Screen-Printed Sensors Coated with Polyaniline/Molecularly Imprinted Polymer Membranes for the Potentiometric Determination of 2,4-Dichlorophenoxyacetic Acid Herbicide in Wastewater and Agricultural Soil
Abstract
1. Introduction
2. Experimental
2.1. Chemicals and Reagents
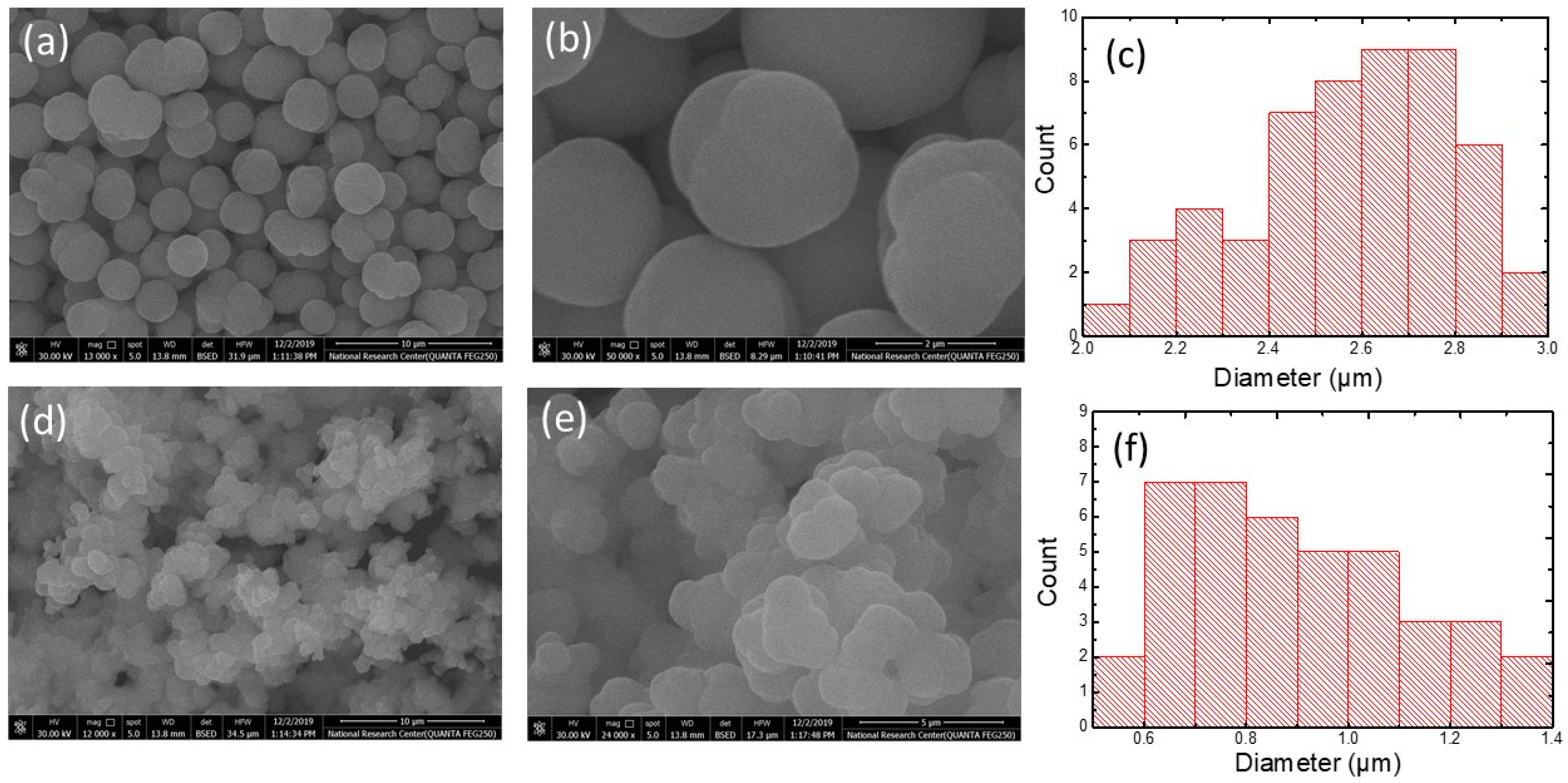
2.2. Preparation and Characterization of MIPs
