A Robust Discriminant Framework Based on Functional Biomarkers of EEG and Its Potential for Diagnosis of Alzheimer’s Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. EEG Data Acquisition
2.2. EEG Preprocessing
2.3. Maximal Overlap Discrete Wavelet Transform (MODWT)
2.4. Feature Extraction and Selection
- Variance (VA):
- Pearson correlation coefficient (PCC):
- Interquartile range (IQR):
- Hoeffding’s D measure (D):
- Permutation entropy (PE):
2.5. Classification and Validation
- Accuracy, or correct rate:
- Precision, or Positive Predictive Value:
- Recall, True Positive Rate (TPR) or sensitivity:
- Specificity, True Negative Rate (TNR):
- F-measure:
- Area under the curve (AUC) for the receiver operating characteristic curve:
3. Results
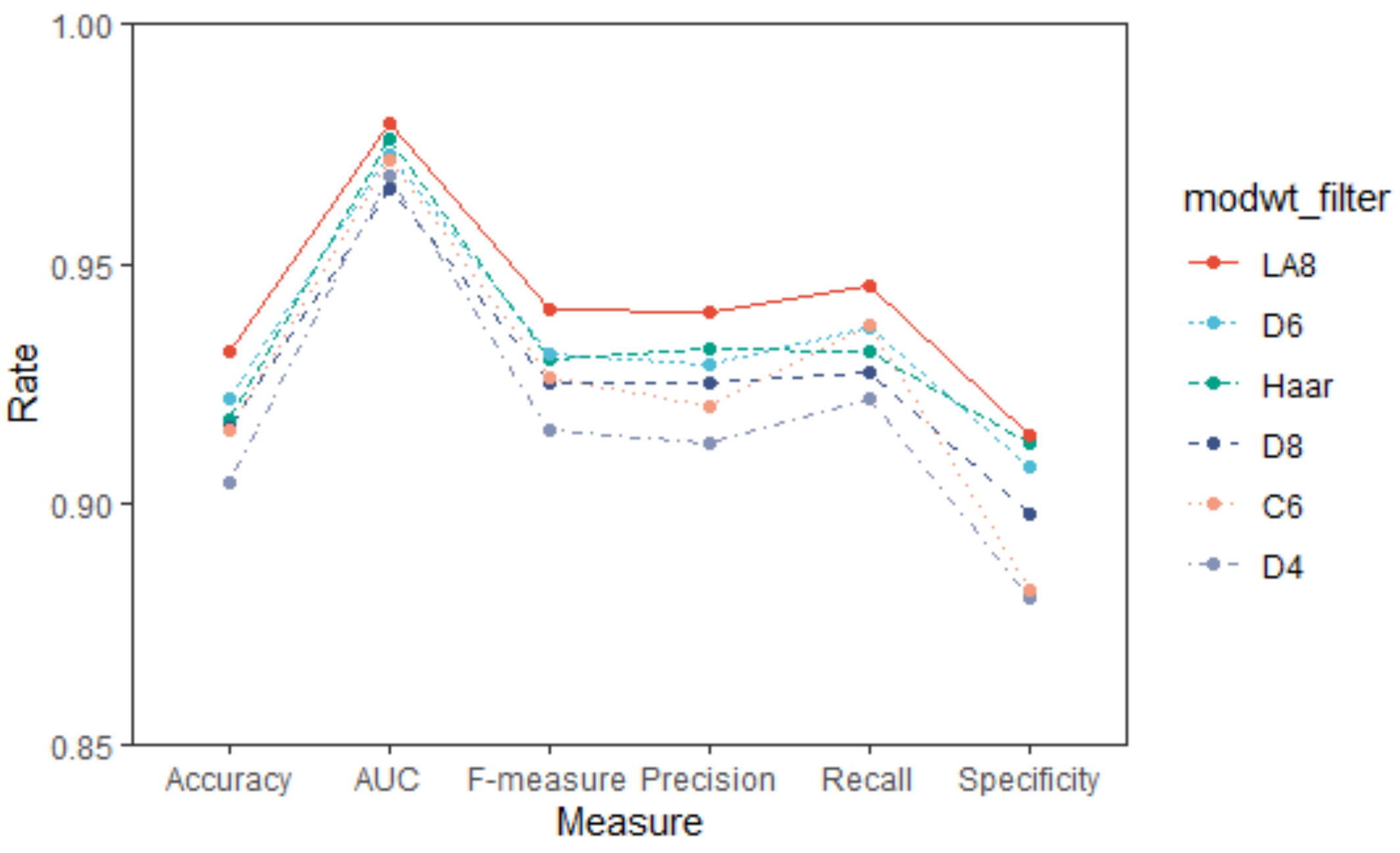
3.1. Selection of Wavelets Filters
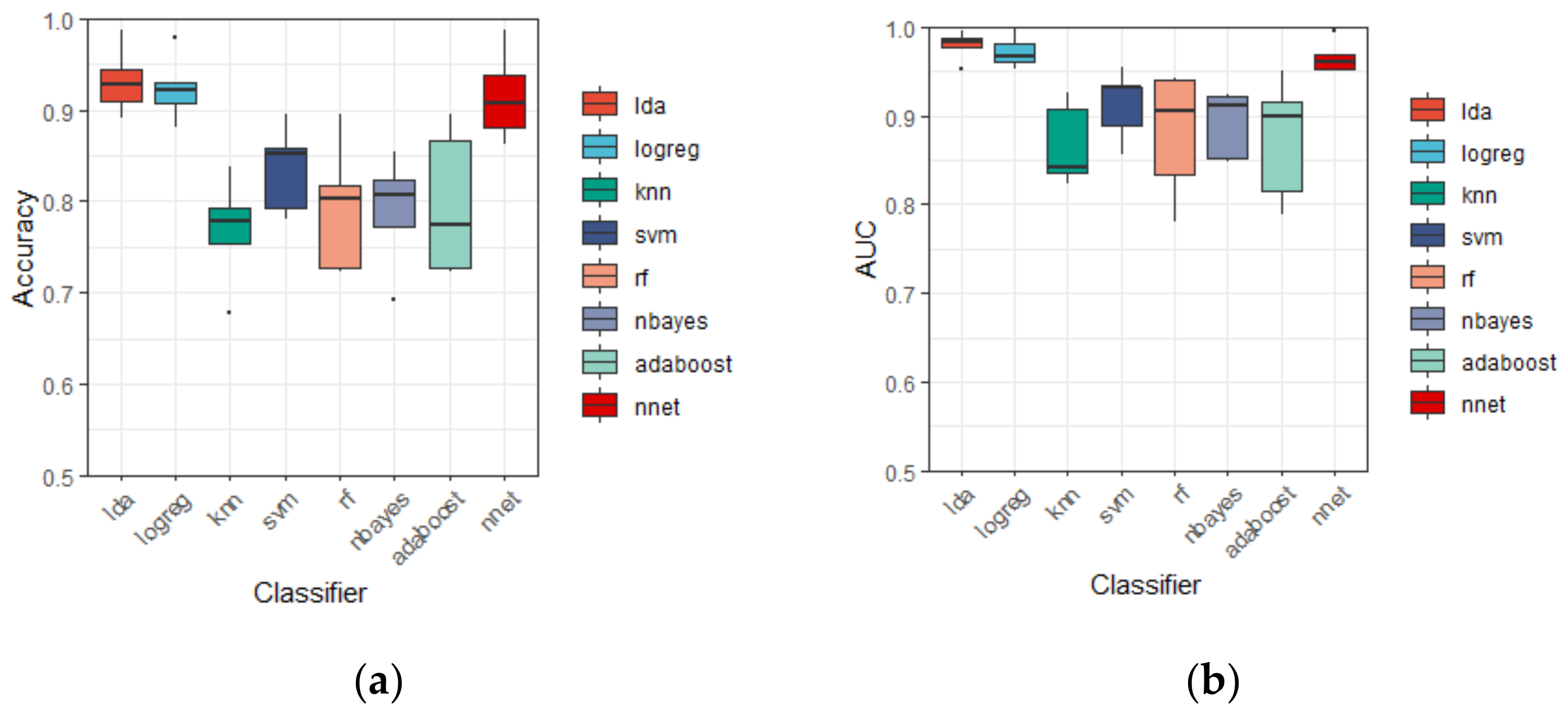
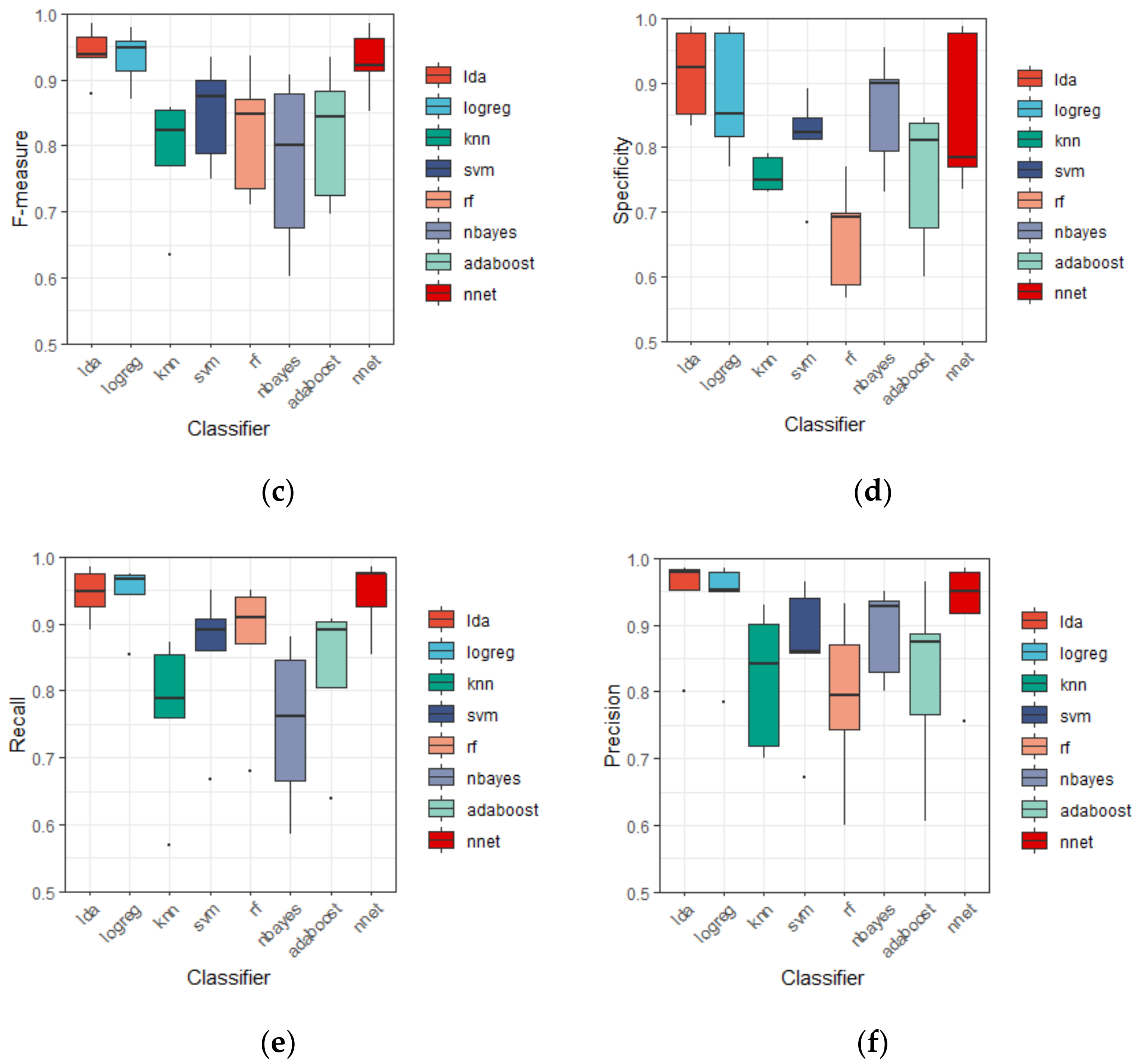
3.2. Discrinimant Performance of Different Classifiers
3.3. Discrinimant Performance of Different Features
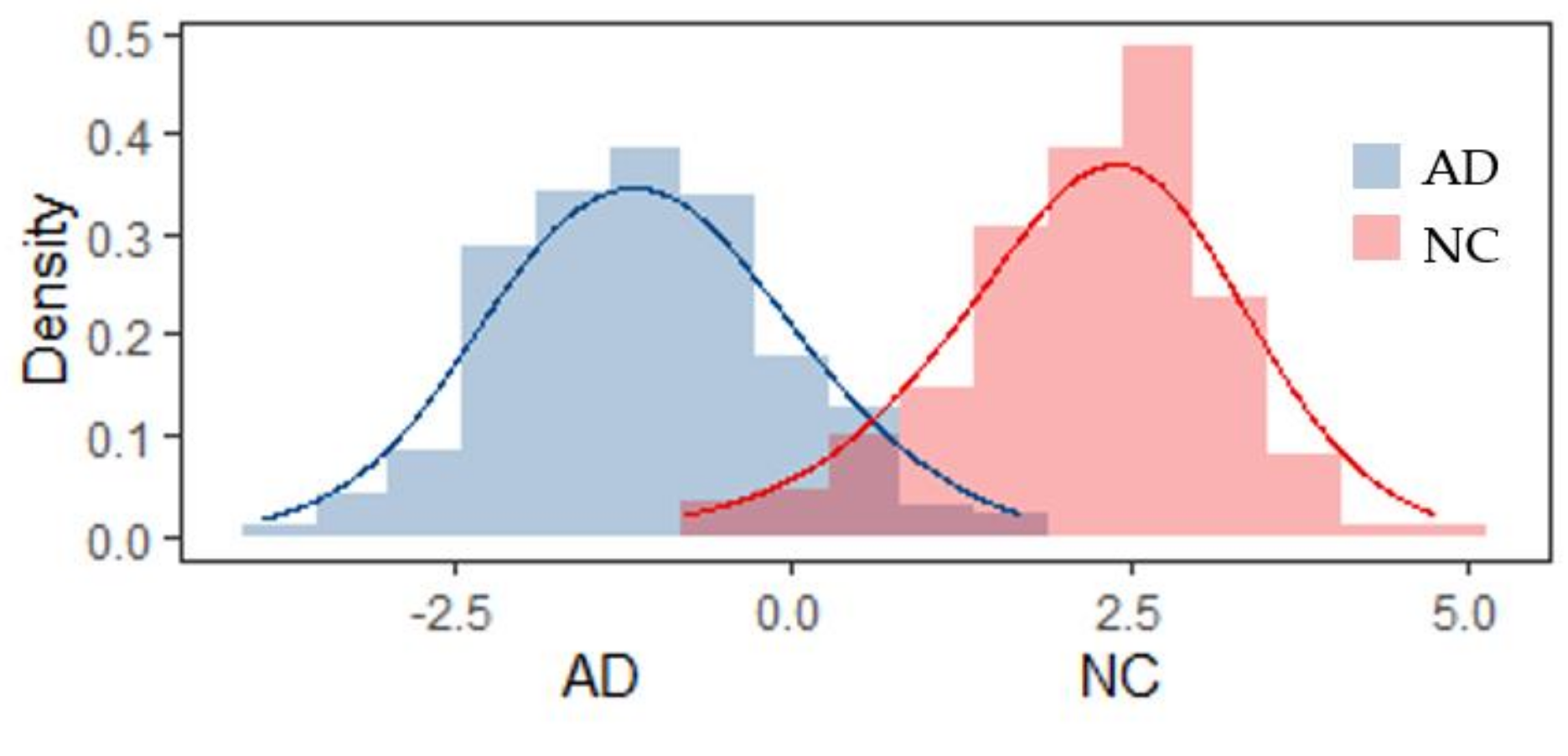
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alzheimer’s Association. 2020 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2020, 16, 391–460. [Google Scholar] [CrossRef] [PubMed]
- Winblad, B.; Amouyel, P.; Andrieu, S.; Ballard, C.; Brayne, C.; Brodaty, H.; Cedazo-Minguez, A.; Dubois, B.; Edvardsson, D.; Feldman, H.; et al. Defeating Alzheimer’s disease and other dementias: A priority for European science and society. Lancet Neurol. 2016, 15, 455–532. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Global Action Plan on the Public Health Response to Dementia 2017–2025; World Health Organization: Geneva, Switzerland, 2017; Volume 52. [Google Scholar]
