini, F. Data Fusion Strategies in Food Analysis. Data Handl. Sci. Technol. 2019, 31, 271–310. [Google Scholar] [CrossRef]
- Doeswijk, T.G.; Smilde, A.K.; Hageman, J.A.; Westerhuis, J.A.; van Eeuwijk, F.A. On the increase of predictive performance with high-level data fusion. Anal. Chim. Acta 2011, 705, 41–47. [Google Scholar] [CrossRef]
- Strani, L.; D’alessandro, A.; Ballestrieri, D.; Durante, C.; Cocchi, M. Fast GC E-Nose and Chemometrics for the Rapid Assessment of Basil Aroma. Chemosensors 2022, 10, 105. [Google Scholar] [CrossRef]
- Rivera-Pérez, A.; Romero-González, R.; Garrido Frenich, A. Application of an innovative metabolomics approach to discriminate geographical origin and processing of black pepper by untargeted UHPLC-Q-Orbitrap-HRMS analysis and mid-level data fusion. Food Res. Int. 2021, 150, 110722. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, F.; Biancolillo, A.; Mazzulli, D.; Rossi, L.; D’Archivio, A.A. HS-SPME/GC–MS volatile fraction determination and chemometrics for the discrimination of typical Italian Pecorino cheeses. Microchem. J. 2021, 165, 106133. [Google Scholar] [CrossRef]
- de Jesus Filho, M.; Klein, B.; Wagner, R.; Godoy, H.T. Key aroma compounds of Canastra cheese: HS-SPME optimization assisted by olfactometry and chemometrics. Food Res. Int. 2021, 150, 110788. [Google Scholar] [CrossRef] [PubMed]
- Biancolillo, A.; Aloia, R.; Rossi, L.; D’Archivio, A.A. Organosulfur volatile profiles in Italian red garlic (Allium Sativum L.) varieties investigated by HS-SPME/GC-MS and chemometrics. Food Control 2022, 131, 108477. [Google Scholar] [CrossRef]
- Herrera, J.G.; Ramos, M.P.; de Lima Albuquerque, B.N.; de Oliveira Farias de Aguiar, J.C.R.; Agra Neto, A.C.; Guedes Paiva, P.M.; do Amaral Ferraz Navarro, D.M.; Pinto, L. Multivariate evaluation of process parameters to obtain essential oil of Piper corcovadensis using supercritical fluid extraction. Microchem. J. 2022, 181, 107747. [Google Scholar] [CrossRef]
- Andruszkiewicz, P.J.; Corno, M.; Kuhnert, N. HPLC-MS-based design of experiments approach on cocoa roasting. Food Chem. 2021, 360, 129694. [Google Scholar] [CrossRef]
- Cumeras, R.; Figueras, E.; Davis, C.E.; Baumbach, J.I.; Gràcia, I. Review on Ion Mobility Spectrometry. Part 1: Current instrumentation. Analyst 2015, 140, 1376–1390. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, X.; Zhao, B.; Chen, X.; Zhang, Q. Discriminant analysis of vegetable oils by thermogravimetric-gas chromatography/mass spectrometry combined with data fusion and chemometrics without sample pretreatment. LWT 2022, 161, 113403. [Google Scholar] [CrossRef]
- Chen, T.; Liu, C.; Meng, L.; Lu, D.; Chen, B.; Cheng, Q. Early warning of rice mildew based on gas chromatography-ion mobility spectrometry technology and chemometrics. J. Food Meas. Charact. 2021, 15, 1939–1948. [Google Scholar] [CrossRef]
- Chen, S.; Lu, J.; Qian, M.; He, H.; Li, A.; Zhang, J.; Shen, X.; Gao, J.; Xu, Y. Untargeted headspace-gas chromatography-ion mobility spectrometry in combination with chemometrics for detecting the age of chinese liquor (Baijiu). Foods 2021, 10, 2888. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qian, M.C.; Deng, Y.; Yuan, H.; Jiang, Y. Insight into aroma dynamic changes during the whole manufacturing process of chestnut-like aroma green tea by combining GC-E-Nose, GC-IMS, and GC × GC-TOFMS. Food Chem. 2022, 387, 132813. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Zhang, X.; Wang, Y.; Zheng, M.; Zhao, J.; Gong, H.; Wang, X. Effect of different genotypes on the fruit volatile profiles, flavonoid composition and antioxidant activities of chilli peppers. Food Chem. 2022, 374, 131751. [Google Scholar] [CrossRef] [PubMed]
- Christmann, J.; Rohn, S.; Weller, P. Finding features—Variable extraction strategies for dimensionality reduction and marker compounds identification in GC-IMS data. Food Res. Int. 2022, 161, 111779. [Google Scholar] [CrossRef] [PubMed]
- Christmann, J.; Rohn, S.; Weller, P. gc-ims-tools—A new Python package for chemometric analysis of GC–IMS data. Food Chem. 2022, 394, 133476. [Google Scholar] [CrossRef]
- Roda, B.; Zhang, N.; Gambari, L.; Grigolo, B.; Eller-Vainicher, C.; Gennari, L.; Zappi, A.; Giordani, S.; Marassi, V.; Zattoni, A.; et al. Optimization of a Monobromobimane (MBB) Derivatization and RP-HPLC-FLD Detection Method for Sulfur Species Measurement in Human Serum after Sulfur Inhalation Treatment. Antioxidants 2022, 11, 939. [Google Scholar] [CrossRef]
- Choi, M.Y.; Chai, C.; Park, J.H.; Lim, J.; Lee, J.; Kwon, S.W. Effects of storage period and heat treatment on phenolic compound composition in dried Citrus peels (Chenpi) and discrimination of Chenpi with different storage periods through targeted metabolomic study using HPLC-DAD analysis. J. Pharm. Biomed. Anal. 2011, 54, 638–645. [Google Scholar] [CrossRef]