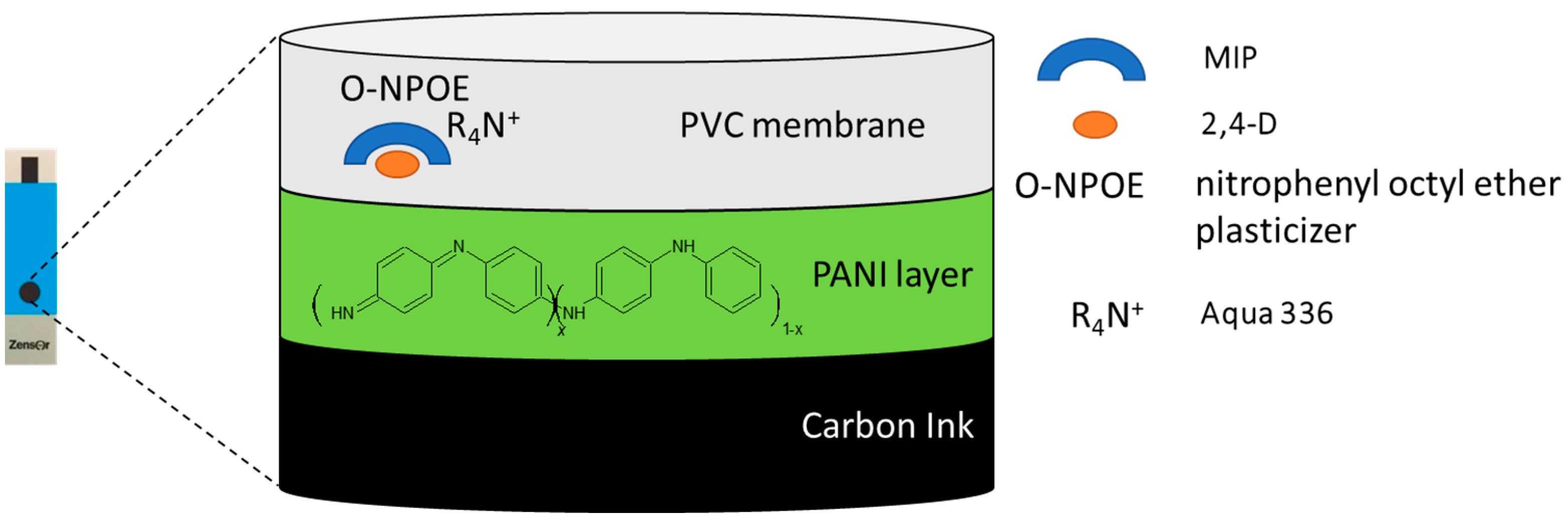
2.3. Preparation of Screen-Printed Electrode
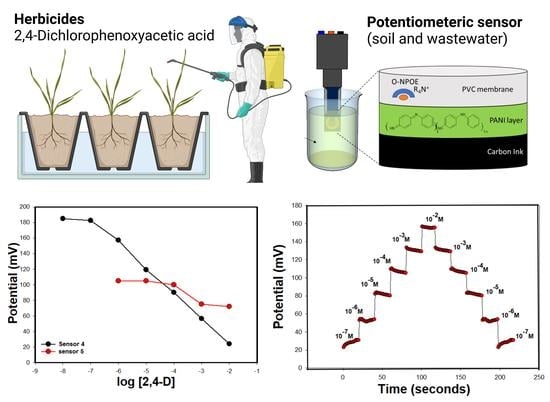
2.4. Potential Measurements
2.5. Water Layer Test and pH Effect
2.6. Selectivity against Interfering Ions
2.7. Analytical Applications and Sample Analysis
3. Results and Discussion
3.1. Molecularly Imprinted Polymers Characterization