- Morris, J.C. Early-stage and preclinical Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2005, 19, 163–165. [Google Scholar] [CrossRef]
- Gordon, B.A.; Blazey, T.M.; Su, Y.; Hari-Raj, A.; Dincer, A.; Flores, S.; Christensen, J.; McDate, E.; Qiao, G.W.; Jie, X.C.; et al. Spatial patterns of neuroimaging biomarker change in individuals from families with autosomal dominant Alzheimer’s disease: A longitudinal study. Lancet Neurol. 2018, 17, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Brooker, D.; Fontaine, J.L.; Evans, S.; Bray, J.; Saad, K. Public health guidance to facilitate timely diagnosis of dementia: Alzheimer’s cooperative valuation in Europe recommendations. Int. J. Geriatr. Psychiatr. 2014, 29, 682–693. [Google Scholar] [CrossRef]
- Alberdi, A.; Aztiria, A.; Basarab, A. On the early diagnosis of Alzheimer’s Disease from multimodal signals: A survey. Artif. Intell. Med. 2016, 71, 1–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Dubois, B.; Feldman, H.H.; Jacova, C.; Hampel, H.; Molinuevo, J.L.; Blennow, K.; DeKosky, S.T.; Gauthier, S.; Selkoe, D.; Bateman, R.; et al. Advancing research diagnostic criteria for Alzheimer’s disease: The IWG-2 criteria. Lancet Neurol. 2014, 13, 614–629. [Google Scholar] [CrossRef]