- Li, J.; Li, X.; Liu, X.; Wang, X.; Li, J.; Lin, K.; Sun, S.; Yue, H.; Dai, Y. Untargeted metabolomic study of acute exacerbation of pediatric asthma via HPLC-Q-Orbitrap-MS. J. Pharm. Biomed. Anal. 2022, 215, 114737. [Google Scholar] [CrossRef]
- Stojanović, J.; Krmar, J.; Protić, A.; Svrkota, B.; Djajić, N.; Otašević, B. DoE Experimental design in HPLC separation of pharmaceuticals; a review. Arch. Pharm. 2021, 71, 279–301. [Google Scholar] [CrossRef]
- Abbas, O.; Zadravec, M.; Baeten, V.; Mikuš, T.; Lešić, T.; Vulić, A.; Prpić, J.; Jemeršić, L.; Pleadin, J. Analytical methods used for the authentication of food of animal origin. Food Chem. 2018, 246, 6–17. [Google Scholar] [CrossRef]
- Herrero, A.; Sanllorente, S.; Reguera, C.; Ortiz, M.C.; Sarabia, L.A. A new multiresponse optimization approach in combination with a D-Optimal experimental design for the determination of biogenic amines in fish by HPLC-FLD. Anal. Chim. Acta 2016, 945, 31–38. [Google Scholar] [CrossRef] [PubMed]
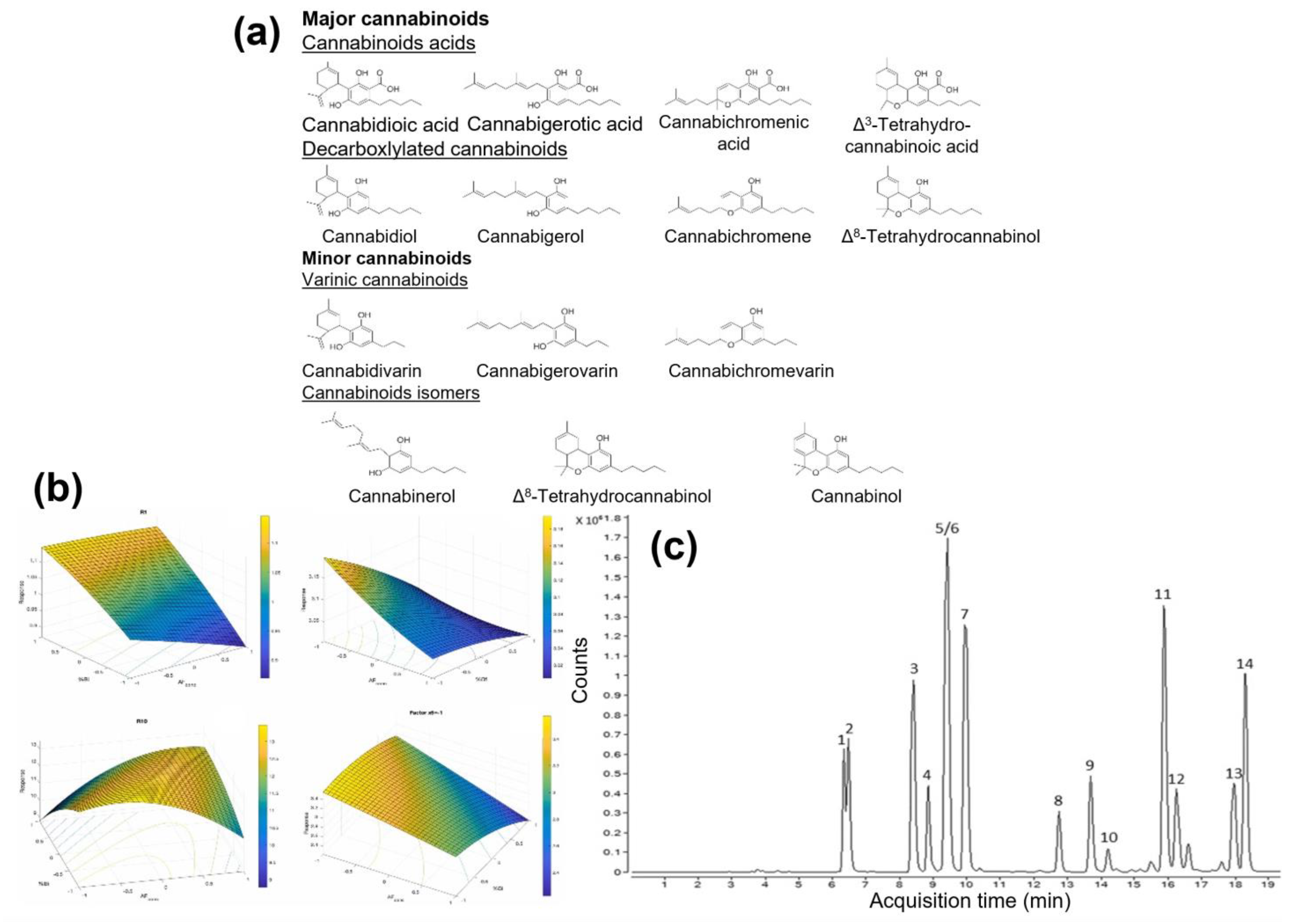
- Durante, C.; Anceschi, L.; Brighenti, V.; Caroli, C.; Afezolli, C.; Marchetti, A.; Cocchi, M.; Salamone, S.; Pollastro, F.; Pellati, F. Application of experimental design in HPLC method optimisation for the simultaneous determination of multiple bioactive cannabinoids. J. Pharm. Biomed. Anal. 2022, 221, 115037. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, P.B.; Bagul, N.; Kalyankar, G. Implementation of DoE and Risk-Based Enhanced Analytical Quality by Design Approach to Stability-Indicating RP-HPLC Method for Stability Study of Bosutinib. J. AOAC Int. 2021, 104, 1742–1753. [Google Scholar] [CrossRef]
- Gopireddy, R.R.; Maruthapillai, A.; Devikala, S.; Tamilselvi, M.; Arockia Selvi, J.; Mahapatra, S. DoE Approach: A validated Stability Indicating RP-HPLC Method Development for the Separation of Diasteromeric Analogs and Process Impurities of Carfilzomib. Mater. Today Proc. 2019, 14, 514–531. [Google Scholar] [CrossRef]
- Sahu, P.K.; Ramisetti, N.R.; Cecchi, T.; Swain, S.; Patro, C.S.; Panda, J. An overview of experimental designs in HPLC method development and validation. J. Pharm. Biomed. Anal. 2018, 147, 590–611. [Google Scholar] [CrossRef]
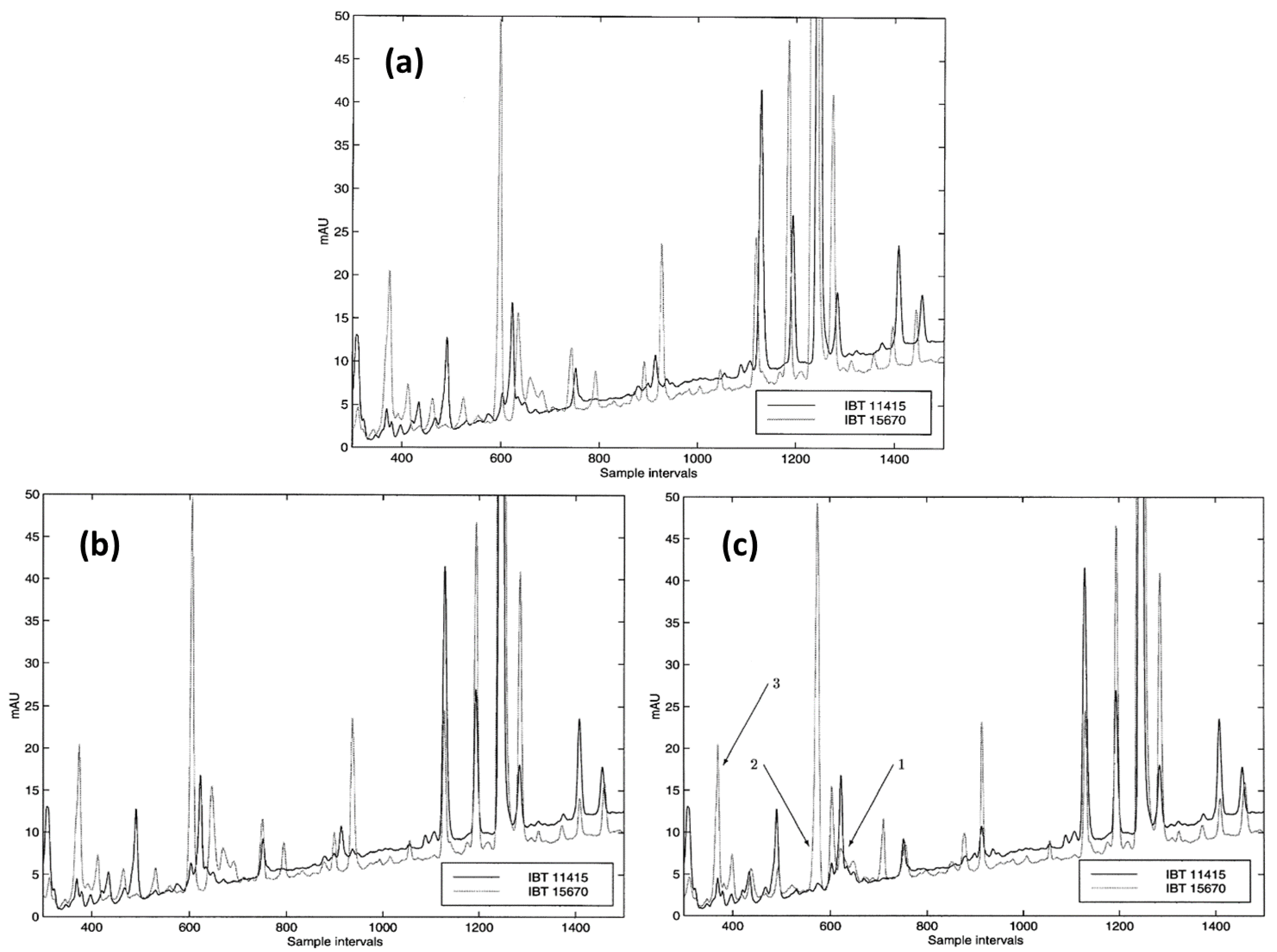
- van Nederkassel, A.M.; Xu, C.J.; Lancelin, P.; Sarraf, M.; MacKenzie, D.A.; Walton, N.J.; Bensaid, F.; Lees, M.; Martin, G.J.; Desmurs, J.R.; et al. Chemometric treatment of vanillin fingerprint chromatograms: Effect of different signal alignments on principal component analysis plots. J. Chromatogr. A 2006, 1120, 291–298. [Google Scholar] [CrossRef]
- Wallace, W.E.; Srivastava, A.; Telu, K.H.; Simón-Manso, Y. Pairwise alignment of chromatograms using an extended Fisher–Rao metric. Anal. Chim. Acta 2014, 841, 10–16. [Google Scholar] [CrossRef]
- Tucker, J.D.; Wu, W.; Srivastava, A. Generative Models for Functional Data using Phase and Amplitude Separation. Comput. Stat. Data Anal. 2012, 61, 50–66. [Google Scholar] [CrossRef]
- Clifford, D.; Stone, G.; Montoliu, I.; Rezzi, S.; Martin, F.P.; Guy, P.; Bruce, S.; Kochhar, S. Alignment using variable penalty dynamic time warping. Anal. Chem. 2009, 81, 1000–1007. [Google Scholar] [CrossRef]
- Bloemberg, T.G.; Gerretzen, J.; Wouters, H.J.P.; Gloerich, J.; van Dael, M.; Wessels, H.J.C.T.; van den Heuvel, L.P.; Eilers, P.H.C.; Buydens, L.M.C.; Wehrens, R. Improved parametric time warping for proteomics. Chemom. Intell. Lab. Syst. 2010, 104, 65–74. [Google Scholar] [CrossRef]
- Korifi, R.; Le Dréau, Y.; Dupuy, N. Comparative study of the alignment method on experimental and simulated chromatographic data. J. Sep. Sci. 2014, 37, 3276–3291. [Google Scholar] [CrossRef]
- Nielsen, N.P.V.; Carstensen, J.M.; Smedsgaard, J. Aligning of single and multiple wavelength chromatographic profiles for chemometric data analysis using correlation optimised warping. J. Chromatogr. A 1998, 805, 17–35. [Google Scholar] [CrossRef]
- Tomasi, G.; Van Den Berg, F.; Andersson, C. Correlation optimized warping and dynamic time warping as preprocessing methods for chromatographic data. J. Chemom. 2004, 18, 231–241. [Google Scholar] [CrossRef]
- De Luca, S.; Ciotoli, E.; Biancolillo, A.; Bucci, R.; Magrì, A.D.; Marini, F. Simultaneous quantification of caffeine and chlorogenic acid in coffee green beans and varietal classification of the samples by HPLC-DAD coupled with chemometrics. Environ. Sci. Pollut. Res. Int. 2018, 25, 28748–28759. [Google Scholar] [CrossRef]
