Optical Technologies for Single-Cell Analysis on Microchips
Abstract
1. Introduction
2. Optical Technologies
2.1. Fluorescence Technique
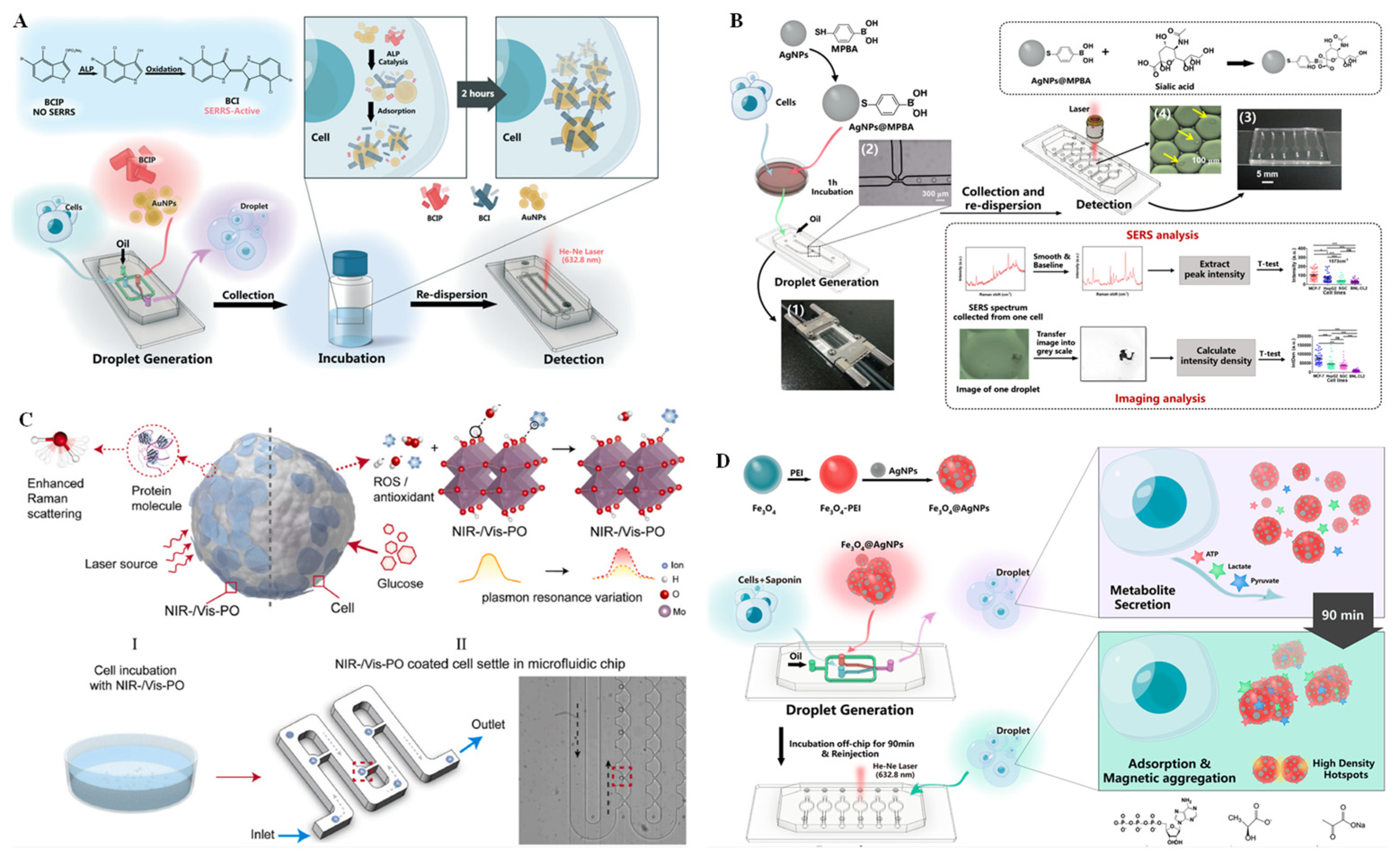
2.2. SERS Technique
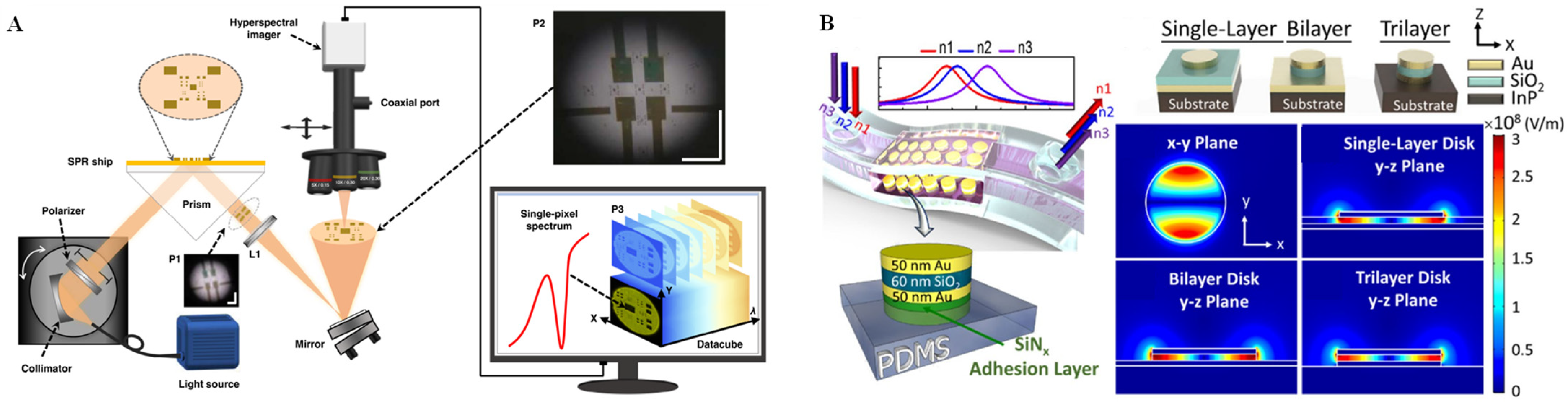
2.3. SPR Technique
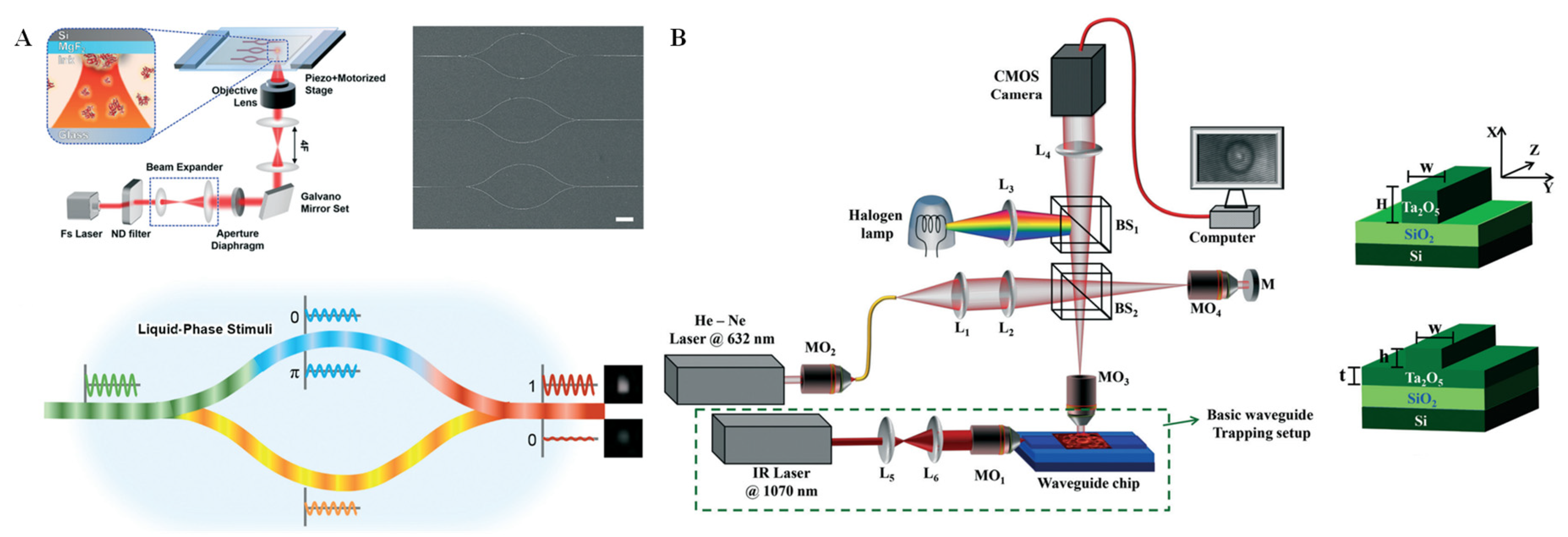
2.4. Interferometry Technique
3. Optical Technologies for Single-Cell Analysis on Microchips
3.1. Fluorescence Technique for Single-Cell Analysis on Microchips
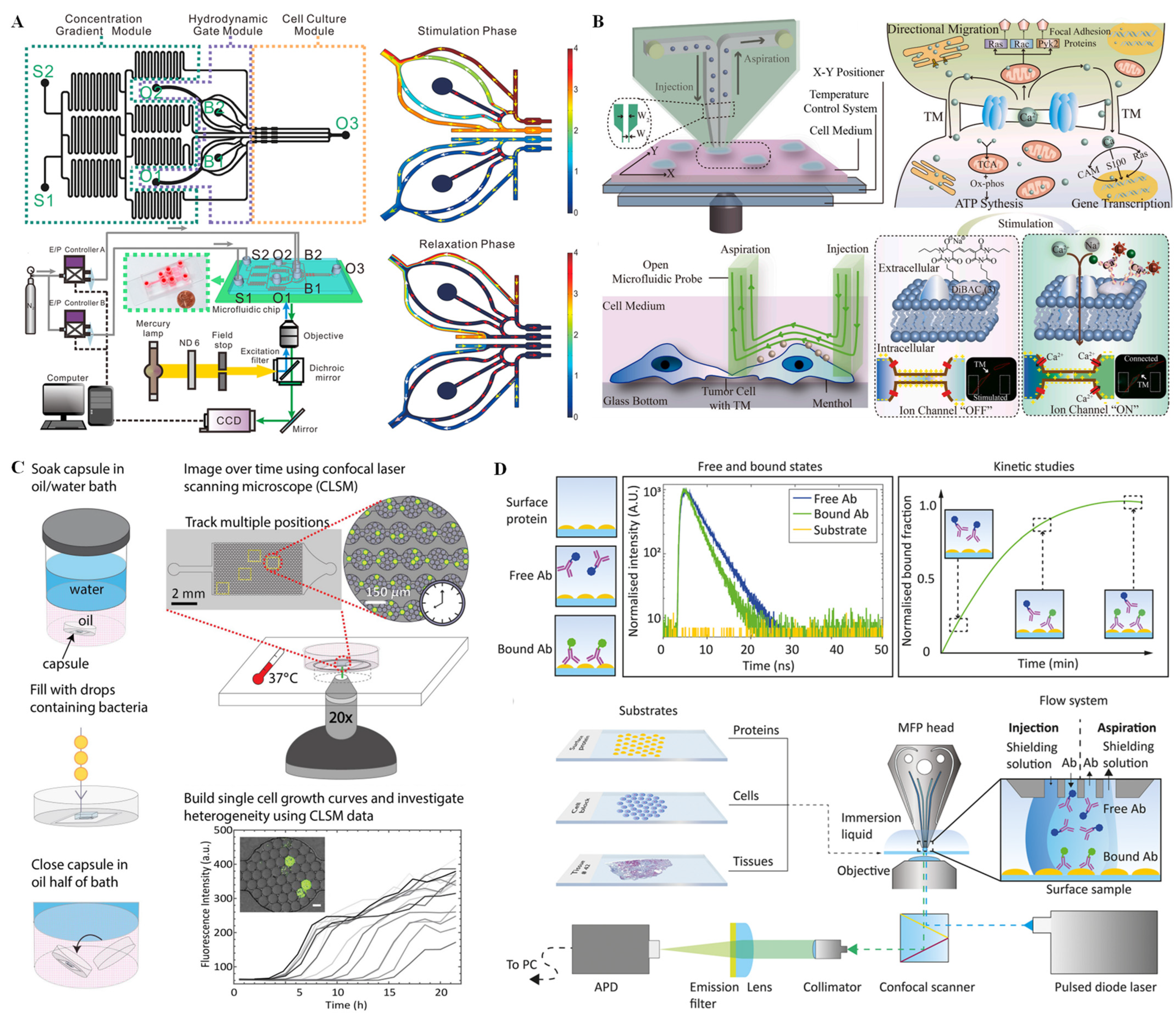
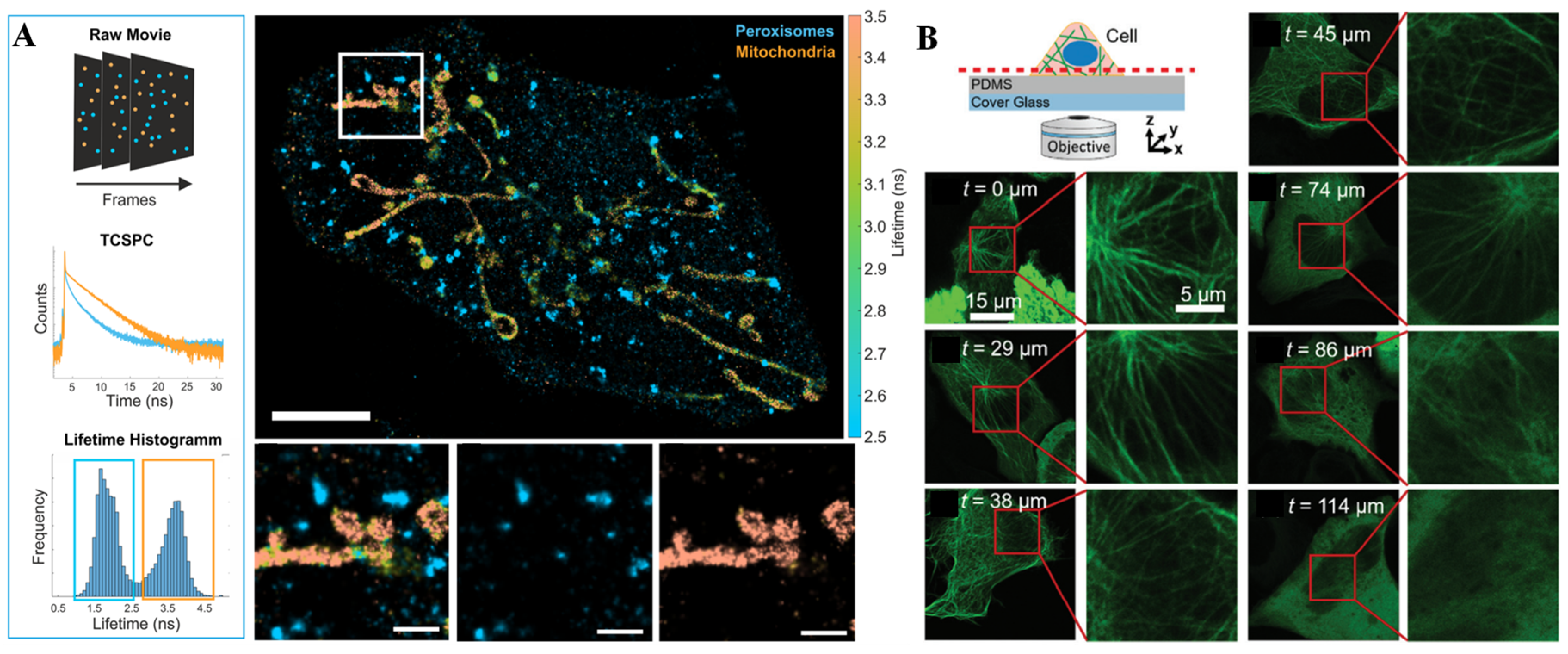
3.2. SERS Technique for Single-Cell Analysis on Microchips
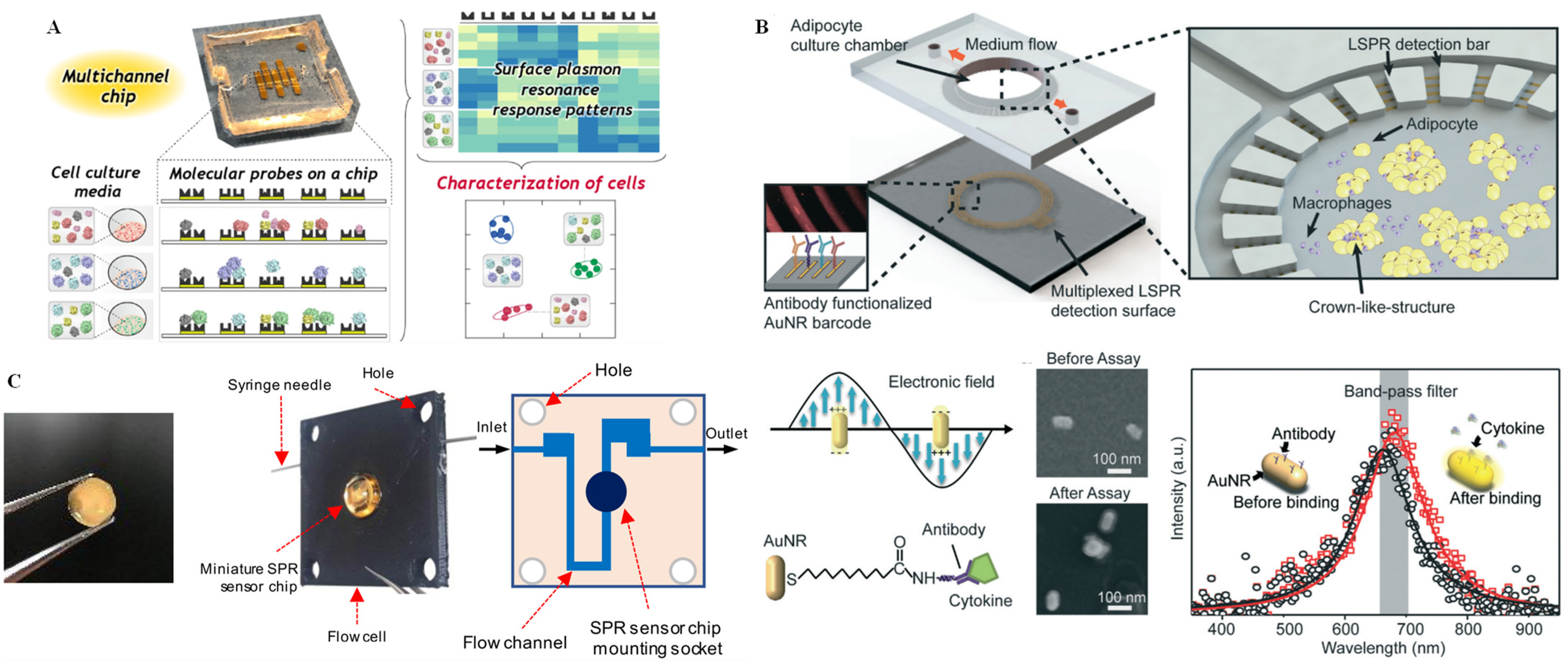
3.3. SPR Technique for Single-Cell Analysis on Microchips
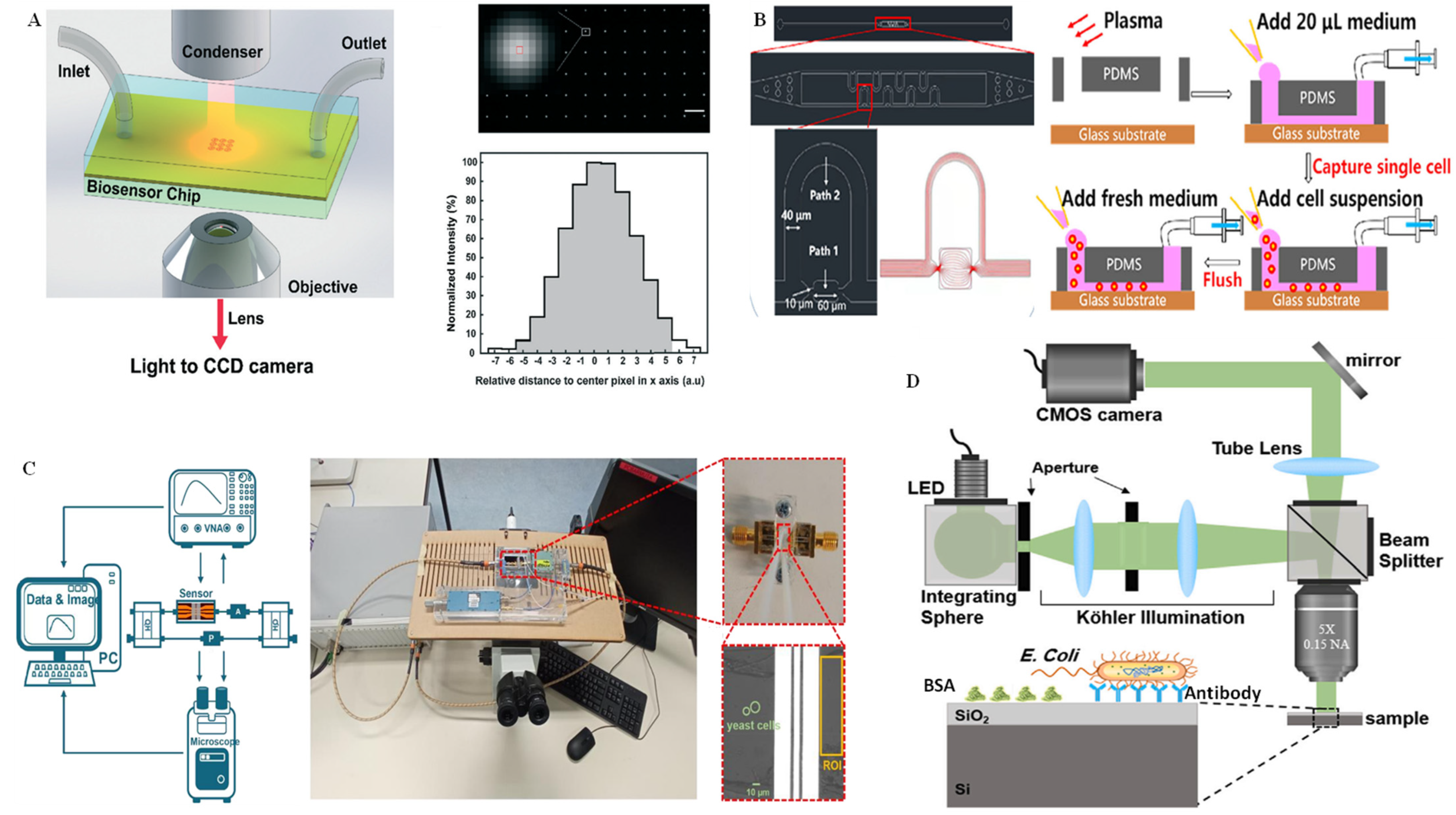
3.4. Interferometry Technique for Single-Cell Analysis on Microchips
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cai, D.; Yi, Q.; Shen, C.; Lan, Y.; Urban, G.; Du, W. Direct enrichment of pathogens from physiological samples of high conductivity and viscosity using H-filter and positive dielectrophoresis. Biomicrofluidics 2018, 12, 014109. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Hughes, A.J.; Spelke, D.P.; Xu, Z.; Kang, C.; Schaffer, D.V.; Herr, A.E. Single-cell western blotting. Nat. Methods 2014, 11, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Wu, H.; Bhaya, D.; Grossman, A.; Granier, S.; Kobilka, B.K.; Zare, R.N. Counting low-copy number proteins in a single cell. Science 2007, 315, 81–84. [Google Scholar] [CrossRef]
- De Silva, I.W.; Kretsch, A.R.; Lewis, H.; Bailey, M.; Verbeck, G.F. True one cell chemical analysis: A review. Analyst 2019, 144, 4733–4749. [Google Scholar] [CrossRef]
- Galler, K.; Brautigam, K.; Grosse, C.; Popp, J.; Neugebauer, U. Making a big thing of a small cell—Recent advances in single cell analysis. Analyst 2014, 139, 1237–1273. [Google Scholar] [CrossRef]
- Lo, S.; Yao, D. Get to understand more from single-cells: Current studies of microfluidic-based techniques for single-cell analysis. Int. J. Mol. Sci. 2015, 16, 16763–16777. [Google Scholar] [CrossRef]
- Volpato, V.; Webber, C. Addressing variability in iPSC-derived models of human disease: Guidelines to promote reproducibility. Dis. Model. Mech. 2020, 13, dmm042317. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef]
- Zheng, H.; Pomyen, Y.; Hernandez, M.O.; Li, C.; Livak, F.; Tang, W.; Dang, H.; Greten, T.F.; Davis, J.L.; Zhao, Y.; et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 2018, 68, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Oomen, P.E.; Aref, M.A.; Kaya, I.; Phan, N.T.N.; Ewing, A.G. Chemical analysis of single cells. Anal. Chem. 2019, 91, 588–621. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ju, R.; Sekine, S.; Zhang, D.; Zhuang, S.; Yamaguchi, Y. All-in-one microfluidic device for on-site diagnosis of pathogens based on an integrated continuous flow PCR and electrophoresis biochip. Lab Chip 2019, 19, 2663–2668. [Google Scholar] [CrossRef] [PubMed]