- Jack, C.R.; Albert, M.S.; Knopman, D.S.; McKhann, G.M.; Sperling, R.A.; Carrillo, M.C.; Thies, B.; Phelps, C.H. Introduction to the recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011, 7, 257–262. [Google Scholar] [CrossRef] [Green Version]
- Hampel, H.; Frank, R.; Broich, K.; Teipel, S.J.; Katz, R.G.; Hardy, J.; Herholz, K.; Bokde, A.L.; Jessen, F.; Hoessler, Y.C.; et al. Biomarkers for Alzheimer’s disease: Academic, industry and regulatory perspectives. Nat. Rev. Drug Discov. 2010, 9, 560–574. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Knopman, D.S.; Jagust, W.J.; Shaw, L.M.; Aisen, P.S.; Weiner, M.W.; Petersen, R.C.; Trojanowski, J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010, 9, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Laske, C.; Sohrabi, H.R.; Frost, S.M.; López-de-Ipiña, K.; Garrard, P.; Buscema, M.; Dauwels, J.; Soekadar, S.R.; Mueller, S.; Linnemann, C.; et al. Innovative diagnostic tools for early detection of Alzheimer’s disease. Alzheimer’s Dement. 2015, 11, 561–578. [Google Scholar] [CrossRef] [PubMed]
- Scheff, S.W.; Price, D.A.; Schmitt, F.A.; DeKosky, S.T.; Mufson, E.J. Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology 2007, 68, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- DeKosky, S.T.; Scheff, S.W. Synapse loss in frontal cortex biopsies in Alzheimer’s disease: Correlation with cognitive severity. Ann. Neurol. 1990, 27, 457–464. [Google Scholar] [CrossRef]
- Smailovic, U.; Jelic, V. Neurophysiological Markers of Alzheimer’s Disease: Quantitative EEG Approach. Neurol. Ther. 2019, 8, 37–55. [Google Scholar] [CrossRef] [Green Version]