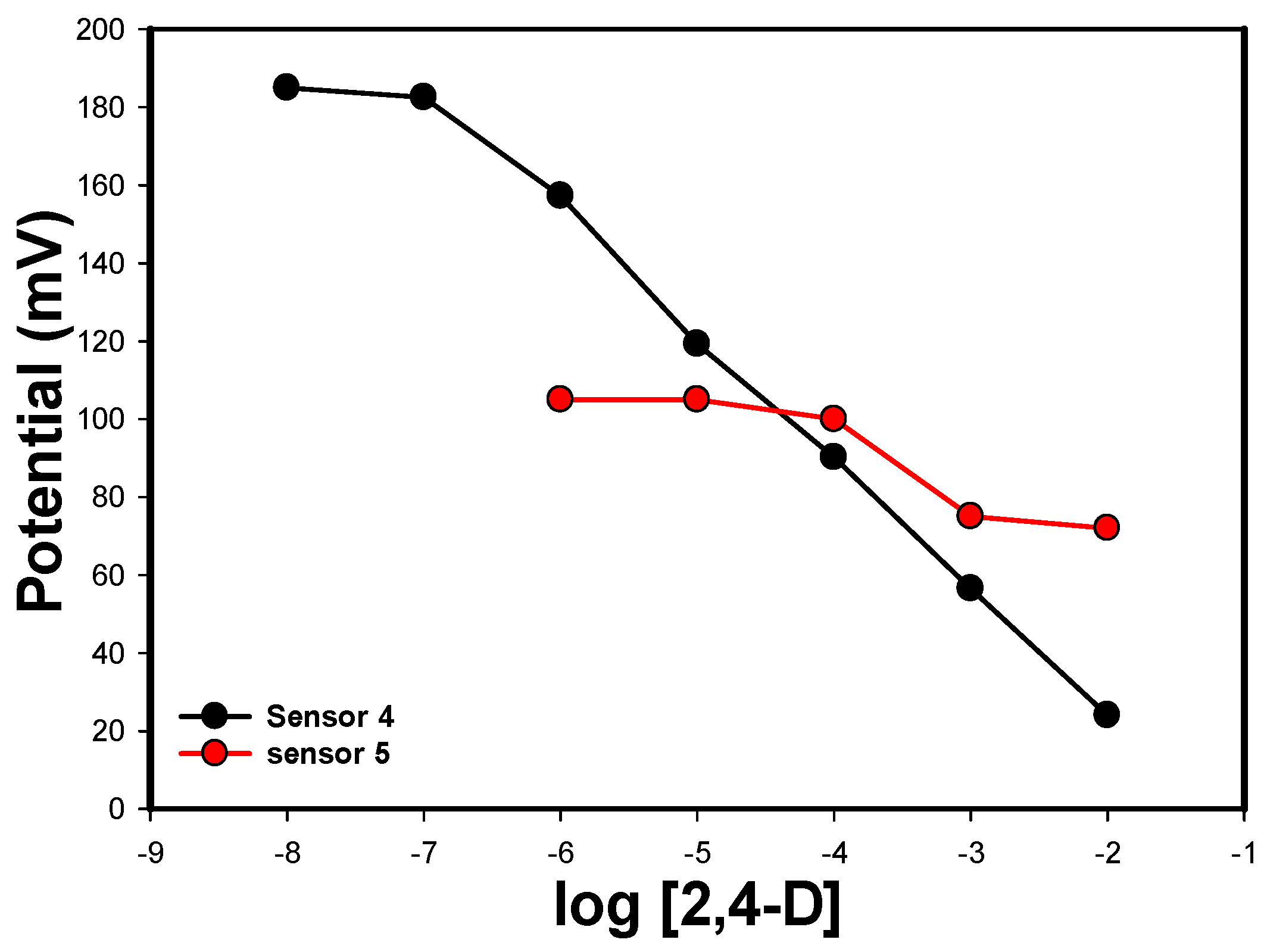
3.2. Characteristics of the Modified Screen-Printed Electrodes
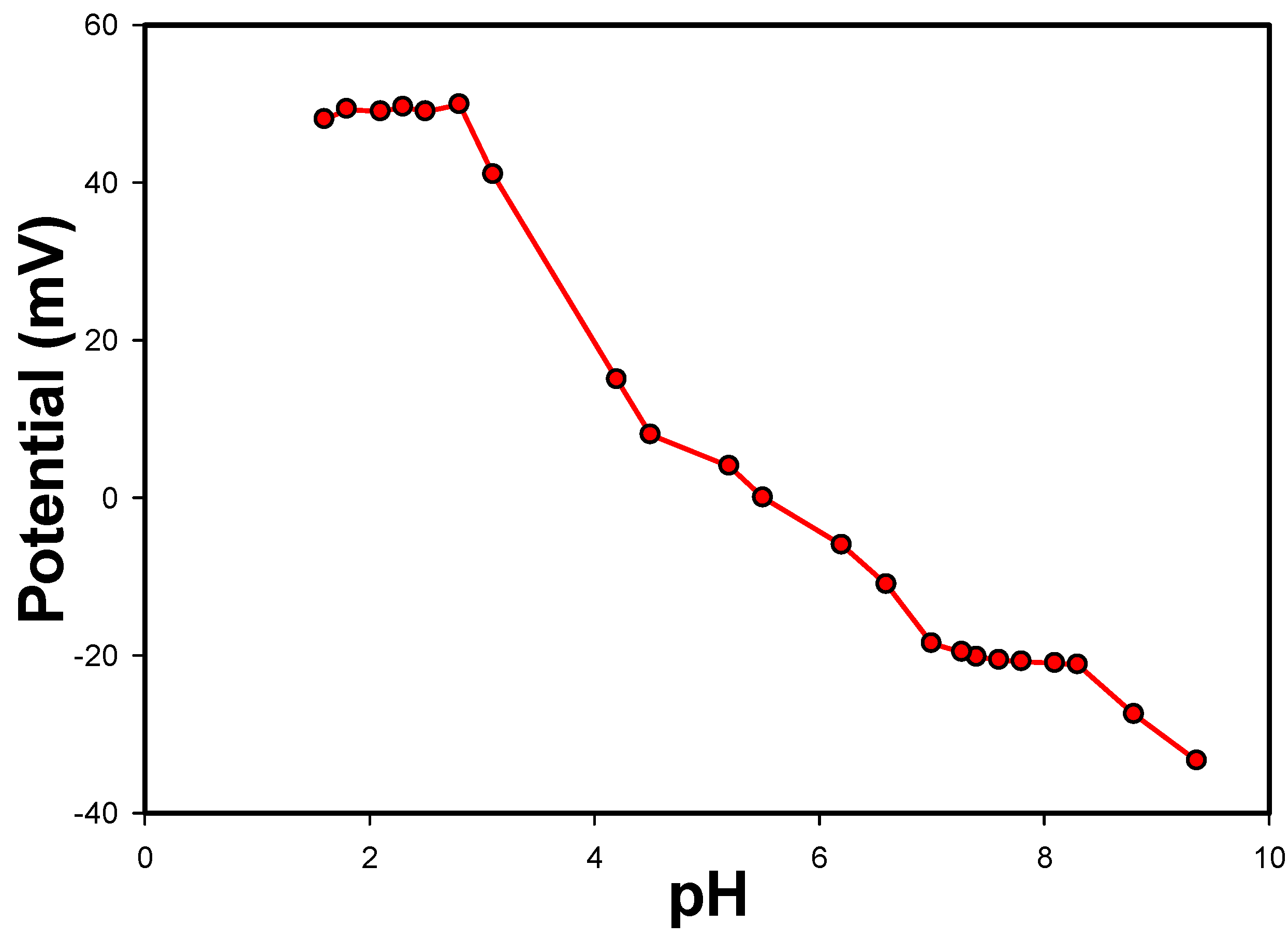
3.3. Effect of pH
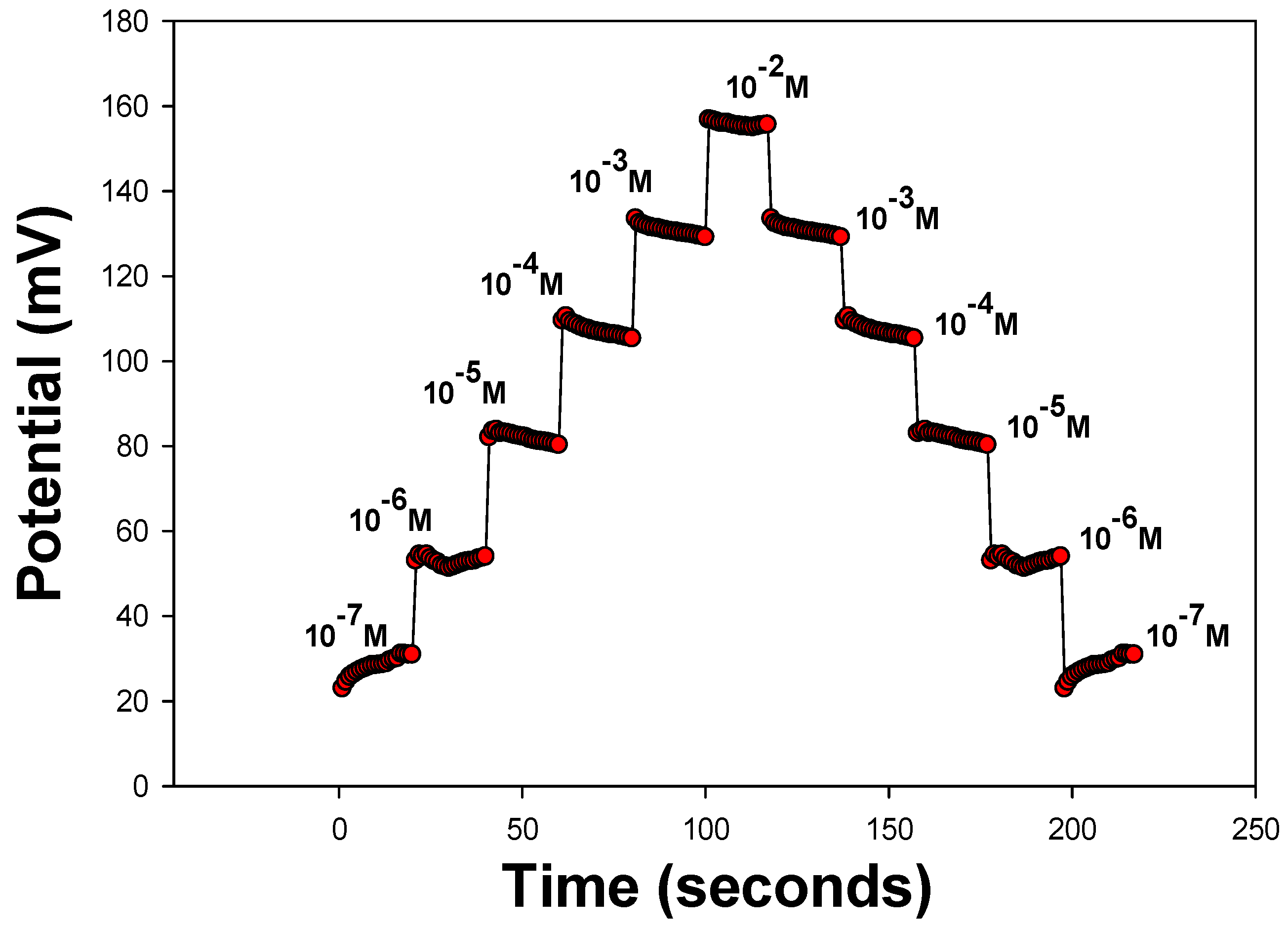
3.4. Stability of Sensor Response
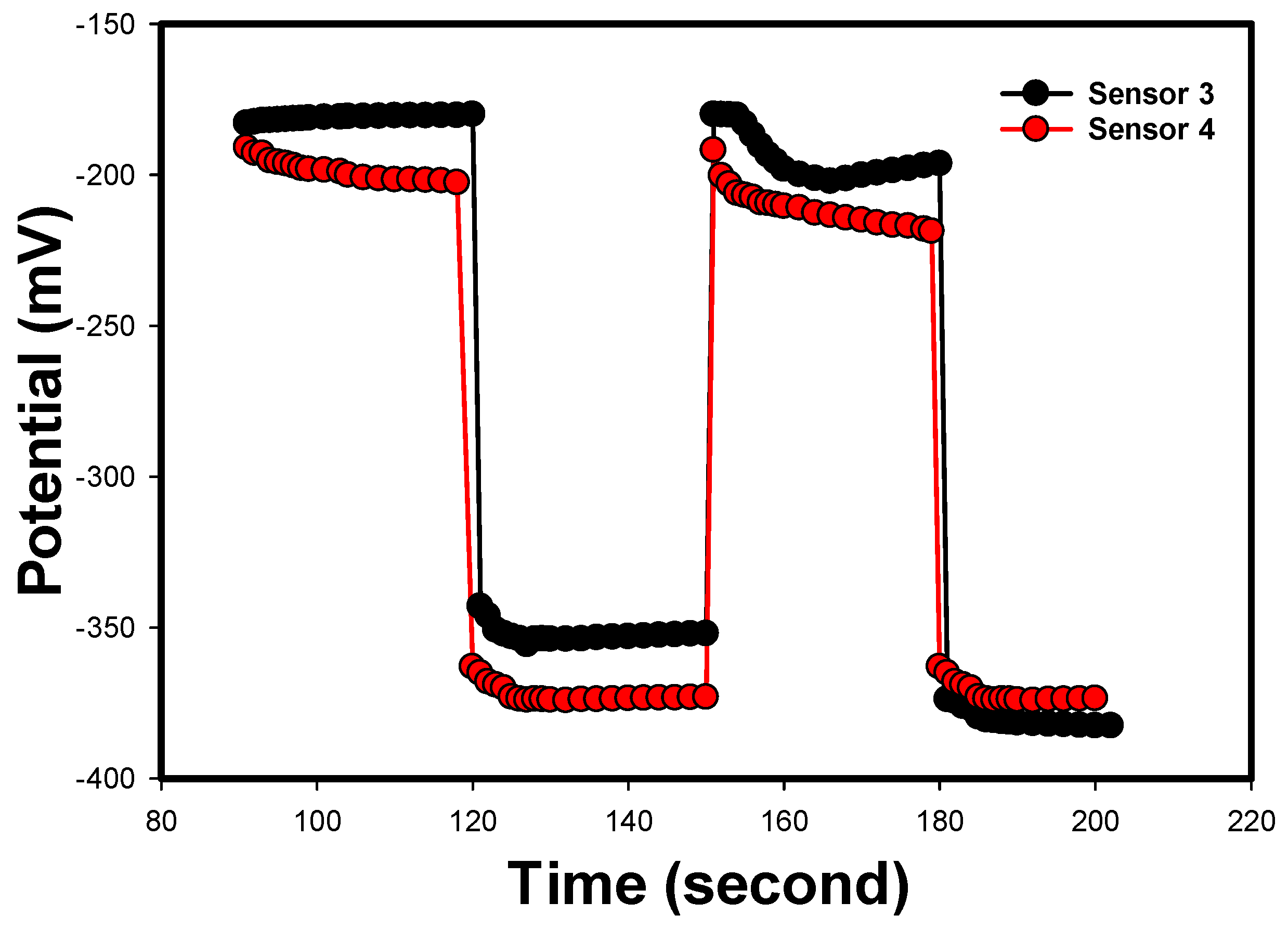
3.5. Reversibility, Response, Soaking, and Lifetimes
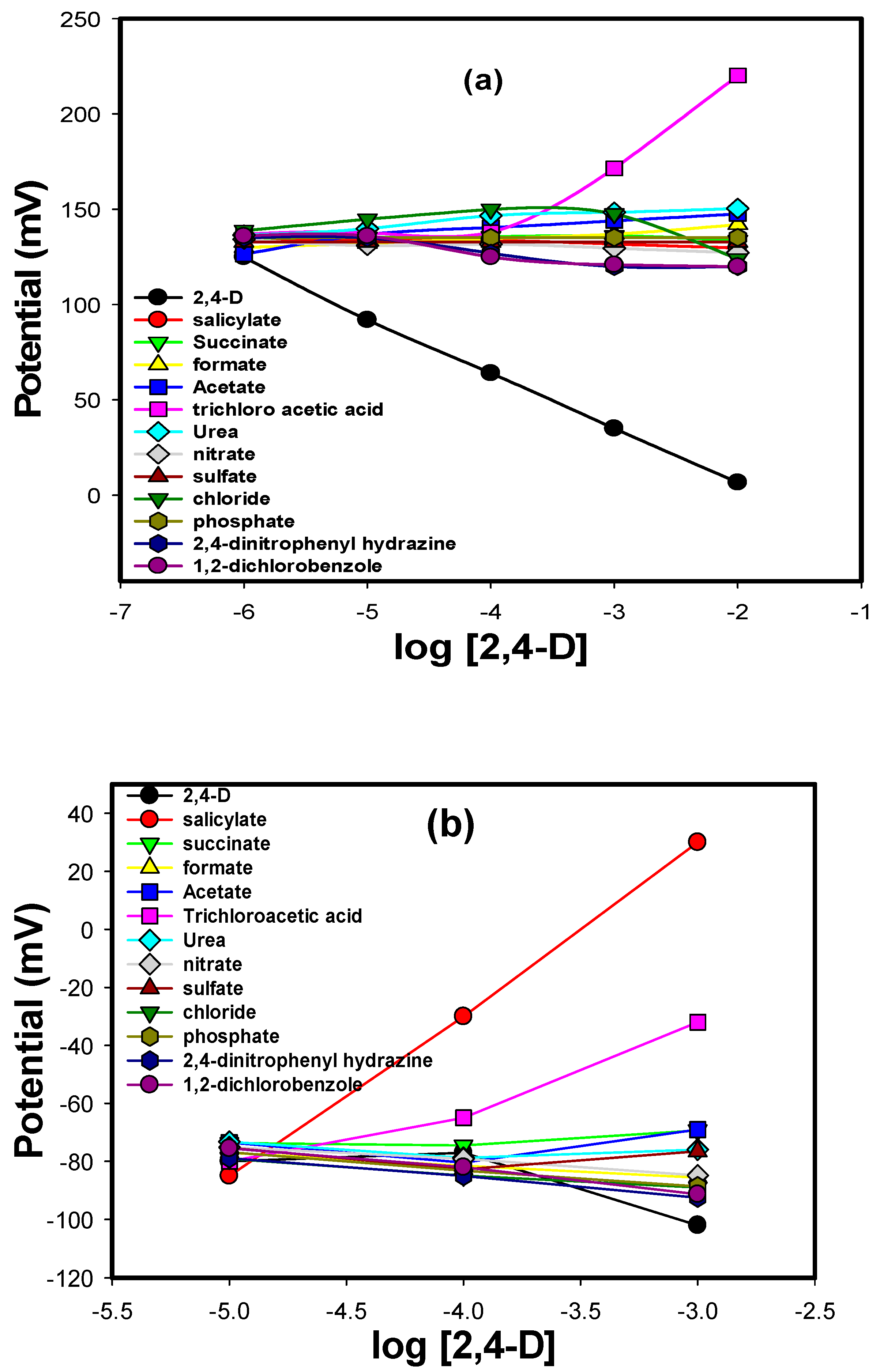
3.6. Interfering Species
3.7. Applications
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. The WHO Recommended Classification of Pesticides by Hazard and Guidelines to Classification 2009; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Tayeb, W.; Nakbi, A.; Trabelsi, M.; Miled, A.; Hammami, M. Biochemical and histological evaluation of kidney damage after sub-acute exposure to 2,4-dichlorophenoxyacetic herbicide in rats: Involvement of oxidative stress. Toxicol. Mech. Methods 2012, 22, 696–704. [Google Scholar] [CrossRef] [PubMed]
- By, A.; Edwards, D. Reregistration Eligibility Decision for 2,4-D, EPA 738-R-05-002; National Service Center for Environmental Publications (NSCEP): Cincinnati, OH, USA, 2005.