- Black, W.A.; Stocks, B.B.; Mellors, J.S.; Engen, J.R.; Ramsey, J.M. Utilizing microchip capillary electrophoresis electrospray ionization for hydrogen exchange mass spectrometry. Anal. Chem. 2015, 87, 6280–6287. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Chen, D.; Li, S.; Ou, X.; Liu, B.F. Microfluidics towards single cell resolution protein analysis. Trends Anal. Chem. 2019, 117, 2–12. [Google Scholar] [CrossRef]
- Murphy, T.W.; Zhang, Q.; Naler, L.B.; Ma, S.; Lu, C. Recent advances in the use of microfluidic technologies for single cell analysis. Analyst 2017, 143, 6–8. [Google Scholar] [CrossRef]
- Cong, L.; Wang, J.; Li, X.; Tian, Y.; Xu, S.; Liang, C.; Xu, W.; Wang, W.; Xu, S. Microfluidic droplet-SERS platform for single-cell cytokine analysis via a cell surface bioconjugation strategy. Anal. Chem. 2022, 94, 10375–10383. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z.; Li, Q. Microfluidic techniques for dynamic single-cell analysis. Microchim. Acta 2010, 168, 177–195. [Google Scholar] [CrossRef]
- Xie, T.; Zhang, Q.; Zhang, W.; Feng, S.; Lin, J. Inkjet-patterned microdroplets as individual microenvironments for adherent single cell culture. Small 2022, 18, 2107992. [Google Scholar] [CrossRef]
- Yan, S.; Yuan, D. Continuous microfluidic 3D focusing enabling microflow cytometry for single-cell analysis. Talanta 2021, 221, 121401. [Google Scholar] [CrossRef]
- Yan, S.; Chen, P.; Zeng, X.; Zhang, X.; Li, Y.; Xia, Y.; Wang, J.; Dai, X.; Feng, X.; Du, W.; et al. Integrated multifunctional electrochemistry microchip for highly efficient capture, release, lysis, and analysis of circulating tumor cells. Anal. Chem. 2017, 89, 12039–12044. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Liu, B.F.; Li, J.; Liu, X. Advances in coupling microfluidic chips to mass spectrometry. Mass Spectrom. Rev. 2015, 34, 535–557. [Google Scholar] [CrossRef] [PubMed]
- Redman, E.A.; Ramos-Payan, M.; Mellors, J.S.; Ramsey, J.M. Analysis of hemoglobin glycation using microfluidic CE-MS: A rapid, mass spectrometry compatible method for assessing diabetes management. Anal. Chem. 2016, 88, 5324–5330. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, Y.Y.; Chan, K.; Hrudey, S.E.; Li, X.F.; Li, J. Analysis of nitrosamines by capillary electrospray-high-field asymmetric waveform ion mobility spectrometry-MS with programmed compensation voltage. Electrocatalysis 2007, 28, 1327–1334. [Google Scholar] [CrossRef]
- Deng, Y.; Henion, J.; Li, J.; Thibault, P.; Wang, C.; Harrison, D.J. Chip-based capillary electrophoresis/mass spectrometry determination of carnitines in human urine. Anal. Chem. 2001, 73, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, A.; Cortés-Salazar, F.; Gheorghiu, M.; Gáspár, S.; Momotenko, D.; Stanica, L.; Lesch, A.; Gheorghiu, E.; Girault, H.H. Electrochemical push–pull probe: From scanning electrochemical microscopy to multimodal altering of cell microenvironment. Anal. Chem. 2015, 87, 4479–4486. [Google Scholar] [CrossRef]