- Cassani, R.; Estarellas, M.; San-Martin, R.; Fraga, F.J.; Falk, T.H. Systematic review on resting-state EEG for Alzheimer’s disease diagnosis and progression assessment. Dis. Markers 2018, 17, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, G.; Arnaldi, D.; Picco, A. Brain functional network in Alzheimer’s disease: Diagnostic markers for diagnosis and monitoring. Int. J. Alzheimer’s Dis. 2011, 2011, 481903. [Google Scholar] [CrossRef] [Green Version]
- Vecchio, F.; Babiloni, C.; Lizio, R.; Fallani, F.D.V.; Blinowska, K.; Verrienti, G.; Frisoni, G.; Rossini, P.M. Resting state cortical EEG rhythms in Alzheimer’s disease: Toward EEG markers for clinical applications: A review. Suppl. Clin. Neurophysiol. 2013, 62, 223–236. [Google Scholar] [CrossRef]
- Latchoumane, C.F.V.; Vialatte, F.; Cichocki, A.; Jeong, J. Multiway analysis of Alzheimer’s disease: Classification based on space-frequency characteristics of EEG time series. Proc. World Congr. Eng. 2008, 2, 2–4. [Google Scholar]
- Bi, X.; Wang, H. Early Alzheimer’s disease diagnosis based on EEG spectral images using deep learning. Neural. Netw. 2019, 114, 119–135. [Google Scholar] [CrossRef]
- Dauwels, J.; Vialatte, F.; Cichocki, A. Diagnosis of Alzheimer’s disease from EEG signals: Where are we standing. Curr. Alzheimer Res. 2010, 7, 487–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durongbhan, P.; Zhao, Y.; Chen, L.; Zis, P.; De Marco, M.; Unwin, Z.C.; Venneri, A.; He, X.; Li, S.; Zhao, Y.; et al. A dementia classification framework using frequency and time-frequency features based on EEG signals. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 826–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadlou, M.; Adeli, H.; Adeli, A. Fractality and a wavelet-chaos-methodology for EEG-based diagnosis of Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2011, 25, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Kolekar, M.H.; Jha, K.; Kumar, Y. EEG and cognitive biomarkers based mild cognitive impairment diagnosis. IRBM 2019, 40, 113–121. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, Z. EEG Signal Processing and Feature Extraction; Springer Nature Singapore Pte Ltd.: Singapore, 2019. [Google Scholar]
- Morabito, F.C.; Campolo, M.; Ieracitano, C.; Ebadi, J.M.; Bonanno, L.; Bramanti, A.; Desalvo, S.; Mammone, N.; Bramanti, P. Deep convolutional neural networks for classification of mild cognitive impaired and Alzheimer’s disease patients from scalp EEG recordings. In Proceedings of the 2016 IEEE 2nd International Forum on Research and Technologies for Society and Industry Leveraging a better tomorrow (RTSI), Bologna, Italy, 7–9 September 2016; pp. 1–6. [Google Scholar]
- Simpraga, S.; Alvarez-Jimenez, R.; Mansvelder, H.D.; Van Gerven, J.M.; Groeneveld, G.J.; Poil, S.S.; Linkenkaer-Hansen, K. EEG machine learning for accurate detection of cholinergic intervention and Alzheimer’s disease. Sci. Rep. 2017, 7, 5775. [Google Scholar] [CrossRef] [PubMed]