- García-Seval, V.; Martínez-Alfaro, C.; Saurina, J.; Núñez, O.; Sentellas, S. Characterization, Classification and Authentication of Spanish Blossom and Honeydew Honeys by Non-Targeted HPLC-UV and Off-Line SPE HPLC-UV Polyphenolic Fingerprinting Strategies. Foods 2022, 11, 2345. [Google Scholar] [CrossRef] [PubMed]
- Núñez, N.; Collado, X.; Martínez, C.; Saurina, J.; Núñez, O. Authentication of the Origin, Variety and Roasting Degree of Coffee Samples by Non-Targeted HPLC-UV Fingerprinting and Chemometrics. Application to the Detection and Quantitation of Adulterated Coffee Samples. Foods 2020, 9, 378. [Google Scholar] [CrossRef] [PubMed]
- Carabetta, S.; Di Sanzo, R.; Campone, L.; Fuda, S.; Rastrelli, L.; Russo, M. High-Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAEC–PAD) and Chemometrics for Geographical and Floral Authentication of Honeys from Southern Italy (Calabria region). Foods 2020, 9, 1625. [Google Scholar] [CrossRef]
- Su, H.; Wu, W.; Wan, X.; Ning, J. Discriminating geographical origins of green tea based on amino acid, polyphenol, and caffeine content through high-performance liquid chromatography: Taking Lu’an guapian tea as an example. Food Sci. Nutr. 2019, 7, 2167–2175. [Google Scholar] [CrossRef] [PubMed]
- Campmajó, G.; Cayero, L.; Saurina, J.; Núñez, O. Classification of Hen Eggs by HPLC-UV Fingerprinting and Chemometric Methods. Foods 2019, 8, 310. [Google Scholar] [CrossRef] [PubMed]
- Biancolillo, A.; Preys, S.; Gaci, B.; Le-Quere, J.L.; Laboure, H.; Deuscher, Z.; Cheynier, V.; Sommerer, N.; Fayeulle, N.; Costet, P.; et al. Multi-block classification of chocolate and cocoa samples into sensory poles. Food Chem. 2021, 340, 127904. [Google Scholar] [CrossRef]
- Ghanavati Nasab, S.; Javaheran Yazd, M.; Marini, F.; Nescatelli, R.; Biancolillo, A. Classification of honey applying high performance liquid chromatography, near-infrared spectroscopy and chemometrics. Chemom. Intell. Lab. Syst. 2020, 202, 104037. [Google Scholar] [CrossRef]
- Mishra, P.; Roger, J.M.; Jouan-Rimbaud-Bouveresse, D.; Biancolillo, A.; Marini, F.; Nordon, A.; Rutledge, D.N. Recent trends in multi-block data analysis in chemometrics for multi-source data integration. TrAC Trends Anal. Chem. 2021, 137, 116206. [Google Scholar] [CrossRef]
- Zhao, L.K.; Zhao, Y.B.; Yu, P.C.; Zhang, P.X. Metabolomics approach based on utra-performance liquid chromatography coupled to mass spectrometry with chemometrics methods for high-throughput analysis of metabolite biomarkers to explore the abnormal metabolic pathways associated with myocardial dysfun. Biomed. Chromatogr. BMC 2020, 34, e4847. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Gu, H.; Zhu, J.; Barding, G.; Cheng, H.; Bao, B.; Zhang, L.; Ding, A.; Li, W. Integrated plasma and urine metabolomics coupled with HPLC/QTOF-MS and chemometric analysis on potential biomarkers in liver injury and hepatoprotective effects of Er-Zhi-Wan. Anal. Bioanal. Chem. 2014, 406, 7367–7378. [Google Scholar] [CrossRef] [PubMed]
- Buttery, R.G.; Ling, L.C. Volatile Components of Tomato Fruit and Plant Parts. Bioact. Volatile Compd. Plants 1993, 3, 23–34. [Google Scholar] [CrossRef]
- Yang, J.; Xu, G.; Kong, H.; Zheng, Y.; Pang, T.; Yang, Q. Artificial neural network classification based on high-performance liquid chromatography of urinary and serum nucleosides for the clinical diagnosis of cancer. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002, 780, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Hakimzadeh, N.; Parastar, H.; Fattahi, M. Combination of multivariate curve resolution and multivariate classification techniques for comprehensive high-performance liquid chromatography-diode array absorbance detection fingerprints analysis of Salvia reuterana extracts. J. Chromatogr. A 2014, 1326, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, B.; Ni, Y.; Kokot, S. Multi-wavelength high-performance liquid chromatography: An improved method for analysis of complex substances such as Radix Paeoniae herbs. Chemom. Intell. Lab. Syst. 2014, 130, 159–165. [Google Scholar] [CrossRef]
- Welsh, W.J.; Lin, W.; Tersigni, S.H.; Collantes, E.; Duta, R.; Carey, M.S.; Zielinski, W.L.; Brower, J.; Spencer, J.A.; Layloff, T.P. Pharmaceutical fingerprinting: Evaluation of neural networks and chemometric techniques for distinguishing among same-product manufacturers. Anal. Chem. 1996, 68, 3473–3482. [Google Scholar] [CrossRef]
- Stasiak, J.; Koba, M.; Bober, L.; Baczek, T. Principal Component Analysis of HPLC Retention Data and Molecular Modeling Structural Parameters of Cardiovascular System Drugs in View of Their Pharmacological Activity. Int. J. Mol. Sci. 2010, 11, 2681. [Google Scholar] [CrossRef]