- Choi, J.; Song, H.; Sung, J.H.; Kim, D.; Kim, K. Microfluidic assay-based optical measurement techniques for cell analysis: A review of recent progress. Biosens. Bioelectron. 2016, 77, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Lu, Y.; Zhang, X.; Chen, M.; Wang, J. Recent advances in single-cell ultra-trace analysis. Trends Anal. Chem. 2020, 127, 115886. [Google Scholar] [CrossRef]
- Huang, Q.; Mao, S.; Khan, M.; Li, W.; Zhang, Q.; Lin, J. Single-cell identification by microfluidic-based in situ extracting and online mass spectrometric analysis of phospholipids expression. Chem. Sci. 2020, 11, 253–256. [Google Scholar] [CrossRef]
- Huo, D.; Liu, Z.; Hou, C.; Yang, J.; Luo, X.; Fa, H.; Dong, J.; Zhang, Y.; Zhang, G.; Li, J. Recent advances on optical detection methods and techniques for cell-based microfluidic systems. Chin. J. Anal. Chem. 2010, 38, 1357–1365. [Google Scholar] [CrossRef]
- Zhong, Q.; Huang, X.; Zhang, R.; Zhang, K.; Liu, B.F. Optical sensing strategies for probing single-cell secretion. ACS Sens. 2022, 7, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Long, Y. Confined nanopipette-a new microfluidic approach for single cell analysis. Trends Anal. Chem. 2019, 117, 39–46. [Google Scholar] [CrossRef]
- Xu, X.; Wang, J.; Wu, L.; Guo, J.; Song, Y.; Tian, T.; Wang, W.; Zhu, Z.; Yang, C. Microfluidic single-cell omics analysis. Small 2019, 16, 1903905. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Liu, Y.; Zhang, M.; Xu, X.; Chen, Y.; Zhang, H.; Yang, C. Microfluidic single-cell transcriptomics: Moving towards multimodal and spatiotemporal omics. Lab Chip 2021, 21, 3829–3849. [Google Scholar] [CrossRef]
- Zhou, W.; Yan, Y.; Guo, Q.; Ji, H.; Wang, H.; Xu, T.; Makabel, B.; Pilarsky, C.; He, G.; Yu, X.; et al. Microfluidics applications for high-throughput single cell sequencing. J. Nanobiotechnol. 2021, 19, 312. [Google Scholar] [CrossRef]
- Anggraini, D.; Ota, N.; Shen, Y.; Tang, T.; Tanaka, Y.; Hosokawa, Y.; Li, M.; Yalikun, Y. Recent advances in microfluidic devices for single-cell cultivation: Methods and applications. Lab Chip 2022, 22, 1438–1468. [Google Scholar] [CrossRef]
- Tavakoli, H.; Zhou, W.; Ma, L.; Perez, S.; Ibarra, A.; Xu, F.; Zhan, S.; Li, X. Recent advances in microfluidic platforms for single-cell analysis in cancer biology, diagnosis and therapy. Trends Anal. Chem. 2019, 117, 13–26. [Google Scholar] [CrossRef]
- Sung, J.; Kim, P.; Saga, S.; Hayashi, S.; Osuka, A.; Kim, D. S2 fluorescence dynamics of meso-aryl-substituted subporphyrins. Angew. Chem. Int. Ed. 2013, 52, 12632–12635. [Google Scholar] [CrossRef]
- Blake, R.C.; Iredale, T. Quenching of fluorescence by van der waals forces. Nature 1946, 3982, 228–229. [Google Scholar]
- Oleksiievets, N.; Sargsyan, Y.; Thiele, J.C.; Mougios, N.; Sograte-Idrissi, S.; Nevskyi, O.; Gregor, I.; Opazo, F.; Thoms, S.; Enderlein, J.; et al. Fluorescence lifetime DNA-PAINT for multiplexed super-resolution imaging of cells. Commun. Biol. 2022, 5, 38. [Google Scholar] [CrossRef]