- Fiscon, G.; Weitschek, E.; Cialini, A.; Felici, G.; Bertolazzi, P.; De Salvo, S.; Bramanti, A.; Bramanti, P.; De Cola, M.C. Combining EEG signal processing with supervised methods for Alzheimer’s patients classification. BMC Med. Inform. Decis Mak. 2018, 18, 35. [Google Scholar] [CrossRef]
- Fiscon, G.; Weitschek, E.; Felici, G.; Bertolazzi, P.; De Salvo, S.; Bramanti, P.; De Cola, M.C. Alzheimer’s disease patients classification through EEG signals processing. In Proceedings of the 2014 IEEE Symposium on Computational Intelligence and Data Mining (CIDM), Orlando, FL, USA, 9–12 December 2014; pp. 105–112. [Google Scholar]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef] [Green Version]
- Ieracitano, C.; Mammone, N.; Bramanti, A.; Hussain, A.; Morabito, F.C. A convolutional neural network approach for classification of dementia stages based on 2D-spectral representation of EEG recordings. Neurocomputing 2019, 323, 96–107. [Google Scholar] [CrossRef]
- Mammone, N.; Bonanno, L.; Salvo, S.; Marino, S.; Bramanti, P.; Bramanti, A.; Morabito, F. Permutation Disalignment Index as an Indirect, EEG-Based, Measure of Brain Connectivity in MCI and AD Patients. Int. J. Neural Syst. 2017, 27, 1750020. [Google Scholar] [CrossRef]
- Bodenstein, G.; Praetorius, H.M. Feature extraction from the electroencephalogram by adaptive segmentation. Proc. IEEE 1977, 65, 642–652. [Google Scholar] [CrossRef]
- Sorokin, J. wICA MATLAB Toolkits. 2020. Available online: https://www.mathworks.com/matlabcentral/fileexchange/55413-wica-data-varargin (accessed on 10 November 2020).