- Zhu, L.; Kee Lee, H. Field-Amplified Sample Injection Combined with Water Removal by Electroosmotic Flow Pump in Acidic Buffer for Analysis of Phenoxy Acid Herbicides by Capillary Electrophoresis. Anal. Chem. 2001, 73, 3065–3072. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.S. Determination of Phenoxy Acid Pesticides in Frog and Fish Tissues by Gas Chromatography-Mass Spectrometry. Chromatographia 2006, 63, 579–583. [Google Scholar] [CrossRef]
- Wu, J.; Kim, H.E.; Hian, K.L. Automated dynamic liquid–liquid–liquid microextraction followed by high-performance liquid chromatography-ultraviolet detection for the determination of phenoxy acid herbicides in environmental waters. J. Chromatogr. A 2005, 1082, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Gao, S.; Guo, Q.; Xu, K. Electrochemical sensor for 2,4-dichlorophenoxy acetic acid using molecularly imprinted polypyrrole membrane as recognition element. Microchim. Acta 2010, 169, 145–152. [Google Scholar] [CrossRef]
- Bakker, E.; Bühlmann, P.; Pretsch, E. Carrier-based ion-selective electrodes and bulk optodes. 1. General characteristics. Chem. Rev. 1997, 97, 3083–3132. [Google Scholar] [CrossRef]
- Labib, M.; Sargent, E.H.; Kelley, S.O. Electrochemical Methods for the Analysis of Clinically Relevant Biomolecules. Chem. Rev. 2016, 116, 9001–9090. [Google Scholar] [CrossRef]
- Belbruno, J.J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119, 94–119. [Google Scholar] [CrossRef]
- Hayat, A.; Marty, J.L. Disposable screen printed electrochemical sensors: Tools for environmental monitoring. Sensors 2014, 14, 10432–10453. [Google Scholar] [CrossRef]
- Wang, J.; Liang, R.; Qin, W. Molecularly imprinted polymer-based potentiometric sensors. TrAC—Trends Anal. Chem. 2020, 130, 115980. [Google Scholar] [CrossRef]
- El-Beshlawy, M.; Abdel-Haleem, F.; Barhoum, A. Molecularly Imprinted Polymer-Based Potentiometric Biosensor for Nanomolar Determination of Pioglitazone Hydrochloride in Pharmaceutical Formulations. Electroanalysis 2021, 33, 1244–1254. [Google Scholar] [CrossRef]
- Kröger, S.; Turner, A.P.F.; Mosbach, K.; Haupt, K. Imprinted Polymer-Based Sensor System for Herbicides Using Differential-Pulse Voltammetry on Screen-Printed Electrodes. Anal. Chem. 1999, 71, 3698–3702. [Google Scholar] [CrossRef] [PubMed]
- Kamel, A.H.; Amr, A.E.G.E.; Abdalla, N.S.; El-naggar, M.; Al-omar, M.A.; Almehizia, A.A. Modified Screen-Printed Potentiometric Sensors based on Man-Tailored Biomimetics for Diquat Herbicide Determination. Int. J. Environ. Res. Public Health 2020, 17, 1138. [Google Scholar] [CrossRef] [PubMed]
- Pringsheim, E.; Terpetschnig, E.; Wolfbeis, O.S. Optical sensing of pH using thin films of substituted polyanilines. Anal. Chim. Acta 1997, 357, 247–252. [Google Scholar] [CrossRef]
- Abdel-Haleem, F.M.; Saad, M.; Barhoum, A.; Bechelany, M.; Rizk, M.S. PVC membrane, coated-wire, and carbon-paste ion-selective electrodes for potentiometric determination of galantamine hydrobromide in physiological fluids. Mater. Sci. Eng. C 2018, 89, 140–148. [Google Scholar] [CrossRef]