- Fan, Y.; Dong, D.; Li, Q.; Si, H.; Pei, H.; Li, L.; Tang, B. Fluorescent analysis of bioactive molecules in single cells based on microfluidic chips. Lab Chip 2018, 18, 1151–1173. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Chen, P.; Dong, Y.; Feng, X.; Liu, B.F. Encapsulation of single cells on a microfluidic device integrating droplet generation with fluorescence-activated droplet sorting. Biomed. Microdevices 2013, 15, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Araki, S.; Nakano, M.; Tsugane, M.; Sunaga, F.; Hattori, M.; Nakano, M.; Nagai, T.; Suzuki, H. A simple microfluidic device for live-imaging of the vertical section of epithelial cells. Analyst 2020, 145, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, Y.; Hu, S.; Liu, Y.; Tsao, S.W.; Lau, D.; Luo, G.; Tsang, C.M.; Lam, R. Nondestructive quantification of single-cell nuclear and cytoplasmic mechanical properties based on large whole-cell deformation. Lab Chip 2020, 20, 4175–4185. [Google Scholar] [CrossRef]
- Sharma, B.; Frontiera, R.R.; Henry, A.; Ringe, E.; Van Duyne, R.P. SERS: Materials, applications, and the future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Fikiet, M.A.; Khandasammy, S.R.; Mistek, E.; Ahmed, Y.; Halámková, L.; Bueno, J.; Lednev, I.K. Surface enhanced raman spectroscopy: A review of recent applications in forensic science. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 197, 255–260. [Google Scholar] [CrossRef]
- Liebel, M.; Pazos-Perez, N.; van Hulst, N.F.; Alvarez-Puebla, R.A. Surface-enhanced raman scattering holography. Nat. Nanotechnol. 2020, 15, 1005–1011. [Google Scholar] [CrossRef]
- Li, L.; Lu, Y.; Qian, Z.; Yang, Z.; Yang, K.; Zong, S.; Wang, Z.; Cui, Y. Ultra-sensitive surface enhanced raman spectroscopy sensor for in-situ monitoring of dopamine release using zipper-like ortho-nanodimers. Biosens. Bioelectron. 2021, 180, 113100. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, L.; Yang, K.; Zong, S.; Wang, Z.; Cui, Y. 2D profiling of tumor chemotactic and molecular phenotype at single cell resolution using a SERS-microfluidic chip. Nano Res. 2022, 15, 4357–4365. [Google Scholar] [CrossRef]
- Preechaburana, P.; Gonzalez, M.C.; Suska, A.; Filippini, D. Surface plasmon resonance chemical sensing on cell phones. Angew. Chem. Int. Ed. 2012, 51, 11585–11588. [Google Scholar] [CrossRef]
- Su, Y.; Ke, Y.; Cai, S.; Yao, Q. Surface plasmon resonance of layer-by-layer gold nanoparticles induced photoelectric current in environmentally-friendly plasmon-sensitized solar cell. Light Sci. Appl. 2012, 1, e14. [Google Scholar] [CrossRef]
- Amendola, V.; Pilot, R.; Frasconi, M.; Maragò, O.M.; Iatì, M.A. Surface plasmon resonance in gold nanoparticles: A review. J. Phys. Condens. Matter 2017, 29, 203002. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Dong, T.; Wang, P.; Zhou, F. Microfluidically partitioned dual channels for accurate background subtraction in cellular binding studies by surface plasmon resonance microcopy. Anal. Chem. 2022, 94, 17303–17311. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, J.; Cai, C.; Yang, B.; Qi, Z.M. Flexible hyperspectral surface plasmon resonance microcopy. Nat. Commun. 2022, 13, 6475. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.L.; Yang, Y.; Wang, S.; Liu, X.W. Surface plasmon resonance microscopy: From single-molecule sensing to single-cell imaging. Angew. Chem. Int. Ed. 2019, 132, 1792–1801. [Google Scholar] [CrossRef]