- Saber, F.R.; Mohsen, E.; El-Hawary, S.; Eltanany, B.M.; Elimam, H.; Sobeh, M.; Elmotayam, A.K. Chemometric-enhanced metabolic profiling of five Pinus species using HPLC-MS/MS spectrometry: Correlation to in vitro anti-aging, anti-Alzheimer and antidiabetic activities. J. Chromatogr. B 2021, 1177, 122759. [Google Scholar] [CrossRef] [PubMed]
- De Luca, M.; Ioele, G.; Grande, F.; Platikanov, S.; Tauler, R.; Ragno, G. Photostability study of multicomponent drug formulations via MCR-ALS: The case of the hydrochlorothiazide-amiloride mixture. J. Pharm. Biomed. Anal. 2020, 186, 113332. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, M.; Borioni, A.; Bartolomei, M.; Mosca, A.; Gostoli, G. Classification of the ibuprofen active pharmaceutical ingredients by chemical patterns combining HPLC, 1H-NMR spectroscopy and chemometrics: Traceability of legal medicines. Ann. Ist. Super. Sanita 2020, 56, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.N.; Kothari, C.S. Multivariate UV-Chemometric and HPLC-QbD Method for Simultaneous Estimation of Vardenafil and Dapoxetine in Active Pharmaceutical Ingredients and its Marketed Formulation. Curr. Anal. Chem. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Palei, N.N.; Vijayaraj, S.; Lathasri, K.; Archana, D.; Rajavel, P. Chemometric Approach to Develop and Validate RP-HPLC Method for Estimation of Erlotinib Hydrochloride in Nano Structured Lipid Carriers. Curr. Pharm. Anal. 2018, 16, 210–219. [Google Scholar] [CrossRef]
- Gad, M.A.; Amer, S.M.; Zaazaa, H.E.; Hassan, S.A. Strategies for stabilizing formulation and QbD assisted development of robust stability indicating method of azilsartan medoxomil/chlorthalidone. J. Pharm. Biomed. Anal. 2020, 178, 112910. [Google Scholar] [CrossRef]
- Carranco, N.; Farrés-Cebrián, M.; Saurina, J.; Núñez, O. Authentication and Quantitation of Fraud in Extra Virgin Olive Oils Based on HPLC-UV Fingerprinting and Multivariate Calibration. Foods 2018, 7, 44. [Google Scholar] [CrossRef]
- Núñez, N.; Saurina, J.; Núñez, O. Non-targeted HPLC-FLD fingerprinting for the detection and quantitation of adulterated coffee samples by chemometrics. Food Control 2021, 124, 107912. [Google Scholar] [CrossRef]
- Dinç-Zor, Ş.; Dönmez, Ö.A.; Bozdoğan, A.E. Application of Chemometrics-assisted HPLC-DAD Strategies for Simultaneous Determination of Paracetamol, Pseudoephedrine HCl, Dextromethorphan HBr, Doxylamine Succinate and Saccharin in Syrup Formulation. Curr. Pharm. Anal. 2020, 17, 1043–1050. [Google Scholar] [CrossRef]
- Frenich, A.G.; Galera, M.M.; García, M.D.G.; Vidal, J.L.M.; Catasús, M.; Marti, L.; Mederos, M.V. Resolution of HPLC-DAD highly overlapping analytical signals for quantitation of pesticide mixtures in groundwater and soil using multicomponent analysis and neural networks. J. Liq. Chromatogr. Relat. Technol. 2007, 24, 651–668. [Google Scholar] [CrossRef]
- Guizellini, F.C.; Marcheafave, G.G.; Rakocevic, M.; Bruns, R.E.; Scarminio, I.S.; Soares, P.K. PARAFAC HPLC-DAD metabolomic fingerprint investigation of reference and crossed coffees. Food Res. Int. 2018, 113, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, M.; Wang, P.; Chen, J.; Yang, S.; Luo, P.; Gao, X. Detection and Quantitation of Adulterated Paprika Samples Using Second-Order HPLC-FLD Fingerprints and Chemometrics. Foods 2022, 11, 2376. [Google Scholar] [CrossRef] [PubMed]
- Arce, M.M.; Castro, D.; Sarabia, L.A.; Ortiz, M.C.; Sanllorente, S. Procedure to explore a ternary mixture diagram to find the appropriate gradient profile in liquid chromatography with fluorescence detector. Application to determine four primary aromatic amines in napkins. J. Chromatogr. A 2022, 1676, 463252. [Google Scholar] [CrossRef] [PubMed]
- Ogemdi, I.K. Properties and Uses of Colloids: A Review. Colloid Surf. Sci. 2019, 4, 24–28. [Google Scholar] [CrossRef]
- Qin, S.J. Process data analytics in the era of big data. AIChE J. 2014, 60, 3092–3100. [Google Scholar] [CrossRef]
- Bos, T.S.; Knol, W.C.; Molenaar, S.R.A.; Niezen, L.E.; Schoenmakers, P.J.; Somsen, G.W.; Pirok, B.W.J. Recent applications of chemometrics in one- and two-dimensional chromatography. J. Sep. Sci. 2020, 43, 1678–1727. [Google Scholar] [CrossRef]
- Mahler, H.C.; Friess, W.; Grauschopf, U.; Kiese, S. Protein aggregation: Pathways, induction factors and analysis. J. Pharm. Sci. 2009, 98, 2909–2934. [Google Scholar] [CrossRef]
- Baunsgaard, D.; Andersson, C.A.; Arndal, A. Multi-way chemometrics for mathematical separation of fluorescent colorants and colour precursors from spectrofluorimetry of beet sugar and beet sugar thick juice as validated by HPLC analysis—Staff of the Department of Food Science. Food Chem. 2000, 70, 113–121. [Google Scholar] [CrossRef]