- Abdel-Haleem, F.M.; Mahmoud, S.; Abdel-Ghani, N.E.T.; El Nashar, R.M.; Bechelany, M.; Barhoum, A. Polyvinyl chloride modified carbon paste electrodes for sensitive determination of levofloxacin drug in serum, urine, and pharmaceutical formulations. Sensors 2021, 21, 3150. [Google Scholar] [CrossRef]
- Kamel, A.H.; Amr, A.E.-G.E.; Abdalla, N.S.; El-Naggar, M.; Al-Omar, M.A.; Alkahtani, H.M.; Sayed, A.Y.A. Novel Solid-State Potentiometric Sensors Using Polyaniline (PANI) as A Solid-Contact Transducer for Flucarbazone Herbicide Assessment. Polymers 2019, 11, 1796. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, H.; Wan, Y.; Chen, X.; Li, Y. Determination of 2,4-Dichlorophenoxyacetic acid (2,4-D) in rat serum for pharmacokinetic studies with a simple HPLC method. PLoS ONE 2018, 13, e0191149. [Google Scholar] [CrossRef]
- Abdel-Haleem, F.M.; Rizk, M.S.; El-Beshlawy, M.M. Molecularly-imprinted polymer-base bulk optode for the determination of ivabradine hydrochloride in Procoralan®. RSC Adv. 2022, 12, 17645–17654. [Google Scholar] [CrossRef]
- Abdel-Haleem, F.M.; Salah, A.; Rizk, M.S.; Moustafa, H.; Bechelany, M.; Barhoum, A. Carbon-based Nanosensors for Salicylate Determination in Pharmaceutical Preparations. Electroanalysis 2019, 31, 778–789. [Google Scholar] [CrossRef]
- Abdel-Haleem, F.M.; Gamal, E.; Rizk, M.S.; Madbouly, A.; El Nashar, R.M.; Anis, B.; Elnabawy, H.M.; Khalil, A.S.G.G.; Barhoum, A. Molecularly Imprinted Electrochemical Sensor-Based Fe2O3@MWCNTs for Ivabradine Drug Determination in Pharmaceutical Formulation, Serum, and Urine Samples. Front. Bioeng. Biotechnol. 2021, 9, 648704. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, B.; Dhand, C.; Lakshminarayanan, R.; Dwivedi, N.; Mishra, S.; Solanki, P.; Venkatesh, M.; Beuerman, R.W.; Ramakrishna, S. Polyaniline-based biosensors. Nanobiosensors Dis. Diagnosis 2015, 4, 25–46. [Google Scholar] [CrossRef]
- Dhand, C.; Das, M.; Datta, M.; Malhotra, B.D. Recent advances in polyaniline based biosensors. Biosens. Bioelectron. 2011, 26, 2811–2821. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Food and Agriculture Organization of the United Nations. Pesticide residues in food 2006 Evaluations Part I-Residues; World Health Organization Food and Agriculture Organization of the United Nations: Rome, Italy, 2006. [Google Scholar]
- Nagy, G.; Nagy, L. Potentiometric Sensors. In Agricultural and Food Electroanalysis; Wiley Online Library: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]

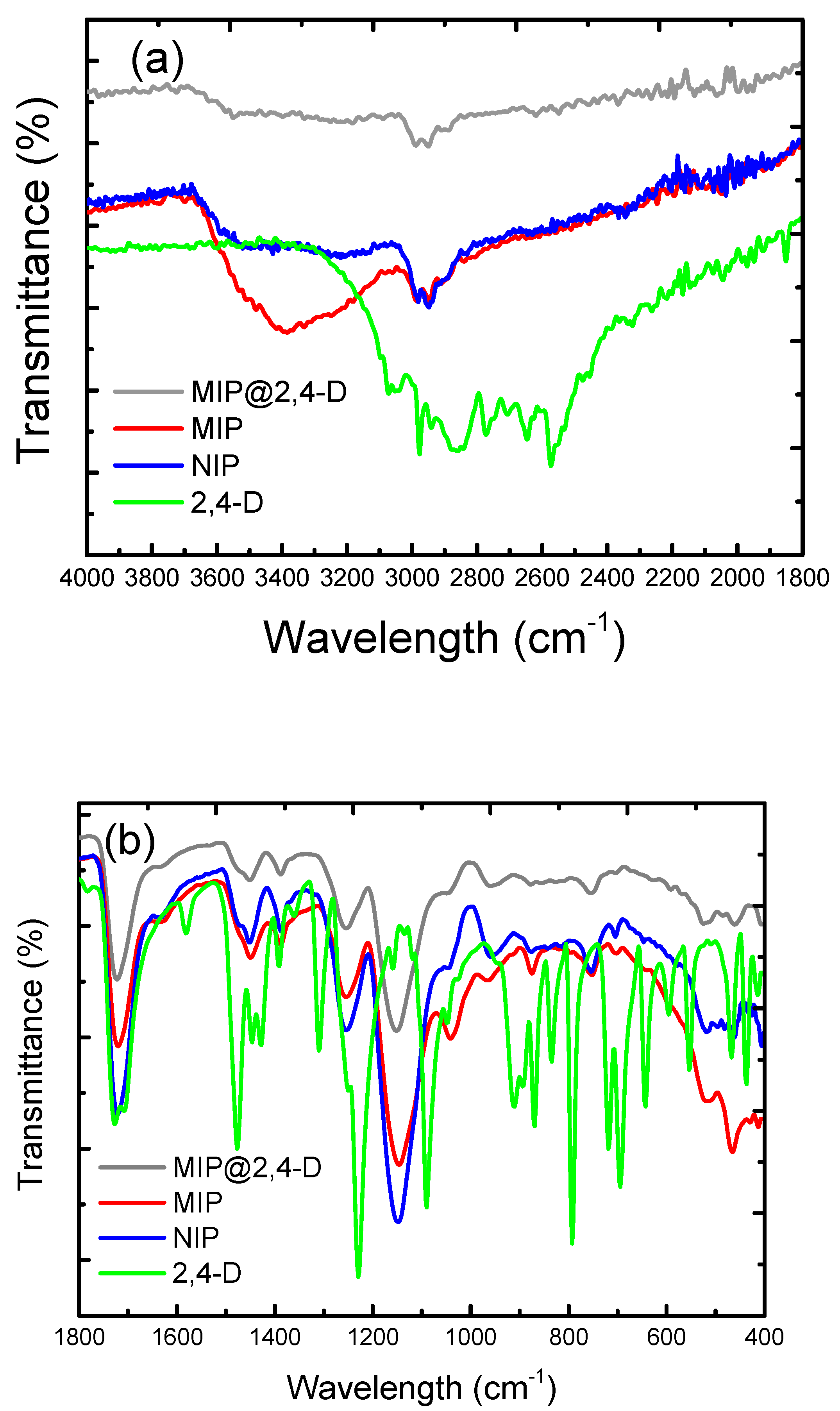
| Band Assignment | 2,4-D | MMA | EGDMA | 2,4-D@MIP | MIP | NIP |
|---|---|---|---|---|---|---|
| OH-stretching | - | - | - | 3443 | disappeared | disappeared |
| CAr-Cl stretching | 693 | - | - | 689 | disappeared | disappeared |
| CAr=CAr | 1435, 1475 | 1637 | 1637 | 1630 | - | - |
| C=O | 1732 | 1718 | 1720 | 1729 | - | - |
| C-O-C symmetric | 1089 | - | - | 1090 | disappeared | disappeared |