- Chang, C.; Lin, H.; Lai, M.; Shieh, T.; Peng, C.; Shih, M.; Tung, Y. Flexible localized surface plasmon resonance sensor with metal–insulator–metal nanodisks on PDMS substrate. Sci. Rep. 2018, 8, 11812. [Google Scholar] [CrossRef]
- Zuo, C.; Li, J.; Sun, J.; Fan, Y.; Zhang, J.; Lu, L.; Zhang, R.; Wang, B.; Huang, L.; Chen, Q. Transport of intensity equation: A tutorial. Opt. Lasers Eng. 2020, 135, 106187. [Google Scholar] [CrossRef]
- Saha, S.K. Modern optical astronomy: Technology and impact of interferometry. Rev. Mod. Phys. 2002, 74, 551–599. [Google Scholar] [CrossRef][Green Version]
- Ricchiuti, G.; Dabrowska, A.; Pinto, D.; Ramer, G.; Lendl, B. Dual-beam photothermal spectroscopy employing a Mach-Zehnder interferometer and an external cavity quantum cascade laser for detection of water traces in organic solvents. Anal. Chem. 2022, 94, 16353–16360. [Google Scholar] [CrossRef]
- Alexander, G. Bose–Einstein and Fermi–Dirac interferometry in particle physics. Rep. Prog. Phys. 2003, 66, 481–522. [Google Scholar] [CrossRef]
- Hou, Z.S.; Sun, Y.L.; Li, Q.S.; Fan, X.; Cheng, R. Smart bio-gel optofluidic Mach-Zehnder interferometers multiphoton-lithographically customized with chemo-mechanical-opto transduction and bio-triggered degradation. Lab Chip 2020, 20, 3815–3823. [Google Scholar] [CrossRef]
- Ahmad, A.; Dubey, V.; Singh, V.R.; Tinguely, J.C.; Oie, C.I.; Wolfson, D.L.; Mehta, D.S.; So, P.; Ahluwalia, B.S. Quantitative phase microscopy of red blood cells during planar trapping and propulsion. Lab Chip 2018, 18, 3025–3036. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chen, P.; Gao, Z.; Li, Y.; Li, S.; Feng, X.; Liu, B.F. A time-coded multi-concentration microfluidic chemical waveform generator for high-throughput probing suspension single-cell signaling. Chin. Chem. Lett. 2022, 33, 3091–3096. [Google Scholar] [CrossRef]
- Guo, Y.; Gao, Z.; Liu, Y.; Li, S.; Zhu, J.; Chen, P.; Liu, B.F. Multichannel synchronous hydrodynamic gating coupling with concentration gradient generator for high-throughput probing dynamic signaling of single cells. Anal. Chem. 2020, 92, 12062–12070. [Google Scholar] [CrossRef]
- Feng, S.; Zhang, Q.; Xie, T.; Hou, Y.; Lin, J. In-situ monitoring calcium signaling through tumor microtubes for single cell-cell communication via an open microfluidic probe. Biosens. Bioelectron. 2022, 206, 114137. [Google Scholar] [CrossRef]
- Lee, B.; Jeong, S.G.; Jin, S.H.; Mishra, R.; Peter, M.; Lee, C.S.; Lee, S.S. Quantitative analysis of yeast MAPK signaling networks and crosstalk using a microfluidic device. Lab Chip 2020, 20, 2646–2655. [Google Scholar] [CrossRef] [PubMed]
- Pratt, S.L.; Zath, G.K.; Akiyama, T.; Williamson, K.S.; Franklin, M.J.; Chang, C.B. DropSOAC: Stabilizing microfluidic drops for time-lapse quantification of single-cell bacterial physiology. Front. Microbiol. 2019, 10, 2112. [Google Scholar] [CrossRef]