- Upadhyay, R.; Sehwag, S.; Niwas Mishra, H. Chemometric approach to develop frying stable sunflower oil blends stabilized with oleoresin rosemary and ascorbyl palmitate. Food Chem. 2017, 218, 496–504. [Google Scholar] [CrossRef]
- Beretta, G.; Fermo, P.; Maffei Facino, R. Simple and rapid simultaneous profiling of minor components of honey by size exclusion chromatography (SEC) coupled to ultraviolet diode array detection (UV-DAD), combined with chemometric methods. J. Pharm. Biomed. Anal. 2012, 58, 193–199. [Google Scholar] [CrossRef]
- Liu, W.; Xu, J.; Zhu, R.; Zhu, Y.; Zhao, Y.; Chen, P.; Pan, C.; Yao, W.; Gao, X. Fingerprinting profile of polysaccharides from Lycium barbarum using multiplex approaches and chemometrics. Int. J. Biol. Macromol. 2015, 78, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gong, X.; Wang, Z.; Pan, C.; Zhao, Y.; Gao, X.; Liu, W. Multiple fingerprint profiles and chemometrics analysis of polysaccharides from Sarcandra glabra. Int. J. Biol. Macromol. 2019, 123, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.j.; Yan, Z.y.; Hong, L.; Li, S.P.; Zhao, J. Quality evaluation of Salvia miltiorrhiza from different geographical origins in China based on qualitative and quantitative saccharide mapping and chemometrics. J. Pharm. Biomed. Anal. 2020, 191, 113583. [Google Scholar] [CrossRef] [PubMed]
- Malkavaara, P.; Alén, R.; Kolehmainen, E. Chemometrics: An Important Tool for the Modern Chemist, an Example from Wood-Processing Chemistry. J. Chem. Inf. Comput. Sci. 2000, 40, 438–441. [Google Scholar] [CrossRef] [PubMed]
- Mol, E.A.; Goumans, M.J.; Doevendans, P.A.; Sluijter, J.P.G.; Vader, P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 2061–2065. [Google Scholar] [CrossRef]
- Burgess, R.R. A brief practical review of size exclusion chromatography: Rules of thumb, limitations, and troubleshooting. Protein Expr. Purif. 2018, 150, 81–85. [Google Scholar] [CrossRef]
- Ricker, R.D.; Sandoval, L.A.; Justice, J.D.; Geiser, F.O. Multivariate visualization in the size-exclusion chromatography and pattern recognition of biological samples. J. Chromatogr. A 1995, 691, 67–79. [Google Scholar] [CrossRef]
- Elshereef, R.; Budman, H.; Moresoli, C.; Legge, R.L. Monitoring the fractionation of a whey protein isolate during dead-end membrane filtration using fluorescence and chemometric methods. Biotechnol. Prog. 2010, 26, 168–178. [Google Scholar] [CrossRef]
- Akhgar, C.K.; Ebner, J.; Alcaraz, M.R.; Kopp, J.; Goicoechea, H.; Spadiut, O.; Schwaighofer, A.; Lendl, B. Application of Quantum Cascade Laser-Infrared Spectroscopy and Chemometrics for In-Line Discrimination of Coeluting Proteins from Preparative Size Exclusion Chromatography. Anal. Chem. 2022, 94, 11192–11200. [Google Scholar] [CrossRef]
- Böing, A.N.; van der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar] [CrossRef]
- Garcia-Ceron, D.; Dawson, C.S.; Faou, P.; Bleackley, M.R.; Anderson, M.A. Size-exclusion chromatography allows the isolation of EVs from the filamentous fungal plant pathogen Fusarium oxysporum f. sp. vasinfectum (Fov). Proteomics 2021, 21, 2000240. [Google Scholar] [CrossRef] [PubMed]
- Vogel, R.; Coumans, F.A.W.; Maltesen, R.G.; Böing, A.N.; Bonnington, K.E.; Broekman, M.L.; Broom, M.F.; Buzás, E.I.; Christiansen, G.; Hajji, N.; et al. A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing. J. Extracell. Vesicles 2016, 5, 31242. [Google Scholar] [CrossRef] [PubMed]
- Vergauwen, G.; Tulkens, J.; Pinheiro, C.; Avila Cobos, F.; Dedeyne, S.; De Scheerder, M.A.; Vandekerckhove, L.; Impens, F.; Miinalainen, I.; Braems, G.; et al. Robust sequential biophysical fractionation of blood plasma to study variations in the biomolecular landscape of systemically circulating extracellular vesicles across clinical conditions. J. Extracell. vesicles 2021, 10, e12122. [Google Scholar] [CrossRef]
- Palviainen, M.; Saraswat, M.; Varga, Z.; Kitka, D.; Neuvonen, M.; Puhka, M.; Joenväärä, S.; Renkonen, R.; Nieuwland, R.; Takatalo, M.; et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo—Implications for biomarker discovery. PLoS ONE 2020, 15, e0236439. [Google Scholar] [CrossRef] [PubMed]
- Zattoni, A.; Roda, B.; Borghi, F.; Marassi, V.; Reschiglian, P. Flow field-flow fractionation for the analysis of nanoparticles used in drug delivery. J. Pharm. Biomed. Anal. 2014, 87, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Contado, C. Field flow fractionation techniques to explore the “nano-world”. Analytical and bioanalytical chemistry 2017, 409, 2501–2518. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Shen, S.; Lee, S.; Dou, H. Field-flow fractionation: A gentle separation and characterization technique in biomedicine. TrAC Trends Anal. Chem. 2018, 108, 231–238. [Google Scholar] [CrossRef]