- Cassani, R.; Falk, T.H.; Fraga, F.J.; Kanda, P.A.; Anghinah, R. The effects of automated artifact removal algorithms on electroencephalography-based Alzheimer’s disease diagnosis. Front. Aging Neurosci. 2014, 6, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nason, G.P.; Sachs, R.V. Wavelets in time series analysis. Philos Trans. R. Soc. Lond. A 1999, 357, 2511–2526. [Google Scholar] [CrossRef]
- Percival, D.B.; Walden, A.T. Wavelet Methods for Time Series Analysis; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Zhu, L.; Wang, Y.; Fan, Q. MODWT-ARMA model for time series prediction. Appl. Math. Model. 2014, 38, 1859–1865. [Google Scholar] [CrossRef]
- Alarcon-Aquino, V. Anomaly Detection and Prediction in Communication Networks Using Wavelet Transforms. Ph.D. Thesis, Imperial College London, University of London, London, UK, 2003. [Google Scholar]
- Strauss, R.E. Statistical Matlab Library; 2004. Available online: http://www.faculty.biol.ttu.edu/Strauss/Matlab/matlab.htm (accessed on 10 November 2020).
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond Accuracy, F-Score and ROC: A Family of Discriminant Measures for Performance Evaluation. In Australasian Joint Conference on Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2006; pp. 1015–1021. [Google Scholar]
- Schreiber, T.; Kantz, H. Nonlinear Time Series Analysis; Cambridge University Press: Cambridge, UK, 1997. [Google Scholar]
- Serroukh, A.; Walden, A.T. Wavelet scale analysisof bivariate time series ii: Statistical properties for linear processes. J. Nonparametr. Stat. 2000, 13, 37–56. [Google Scholar] [CrossRef]
- Houmani, N.; Dreyfus, G.; Vialatte, F.B. Epoch-based Entropy for Early Screening of Alzheimer’s Disease. Int. J. Neural Syst. 2015, 25, 1550032. [Google Scholar] [CrossRef] [PubMed]
- Kakizawa, Y.; Shumway, R.H.; Taniguchi, M. Discrimination and clustering for multivariate time series. J. Am. Stat. Assoc. 1998, 93, 328–340. [Google Scholar] [CrossRef]
- Discriminant Analysis of Multivariate Time Series Using Wavelets. Available online: https://e-archivo.uc3m.es/bitstream/handle/10016/13359/ws120603.pdf?sequence=1&isAllowed=y (accessed on 10 November 2020).
- Cornish, C. WMTSA Wavelet Toolkit for MATLAB. 2004. Available online: http://staff.washington.edu/dbp/wmtsa.html (accessed on 10 November 2020).


| Subjects | N | Sex/N (%) | Age/Years | ||
|---|---|---|---|---|---|
| Female | Male | Median (QL, QU) | Range | ||
| HC | 23 | 14 (60.9) | 9 (39.1) | 59 (50, 74) | (44, 83) |
| AD | 23 | 12 (52.2) | 11 (47.8) | 73 (56, 80) | (49, 85) |
| Wavelet Filters | Haar | D4 | D6 | D8 | LA8 | C6 |
|---|---|---|---|---|---|---|
| Maximum decompose layer | 10 | 8 | 7 | 7 | 7 | 7 |
| The number of layers used | 6 | 4 | 3 | 3 | 3 | 3 |
| The number of features extracted | 1148 | 1104 | 832 | 832 | 832 | 832 |