- Mandracchia, B.; Son, J.; Jia, S. Super-resolution optofluidic scanning microscopy. Lab Chip 2021, 21, 489–493. [Google Scholar] [CrossRef]
- Mathur, P.; Fomitcheva Khartchenko, A.; Stavrakis, S.; Kaigala, G.V.; DeMello, A.J. Quantifying antibody binding kinetics on fixed cells and tissues via fluorescence lifetime imaging. Anal. Chem. 2022, 94, 10967–10975. [Google Scholar] [CrossRef]
- Lee, D.; Li, X.; Ma, N.; Digman, M.A.; Lee, A.P. Rapid and label-free identification of single leukemia cells from blood in a high-density microfluidic trapping array by fluorescence lifetime imaging microscopy. Lab Chip 2018, 18, 1349–1358. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wu, L.; Zhu, D.; Cui, Y. SERS-activated platforms for immunoassay: Probes, encoding methods, and applications. Chem. Rev. 2017, 117, 7910–7963. [Google Scholar] [CrossRef] [PubMed]
- Joensson, H.N.; Andersson Svahn, H. Droplet microfluidics-a tool for single-cell Analysis. Angew. Chem. Int. Ed. 2012, 51, 12176–12192. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Cao, F.; Cong, L.; Xu, W.; Chen, Q.; Shi, W.; Xu, S. Cellular heterogeneity identified by single-cell alkaline phosphatase (ALP) via a SERRS-microfluidic droplet platform. Lab Chip 2019, 19, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Qi, G.; Du, X.; Yi, X.; Wang, M.; Zhu, H.; Sun, D. A SERS and fluorescence dual-channel microfluidic droplet platform for exploring telomerase activity at the single-cell level. Analyst 2022, 147, 5062–5067. [Google Scholar] [CrossRef]
- Cong, L.; Liang, L.; Cao, F.; Sun, D.; Yue, J.; Xu, W.; Liang, C.; Xu, S. Distinguishing cancer cell lines at a single living cell level via detection of sialic acid by dual-channel plasmonic imaging and by using a SERS-microfluidic droplet platform. Microchim. Acta 2019, 186, 367. [Google Scholar] [CrossRef]
- Zhang, B.Y.; Yin, P.; Hu, Y.; Szydzik, C.; Khan, M.W.; Xu, K.; Thurgood, P.; Mahmood, N.; Dekiwadia, C.; Afrin, S.; et al. Highly accurate and label-free discrimination of single cancer cell using a plasmonic oxide-based nanoprobe. Biosens. Bioelectron. 2022, 198, 113814. [Google Scholar] [CrossRef]
- Sun, D.; Cao, F.; Tian, Y.; Li, A.; Xu, W.; Chen, Q.; Shi, W.; Xu, S. Label-free detection of multiplexed metabolites at single-cell level via a SERS-microfluidic droplet Platform. Anal. Chem. 2019, 91, 15484–15490. [Google Scholar] [CrossRef]
- Sugai, H.; Tomita, S.; Ishihara, S.; Yoshioka, K.; Kurita, R. Microfluidic sensing system with a multichannel surface plasmon resonance chip: Damage-free characterization of cells by pattern recognition. Anal. Chem. 2020, 92, 14939–14946. [Google Scholar] [CrossRef]
- Borile, G.; Rossi, S.; Filippi, A.; Gazzola, E.; Capaldo, P.; Tregnago, C.; Pigazzi, M.; Romanato, F. Label-free, real-time on-chip sensing of living cells via grating-coupled surface plasmon resonance. Biophys. Chem. 2019, 254, 106262. [Google Scholar] [CrossRef]