- Coelho, C.; Parot, J.; Gonsior, M.; Nikolantonaki, M.; Schmitt-Kopplin, P.; Parlanti, E.; Gougeon, R.D. Asymmetrical flow field-flow fractionation of white wine chromophoric colloidal matter. Anal. Bioanal. Chem. 2017, 409, 2757–2766. [Google Scholar] [CrossRef]
- Yang, J.S.; Lee, J.C.; Byeon, S.K.; Rha, K.H.; Moon, M.H. Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. Anal. Chem. 2017, 89, 2488–2496. [Google Scholar] [CrossRef]
- Leeman, M.; Choi, J.; Hansson, S.; Storm, M.U.; Nilsson, L. Proteins and antibodies in serum, plasma, and whole blood-size characterization using asymmetrical flow field-flow fractionation (AF4). Anal. Bioanal. Chem. 2018, 410, 4867–4873. [Google Scholar] [CrossRef]
- Marassi, V.; Maggio, S.; Battistelli, M.; Stocchi, V.; Zattoni, A.; Reschiglian, P.; Guescini, M.; Roda, B. An ultracentrifugation—hollow-fiber flow field-flow fractionation orthogonal approach for the purification and mapping of extracellular vesicle subtypes. J. Chromatogr. A 2021, 1638, 461861. [Google Scholar] [CrossRef]
- Roda, B.; Marassi, V.; Zattoni, A.; Borghi, F.; Anand, R.; Agostoni, V.; Gref, R.; Reschiglian, P.; Monti, S. Flow field-flow fractionation and multi-angle light scattering as a powerful tool for the characterization and stability evaluation of drug-loaded metal-organic framework nanoparticles. Anal. Bioanal. Chem. 2018, 410, 5245–5253. [Google Scholar] [CrossRef] [PubMed]
- Caputo, F.; Mehn, D.; Clogston, J.D.; Rösslein, M.; Prina-Mello, A.; Borgos, S.E.; Gioria, S.; Calzolai, L. Asymmetric-flow field-flow fractionation for measuring particle size, drug loading and (in)stability of nanopharmaceuticals. The joint view of European Union Nanomedicine Characterization Laboratory and National Cancer Institute—Nanotechnology Characterization Laboratory. J. Chromatogr. A 2021, 1635, 461767. [Google Scholar] [CrossRef] [PubMed]
- Iavicoli, P.; Urbán, P.; Bella, A.; Ryadnov, M.G.; Rossi, F.; Calzolai, L. Application of Asymmetric Flow Field-Flow Fractionation hyphenations for liposome-antimicrobial peptide interaction. J. Chromatogr. A 2015, 1422, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Marassi, V.; Casolari, S.; Panzavolta, S.; Bonvicini, F.; Gentilomi, G.A.; Giordani, S.; Zattoni, A.; Reschiglian, P.; Roda, B. Synthesis Monitoring, Characterization and Cleanup of Ag-Polydopamine Nanoparticles Used as Antibacterial Agents with Field-Flow Fractionation. Antibiotics 2022, 11, 358. [Google Scholar] [CrossRef]
- Qureshi, R.N.; Kok, W.T. Application of flow field-flow fractionation for the characterization of macromolecules of biological interest: A review. Anal. Bioanal. Chem. 2011, 399, 1401. [Google Scholar] [CrossRef]
- Marassi, V.; Giordani, S.; Reschiglian, P.; Roda, B.; Zattoni, A. Tracking Heme-Protein Interactions in Healthy and Pathological Human Serum in Native Conditions by Miniaturized FFF-Multidetection. Appl. Sci. 2022, 12, 6762. [Google Scholar] [CrossRef]
- Marassi, V.; Mattarozzi, M.; Toma, L.; Giordani, S.; Ronda, L.; Roda, B.; Zattoni, A.; Reschiglian, P.; Careri, M. FFF-based high-throughput sequence shortlisting to support the development of aptamer-based analytical strategies. Anal. Bioanal. Chem. 2022, 414, 5519–5527. [Google Scholar] [CrossRef]
- Lou, J.; Myers, M.N.; Giddings, J.C. Separation of Polysaccharides by Thermal Field-Flow Fractionation. J. Liq. Chromatogr. Relat. Technol. 1994, 17, 3239–3260. [Google Scholar] [CrossRef]
- Marassi, V.; De Marchis, F.; Roda, B.; Bellucci, M.; Capecchi, A.; Reschiglian, P.; Pompa, A.; Zattoni, A. Perspectives on protein biopolymers: Miniaturized flow field-flow fractionation-assisted characterization of a single-cysteine mutated phaseolin expressed in transplastomic tobacco plants. J. Chromatogr. A 2021, 1637, 461806. [Google Scholar] [CrossRef]
- Duthen, S.; Rochat, C.; Kleiber, D.; Violleau, F.; Daydé, J.; Raynaud, C.; Levasseur-Garcia, C. Physicochemical characterization and study of molar mass of industrial gelatins by AsFlFFF-UV/MALS and chemometric approach. PLoS ONE 2018, 13, e0203595. [Google Scholar] [CrossRef] [PubMed]
- Novikov, E.A.; Sergeev, Y.A.; Sanzharov, V.V.; Safieva, R.Z.; Vinokurov, V.A. Application of Multidimensional Analysis Methods to Dead Oil Characterization on the Basis of Data on Thermal Field-Flow Fractionation of Native Asphaltene Nanoparticles. Pet. Chem. 2019, 59, 34–47. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Shim, S.; Noshari, J.; Becker, F.F.; Stemke-Hale, K. Correlations between the dielectric properties and exterior morphology of cells revealed by dielectrophoretic field-flow fractionation. Electrophoresis 2013, 34, 1042–1050. [Google Scholar] [CrossRef]