| No. | Solution | Membrane Composition (wt %) | Response Characteristics | |||||
|---|---|---|---|---|---|---|---|---|
| Recognition Element | Anion Exchanger | Plasticizer | Modifier | Slope | LDR, M | LOD, M | ||
| Sensor 1 | Water | 0.006 mg MIP | 1 mg TDMAC | 137 mg DOP | - | 26.4 | 10−4–10−5 | 1.0 × 10−5 |
| Sensor 2 | Water | 0.006 mg MIP | 1 mg Aliquate | 137 mg DOP | - | 26.0 | 10−2–10−5 | 1.0 × 10−5 |
| Sensor 3 | Water | 0.006 mg MIP | 6 mg Aliquate | 135 mg o-NPOE | - | 24.2 | 10−2–10−5 | 9.1 × 10−6 |
| Sensor 4 | Water | 0.006 mg MIP | 6 mg Aliquate | 135 mg o-NPOE | PANI | 29.9 | 10−2–10−6 | 8.8 × 10−7 |
| Sensor 4 | Tris | 0.006 mg MIP | 6 mg Aliquate | 135 mg o-NPOE | PANI | 32.9 | 10−2–10−7 | 6.1 × 10−8 |
| Sensor 5 | Tris | 0.006 mg NIP | 6 mg Aliquate | 135 mg o-NPOE | PANI | 25.8 | 10−3–10−4 | 1.0 × 10−4 |
| Selectivity Coefficients | MIP (Sensor 4) | NIP (Sensor 5) |
|---|---|---|
| 2,4 dinitrophenylhydrazine | −5.30 | −1.61 |
| Salicylate | −4.11 | −2.50 |
| Formate | −4.53 | −1.89 |
| Urea | −4.81 | −2.29 |
| Nitrate | −4.04 | −1.93 |
| Sulfate | −4.22 | −1.76 |
| Chloride | −3.92 | −1.78 |
| Phosphate | −4.29 | −1.77 |
| 1,2 dichlorobenzole | −3.10 | −1.66 |
| Trichloroacetic acid | −7.34 | −4.60 |
| Spiked Amount | Pure Sample Sensor 4 | Wastewater Sensor 4 | Real Soil Sample | |
|---|---|---|---|---|
| Sensor 4 | Reference Method [20] | |||
| 2.21 ppm | 2.21 (100.10%) | 2.13 (96.58%) | 2.29 (103.79%) | 2.22 ppm |
| 22.10 ppm | 22.17 (100.32%) | 25.01 (112.83%) | 24.37 (110.30%) | 22.19 ppm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Beshlawy, M.M.; Abdel-Haleem, F.M.; Kamel, A.H.; Barhoum, A. Screen-Printed Sensors Coated with Polyaniline/Molecularly Imprinted Polymer Membranes for the Potentiometric Determination of 2,4-Dichlorophenoxyacetic Acid Herbicide in Wastewater and Agricultural Soil. Chemosensors 2023, 11, 3. https://doi.org/10.3390/chemosensors11010003
El-Beshlawy MM, Abdel-Haleem FM, Kamel AH, Barhoum A. Screen-Printed Sensors Coated with Polyaniline/Molecularly Imprinted Polymer Membranes for the Potentiometric Determination of 2,4-Dichlorophenoxyacetic Acid Herbicide in Wastewater and Agricultural Soil. Chemosensors. 2023; 11(1):3. https://doi.org/10.3390/chemosensors11010003
Chicago/Turabian StyleEl-Beshlawy, Menna M., Fatehy M. Abdel-Haleem, Ayman H. Kamel, and Ahmed Barhoum. 2023. "Screen-Printed Sensors Coated with Polyaniline/Molecularly Imprinted Polymer Membranes for the Potentiometric Determination of 2,4-Dichlorophenoxyacetic Acid Herbicide in Wastewater and Agricultural Soil" Chemosensors 11, no. 1: 3. https://doi.org/10.3390/chemosensors11010003
APA StyleEl-Beshlawy, M. M., Abdel-Haleem, F. M., Kamel, A. H., & Barhoum, A. (2023). Screen-Printed Sensors Coated with Polyaniline/Molecularly Imprinted Polymer Membranes for the Potentiometric Determination of 2,4-Dichlorophenoxyacetic Acid Herbicide in Wastewater and Agricultural Soil. Chemosensors, 11(1), 3. https://doi.org/10.3390/chemosensors11010003