- Nootchanat, S.; Jaikeandee, W.; Yaiwong, P.; Lertvachirapaiboon, C.; Shinbo, K.; Kato, K.; Ekgasit, S.; Baba, A. Fabrication of miniature surface plasmon resonance sensor chips by using confined sessile drop technique. ACS Appl. Mater. Interfaces 2019, 11, 11954–11960. [Google Scholar] [CrossRef]
- Zeng, S.; Baillargeat, D.; Ho, H.P.; Yong, K.T. Nanomaterials enhanced surface plasmon resonance for biological and chemical sensing applications. Chem. Soc. Rev. 2014, 43, 3426–3452. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; He, J.; Verano, M.; Brimmo, A.T.; Glia, A.; Qasaimeh, M.A.; Chen, P.; Aleman, J.O.; Chen, W. An integrated adipose-tissue-on-chip nanoplasmonic biosensing platform for investigating obesity-associated inflammation. Lab Chip 2018, 18, 3550–3560. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Feng, H.; Wang, Y.; Zhou, C.; Jiang, W.; Zhong, M.; Zhou, J. Capture of red blood cells onto optical sensor for rapid ABO blood group typing and erythrocyte counting. Sens. Actuators B Chem. 2018, 262, 411–417. [Google Scholar] [CrossRef]
- Duval, D.; González-Guerrero, A.B.; Dante, S.; Osmond, J.; Monge, R.; Fernández, L.J.; Zinoviev, K.E.; Domínguez, C.; Lechuga, L.M. Nanophotonic lab-on-a-chip platforms including novel bimodal interferometers, microfluidics and grating couplers. Lab Chip 2012, 12, 1987. [Google Scholar] [CrossRef] [PubMed]
- Ferrier, G.A.; Romanuik, S.F.; Thomson, D.J.; Bridges, G.E.; Freeman, M.R. A microwave interferometric system for simultaneous actuation and detection of single biological cells. Lab Chip 2009, 9, 3406. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Zeng, X.; Gao, Y.; Li, H.; Kumar, S.; Gan, Q.; Cheng, X.; Bartoli, F.J. Intensity-modulated nanoplasmonic interferometric sensor for MMP-9 detection. Lab Chip 2019, 19, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Su, Y.; Wang, R.; Lin, X.; Jin, X.; Yang, H.; Du, W.; Shan, X.; Lv, W.; Huang, G. Single cell capture, isolation, and long-term in-situ imaging using quantitative self-interference spectroscopy. Cytom. A 2021, 99, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Fu, R.; Du, W.; Yang, H.; Ma, L.; Luo, X.; Wang, R.; Lin, X.; Jin, X.; Shan, X.; et al. Label-free and quantitative dry mass monitoring for single cells during in situ culture. Cells 2021, 10, 1635. [Google Scholar] [CrossRef]
- Zhang, M.; Huo, G.; Bao, J.; Markovic, T.; Dijck, P.V.; Nauwelaers, B. Microwave interferometric cytometry for signal analysis of single yeast cells. Chemosensors 2022, 10, 318. [Google Scholar] [CrossRef]
- Zaraee, N.; Kanik, F.E.; Bhuiya, A.M.; Gong, E.S.; Geib, M.T.; Lortlar Ünlü, N.; Ozkumur, A.Y.; Dupuis, J.R.; Ünlü, M.S. Highly sensitive and label-free digital detection of whole cell E. coli with interferometric reflectance imaging. Biosens. Bioelectron. 2020, 162, 112258. [Google Scholar] [CrossRef]

| Optical Method | Device | Analysis Category | Detection Instrument | Label-Free | Resolution | Ref |
|---|---|---|---|---|---|---|