- Roda, A.; Mirasoli, M.; Roda, B.; Bonvicini, F.; Colliva, C.; Reschiglian, P. Recent developments in rapid multiplexed bioanalytical methods for foodborne pathogenic bacteria detection. Microchim. Acta 2012, 178, 7–28. [Google Scholar] [CrossRef]
- Vernhet, A. Red Wine Clarification and Stabilization; Academic Press: Cambridge, MA, USA, 2019; pp. 237–251. [Google Scholar] [CrossRef]
- Martínez-Lapuente, L.; Guadalupe, Z.; Ayestarán, B.; Martínez-Lapuente, L.; Guadalupe, Z.; Ayestarán, B. Properties of Wine Polysaccharides; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Marassi, V.; Marangon, M.; Zattoni, A.; Vincenzi, S.; Versari, A.; Reschiglian, P.; Roda, B.; Curioni, A. Characterization of red wine native colloids by asymmetrical flow field-flow fractionation with online multidetection. Food Hydrocoll/ 2021, 110, 106204. [Google Scholar] [CrossRef]
- Pascotto, K.; Leriche, C.; Caillé, S.; Violleau, F.; Boulet, J.C.; Geffroy, O.; Levasseur-Garcia, C.; Cheynier, V. Study of the relationship between red wine colloidal fraction and astringency by asymmetrical flow field-flow fractionation coupled with multi-detection. Food Chem. 2021, 361, 130104. [Google Scholar] [CrossRef]
- Roger, J.M.; Palagos, B.; Bertrand, D.; Fernandez-Ahumada, E. CovSel: Variable selection for highly multivariate and multi-response calibration: Application to IR spectroscopy. Chemom. Intell. Lab. Syst. 2011, 106, 216–223. [Google Scholar] [CrossRef]
- Osorio-Macías, D.E.; Bolinsson, H.; Linares-Pastén, J.A.; Ferrer-Gallego, R.; Choi, J.; Peñarrieta, J.M.; Bergenståhl, B. Characterization on the impact of different clarifiers on the white wine colloids using Asymmetrical Flow Field-Flow Fractionation. Food Chem. 2022, 381, 132123. [Google Scholar] [CrossRef]
- Krebs, G.; Gastl, M.; Becker, T. Chemometric modeling of palate fullness in lager beers. Food Chem. 2021, 342, 128253. [Google Scholar] [CrossRef]
- Roda, B.; Mirasoli, M.; Zattoni, A.; Casale, M.; Oliveri, P.; Bigi, A.; Reschiglian, P.; Simoni, P.; Roda, A. A new analytical platform based on field-flow fractionation and olfactory sensor to improve the detection of viable and non-viable bacteria in food. Anal. Bioanal. Chem. 2016, 408, 7367–7377. [Google Scholar] [CrossRef]
- Austin, M.A.; Rodriguez, B.L.; McKnight, B.; McNeely, M.J.; Edwards, K.L.; Curb, J.D.; Sharp, D.S. Low-density lipoprotein particle size, triglycerides, and high-density lipoprotein cholesterol as risk factors for coronary heart disease in older Japanese-American men. Am. J. Cardiol. 2000, 86, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yang, J.S.; Lee, J.C.; Lee, J.Y.; Lee, J.Y.; Kim, E.; Moon, M.H. Lipidomic alterations in lipoproteins of patients with mild cognitive impairment and Alzheimer’s disease by asymmetrical flow field-flow fractionation and nanoflow ultrahigh performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2018, 1568, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Marassi, V.; Beretti, F.; Roda, B.; Alessandrini, A.; Facci, P.; Maraldi, T.; Zattoni, A.; Reschiglian, P.; Portolani, M. A new approach for the separation, characterization and testing of potential prionoid protein aggregates through hollow-fiber flow field-flow fractionation and multi-angle light scattering. Anal. Chim. Acta 2019, 1087, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Yang, J.S.; Lee, G.B.; Moon, M.H. Evaluation of exosome separation from human serum by frit-inlet asymmetrical flow field-flow fractionation and multiangle light scattering. Anal. Chim. Acta 2020, 1124, 137–145. [Google Scholar] [CrossRef]
- de Oliveira, M.A.; Pound-Lana, G.; Capelari-Oliveira, P.; Pontífice, T.G.; Silva, S.E.D.; Machado, M.G.C.; Postacchini, B.B.; Mosqueira, V.C.F. Release, transfer and partition of fluorescent dyes from polymeric nanocarriers to serum proteins monitored by asymmetric flow field-flow fractionation. J. Chromatogr. A 2021, 1641, 461959. [Google Scholar] [CrossRef]
- Ashby, J.; Flack, K.; Jimenez, L.A.; Duan, Y.; Khatib, A.K.; Somlo, G.; Wang, S.E.; Cui, X.; Zhong, W. Distribution profiling of circulating MicroRNAs in serum. Anal. Chem. 2014, 86, 9343–9349. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Meyer, J.L.; Wallace, J.B.; Eggert, S.L. Leaf Litter as a Source of Dissolved Organic Carbon in Streams. Ecosystems 1998, 1, 240–249. [Google Scholar] [CrossRef]
- Abelho, M. From litterfall to breakdown in streams: A review. TheScientificWorldJournal 2001, 1, 656–680. [Google Scholar] [CrossRef]