| Classifier | Accuracy | AUC | F-Measure | Specificity | Recall | Precision |
|---|---|---|---|---|---|---|
| LDA | 93.18 ± 3.65 | 97.92 ± 1.66 | 94.06 ± 4.04 | 91.45 ± 6.98 | 94.55 ± 3.85 | 94.02 ± 7.95 |
| Logreg | 92.44 ± 3.61 | 97.18 ± 1.77 | 93.36 ± 4.31 | 88.04 ± 9.71 | 94.26 ± 5.07 | 93.05 ± 8.32 |
| KNN | 76.76 ± 5.85 | 86.69 ± 4.61 | 78.79 ± 9.21 | 75.81 ± 2.76 | 76.86 ± 12.06 | 81.85 ± 10.45 |
| SVM | 83.56 ± 4.81 | 91.28 ± 3.99 | 84.92 ± 7.74 | 81.18 ± 7.78 | 85.53 ± 11.04 | 85.85 ± 11.43 |
| RF | 79.28 ± 7.18 | 88.05 ± 7.12 | 82.01 ± 9.49 | 66.30 ± 8.43 | 87.04 ± 11.07 | 78.78 ± 12.77 |
| Nbayes | 78.97 ± 6.22 | 89.12 ± 3.81 | 77.28 ± 13.11 | 85.68 ± 9.14 | 69.26 ± 16.99 | 88.88 ± 6.89 |
| Adaboost | 79.73 ± 8.03 | 87.40 ± 6.85 | 81.69 ± 10.21 | 75.41 ± 11.00 | 82.89 ± 11.40 | 82.00 ± 13.85 |
| NNet | 91.47 ± 4.93 | 96.60 ± 1.84 | 92.68 ± 5.18 | 85.02 ± 12.13 | 94.34 ± 5.49 | 91.76 ± 9.50 |
| Feature | Accuracy | AUC | F-Measure | Specificity | Recall | Precision |
|---|---|---|---|---|---|---|
| VA | 71.32 ± 7.25 | 84.13 ± 9.55 | 75.67 ± 8.57 | 70.04 ± 18.85 | 78.26 ± 12.57 | 77.32 ± 19.06 |
| PCC | 88.18 ± 5.28 | 95.65 ± 2.82 | 89.87 ± 3.99 | 86.67 ± 5.45 | 90.66 ± 7.79 | 89.89 ± 7.35 |
| IQR | 70.32 ± 9.38 | 77.20 ± 14.69 | 73.61 ± 13.37 | 59.51 ± 19.71 | 76.99 ± 17.65 | 73.26 ± 15.15 |
| D | 84.75 ± 3.77 | 92.58 ± 5.53 | 87.03 ± 2.81 | 85.18 ± 6.48 | 87.05 ± 7.38 | 88.02 ± 8.46 |
| PE | 57.09 ± 19.75 | 56.82 ± 21.36 | 59.99 ± 21.81 | 52.65 ± 23.68 | 61.36 ± 34.15 | 65.82 ± 18.38 |
| VA + PCC | 89.52 ± 6.32 | 96.43 ± 2.30 | 91.03 ± 5.74 | 87.80 ± 6.66 | 91.65 ± 6.44 | 90.98 ± 8.77 |
| IQR + HCC + PE | 88.83 ± 4.18 | 96.16 ± 2.29 | 89.97 ± 5.30 | 84.70 ± 4.31 | 92.45 ± 6.13 | 88.25 ± 9.06 |
| ALL | 93.18 ± 3.65 | 97.92 ± 1.66 | 94.06 ± 4.04 | 91.45 ± 6.98 | 94.55 ± 3.85 | 94.02 ± 7.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Q.; Lin, Z.-C.; Gao, Y.-X.; Zhang, J.-X. A Robust Discriminant Framework Based on Functional Biomarkers of EEG and Its Potential for Diagnosis of Alzheimer’s Disease. Healthcare 2020, 8, 476. https://doi.org/10.3390/healthcare8040476
Ge Q, Lin Z-C, Gao Y-X, Zhang J-X. A Robust Discriminant Framework Based on Functional Biomarkers of EEG and Its Potential for Diagnosis of Alzheimer’s Disease. Healthcare. 2020; 8(4):476. https://doi.org/10.3390/healthcare8040476
Chicago/Turabian StyleGe, Qi, Zhuo-Chen Lin, Yong-Xiang Gao, and Jin-Xin Zhang. 2020. "A Robust Discriminant Framework Based on Functional Biomarkers of EEG and Its Potential for Diagnosis of Alzheimer’s Disease" Healthcare 8, no. 4: 476. https://doi.org/10.3390/healthcare8040476
APA StyleGe, Q., Lin, Z.-C., Gao, Y.-X., & Zhang, J.-X. (2020). A Robust Discriminant Framework Based on Functional Biomarkers of EEG and Its Potential for Diagnosis of Alzheimer’s Disease. Healthcare, 8(4), 476. https://doi.org/10.3390/healthcare8040476