| Fluorescence | PDMS chip | Shows intracellular organelle distribution | CLSM | No | High | [43] |
| PDMS chip | Probes dynamic signaling of single cells | CCD camera | No | General | [61] | |
| Open microfluidic probe | Monitors single-cell calcium signaling | FV 5000 Spinning disk | No | General | [62] | |
| PDMS chip | Determines single-cell bacterial physiology | CLSM | No | High | [67] | |
| 96-well plates | quantifies antibody binding kinetics on fixed cells | FLIM | No | High | [69] | |
| SERS | PDMS chip | 2D profiling of the chemotactic and molecular phenotypes of single tumor cells | Duo-Scan mode by 50× objective | No | General | [49] |
| Microfluidic droplet platform | Identifies cellular heterogeneity according to single-cell ALP expression | Horiba Jobin Yvon Aramis spectrometer | Yes | High | [73] | |
| Microfluidic droplet platform | Detects single-cell sialic acid to distinguish cancer cell lines | Confocal Raman microscope system | no | high | [75] | |
| Microfluidic chip | Discriminates single cancer cells | Raman spectrometer equipped with a 10× objective | yes | high | [76] | |
| Microfluidic droplet platform | Detects multiplexed metabolites at the single-cell level | confocal Raman system | yes | general | [77] | |
| SPR | PDMS chip containing Au films | Characterizes cultured cells by detection of cell-secreted molecules | SPR SS-1001 system | yes | general | [78] |
| Confined sessile drop platform | Detects human immunoglobulin G | A home-built SPR setup | no | general | [80] | |
| Microfluidic ‘adipose-tissue-on-chip’ platform | Multiplexed detection of cytokines secreted by the adipocytes and macrophages | A dark-field microscope with an EMCCD camera | yes | general | [82] | |
| Interferometry | PDMS chip with gold substrate | Detects MMP-9 secretion from human monocytic cells | Narrow-band LED source and a CCD camera | yes | general | [86] |
| PDMS chip | Single-cell capture, isolation, and long-term in situ monitoring | Inverted bright-field microscope with LED light source | yes | general | [87] | |
| PDMS chip with quartz substrate | Signal analysis of single yeast cells | Optical microscopy | yes | general | [89] | |
| IRIS chip with layered Si/SiO2 substrate | Detects individual E. coil | Single-particle interferometric reflectance microscope | yes | high | [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ou, X.; Chen, P.; Liu, B.-F. Optical Technologies for Single-Cell Analysis on Microchips. Chemosensors 2023, 11, 40. https://doi.org/10.3390/chemosensors11010040
Ou X, Chen P, Liu B-F. Optical Technologies for Single-Cell Analysis on Microchips. Chemosensors. 2023; 11(1):40. https://doi.org/10.3390/chemosensors11010040
Chicago/Turabian StyleOu, Xiaowen, Peng Chen, and Bi-Feng Liu. 2023. "Optical Technologies for Single-Cell Analysis on Microchips" Chemosensors 11, no. 1: 40. https://doi.org/10.3390/chemosensors11010040
APA StyleOu, X., Chen, P., & Liu, B.-F. (2023). Optical Technologies for Single-Cell Analysis on Microchips. Chemosensors, 11(1), 40. https://doi.org/10.3390/chemosensors11010040