- Freeman, C.; Fenner, N.; Ostle, N.J.; Kang, H.; Dowrick, D.J.; Reynolds, B.; Lock, M.A.; Sleep, D.; Hughes, S.; Hudson, J. Export of dissolved organic carbon from peatlands under elevated carbon dioxide levels. Nature 2004, 430, 195–198. [Google Scholar] [CrossRef]
- Contribution of dissolved organic C to stream metabolism: A mesocosm study using 13C-enriched tree-tissue leachate. J. N. Am. Benthol. Soc. 2005, 24, 48–67. [CrossRef]
- Guéguen, C.; Dominik, J. Partitioning of trace metals between particulate, colloidal and truly dissolved fractions in a polluted river: The Upper Vistula River (Poland). Appl. Geochem. 2003, 18, 457–470. [Google Scholar] [CrossRef]
- Beggs, K.M.H.; Summers, R.S. Character and chlorine reactivity of dissolved organic matter from a mountain pine beetle impacted watershed. Environ. Sci. Technol. 2011, 45, 5717–5724. [Google Scholar] [CrossRef] [PubMed]
- Weishaar, J.L.; Aiken, G.R.; Bergamaschi, B.A.; Fram, M.S.; Fujii, R.; Mopper, K. Evaluation of specific ultraviolet absorbance as an indicator of the chemical composition and reactivity of dissolved organic carbon. Environ. Sci. Technol. 2003, 37, 4702–4708. [Google Scholar] [CrossRef]
- Miller, M.P.; McKnight, D.M. Comparison of seasonal changes in fluorescent dissolved organic matter among aquatic lake and stream sites in the Green Lakes Valley. J. Geophys. Res. Biogeosci. 2010, 115, 1–14. [Google Scholar] [CrossRef]
- Pifer, A.D.; Fairey, J.L. Improving on SUVA 254 using fluorescence-PARAFAC analysis and asymmetric flow-field flow fractionation for assessing disinfection byproduct formation and control. Water Res. 2012, 46, 2927–2936. [Google Scholar] [CrossRef]
- Coble, P.G.; Green, S.A.; Blough, N.V.; Gagosian, R.B. Characterization of dissolved organic matter in the Black Sea by fluorescence spectroscopy. Nature 1990, 348, 432–435. [Google Scholar] [CrossRef]
- Guéguen, C.; Cuss, C.W. Characterization of aquatic dissolved organic matter by asymmetrical flow field-flow fractionation coupled to UV-Visible diode array and excitation emission matrix fluorescence. J. Chromatogr. A 2011, 1218, 4188–4198. [Google Scholar] [CrossRef]
- Cuss, C.W.; Guéguen, C. Determination of relative molecular weights of fluorescent components in dissolved organic matter using asymmetrical flow field-flow fractionation and parallel factor analysis. Anal. Chim. Acta 2012, 733, 98–102. [Google Scholar] [CrossRef]
- Cuss, C.W.; Guéguen, C. Distinguishing dissolved organic matter at its origin: Size and optical properties of leaf-litter leachates. Chemosphere 2013, 92, 1483–1489. [Google Scholar] [CrossRef]
- Cuss, C.W.; Guéguen, C. Relationships between molecular weight and fluorescence properties for size-fractionated dissolved organic matter from fresh and aged sources. Water Res. 2015, 68, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Coble, P.G. Characterization of marine and terrestrial DOM in seawater using excitation-emission matrix spectroscopy. Mar. Chem. 1996, 51, 325–346. [Google Scholar] [CrossRef]
- Mangal, V.; Stenzler, B.R.; Poulain, A.J.; Guéguen, C. Aerobic and Anaerobic Bacterial Mercury Uptake is Driven by Algal Organic Matter Composition and Molecular Weight. Environ. Sci. Technol. 2019, 53, 157–165. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zappi, A.; Marassi, V.; Giordani, S.; Kassouf, N.; Roda, B.; Zattoni, A.; Reschiglian, P.; Melucci, D. Extracting Information and Enhancing the Quality of Separation Data: A Review on Chemometrics-Assisted Analysis of Volatile, Soluble and Colloidal Samples. Chemosensors 2023, 11, 45. https://doi.org/10.3390/chemosensors11010045
Zappi A, Marassi V, Giordani S, Kassouf N, Roda B, Zattoni A, Reschiglian P, Melucci D. Extracting Information and Enhancing the Quality of Separation Data: A Review on Chemometrics-Assisted Analysis of Volatile, Soluble and Colloidal Samples. Chemosensors. 2023; 11(1):45. https://doi.org/10.3390/chemosensors11010045
Chicago/Turabian StyleZappi, Alessandro, Valentina Marassi, Stefano Giordani, Nicholas Kassouf, Barbara Roda, Andrea Zattoni, Pierluigi Reschiglian, and Dora Melucci. 2023. "Extracting Information and Enhancing the Quality of Separation Data: A Review on Chemometrics-Assisted Analysis of Volatile, Soluble and Colloidal Samples" Chemosensors 11, no. 1: 45. https://doi.org/10.3390/chemosensors11010045
APA StyleZappi, A., Marassi, V., Giordani, S., Kassouf, N., Roda, B., Zattoni, A., Reschiglian, P., & Melucci, D. (2023). Extracting Information and Enhancing the Quality of Separation Data: A Review on Chemometrics-Assisted Analysis of Volatile, Soluble and Colloidal Samples. Chemosensors, 11(1), 45. https://doi.org/10.3390/chemosensors11010045










