Cancer Survival Data Representation for Improved Parametric and Dynamic Lifetime Analysis
Abstract
1. Motivation and Objective
2. Dynamic Lifetime Analysis
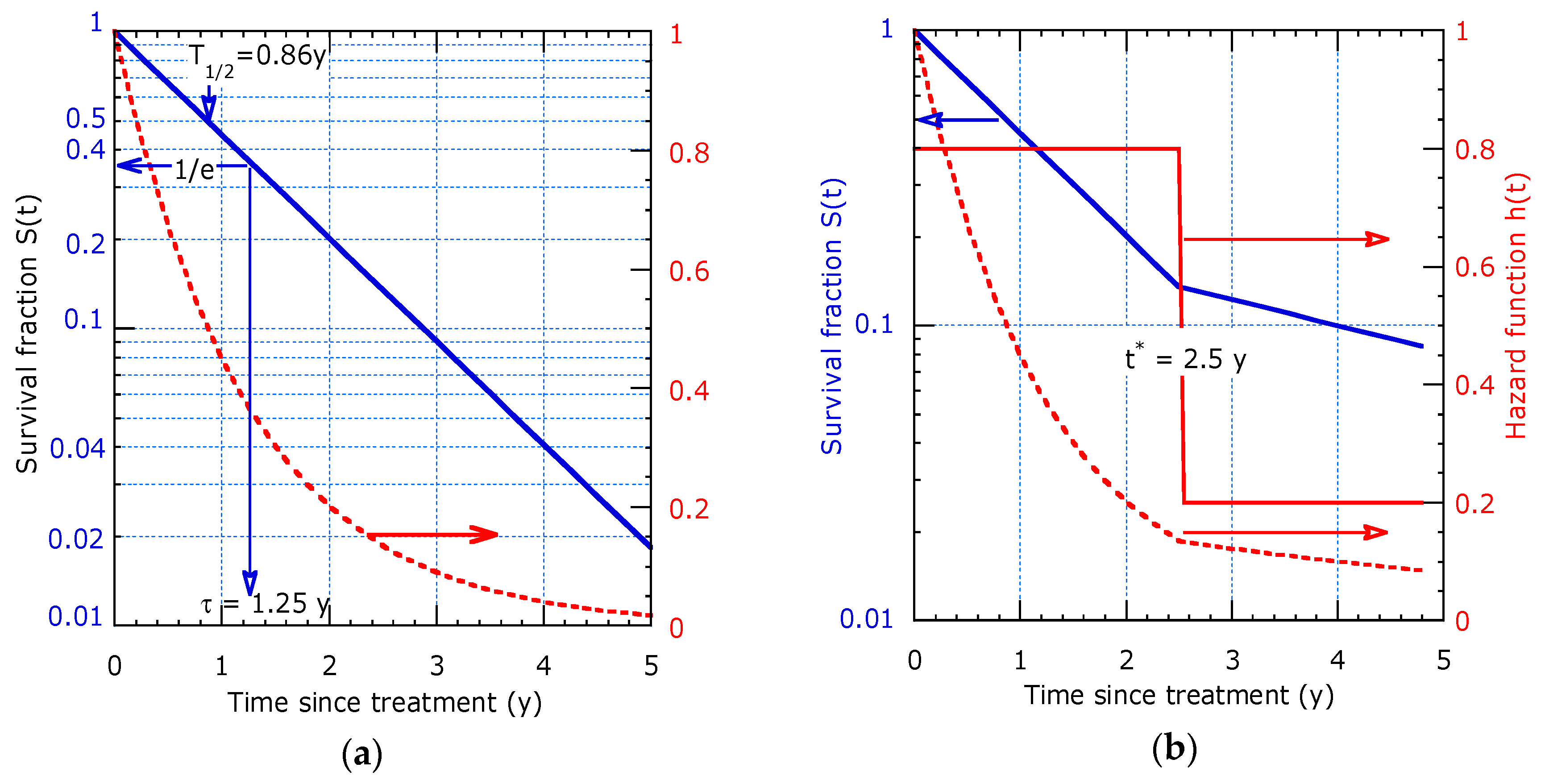
2.1. The Survival Function
2.2. Modeling of Dynamic Lifetime
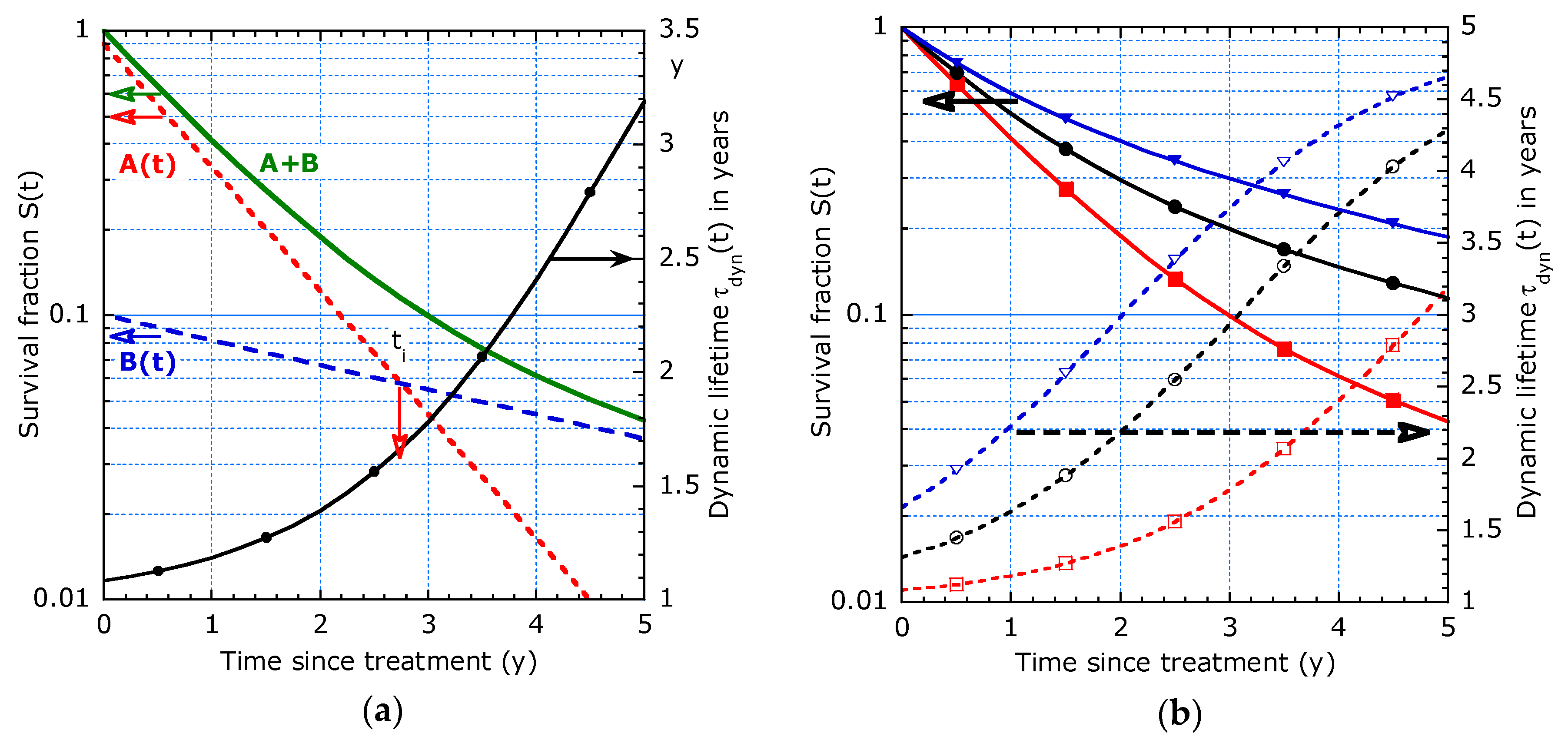
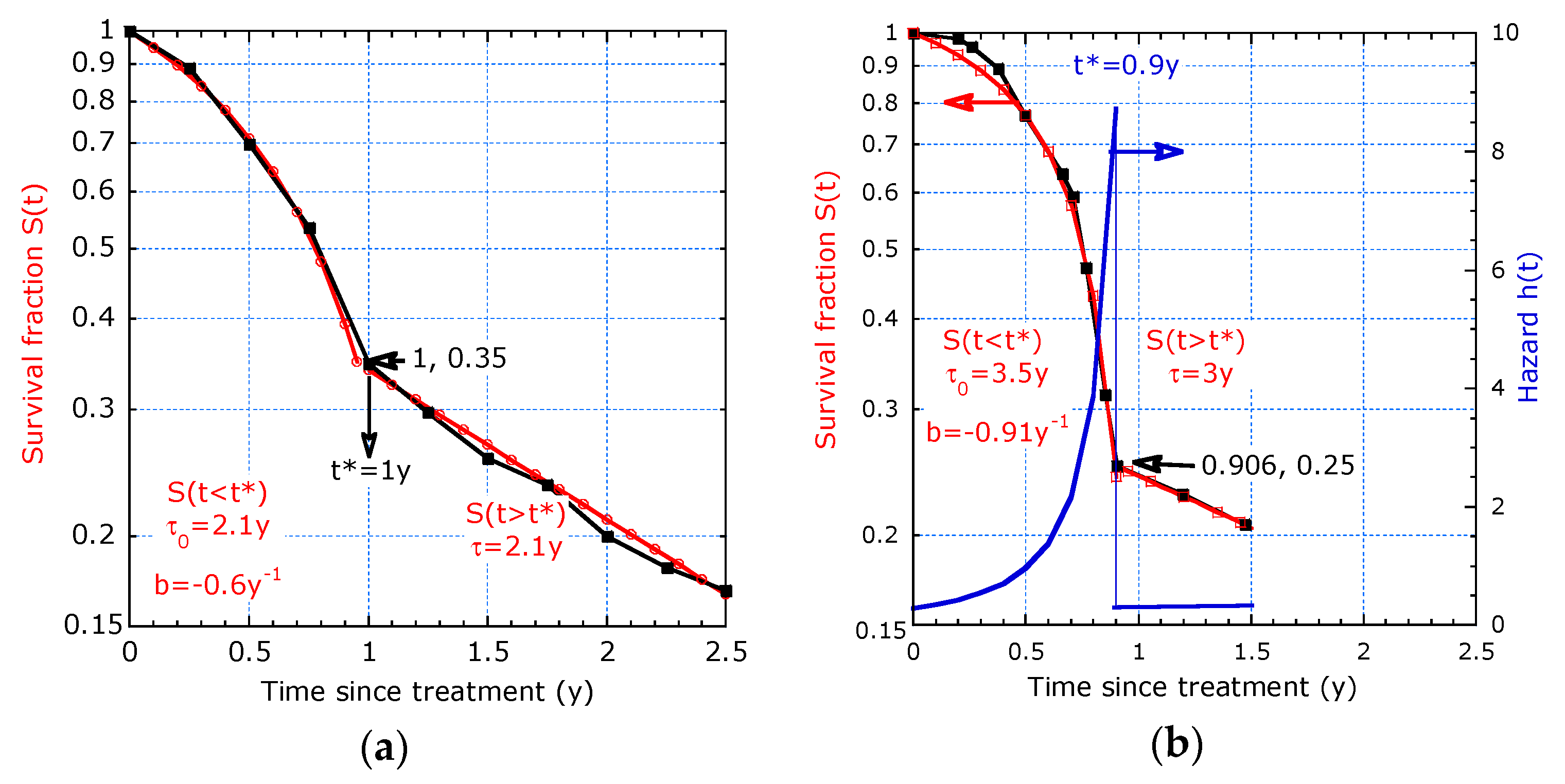
2.2.1. Model with Two Distinct Decay Times
2.2.2. Model with Continuous Evolving Decay Time
3. Re-Analysis of Cancer Survival Data from Literature
3.1. Case Studies from Literature
- 1.
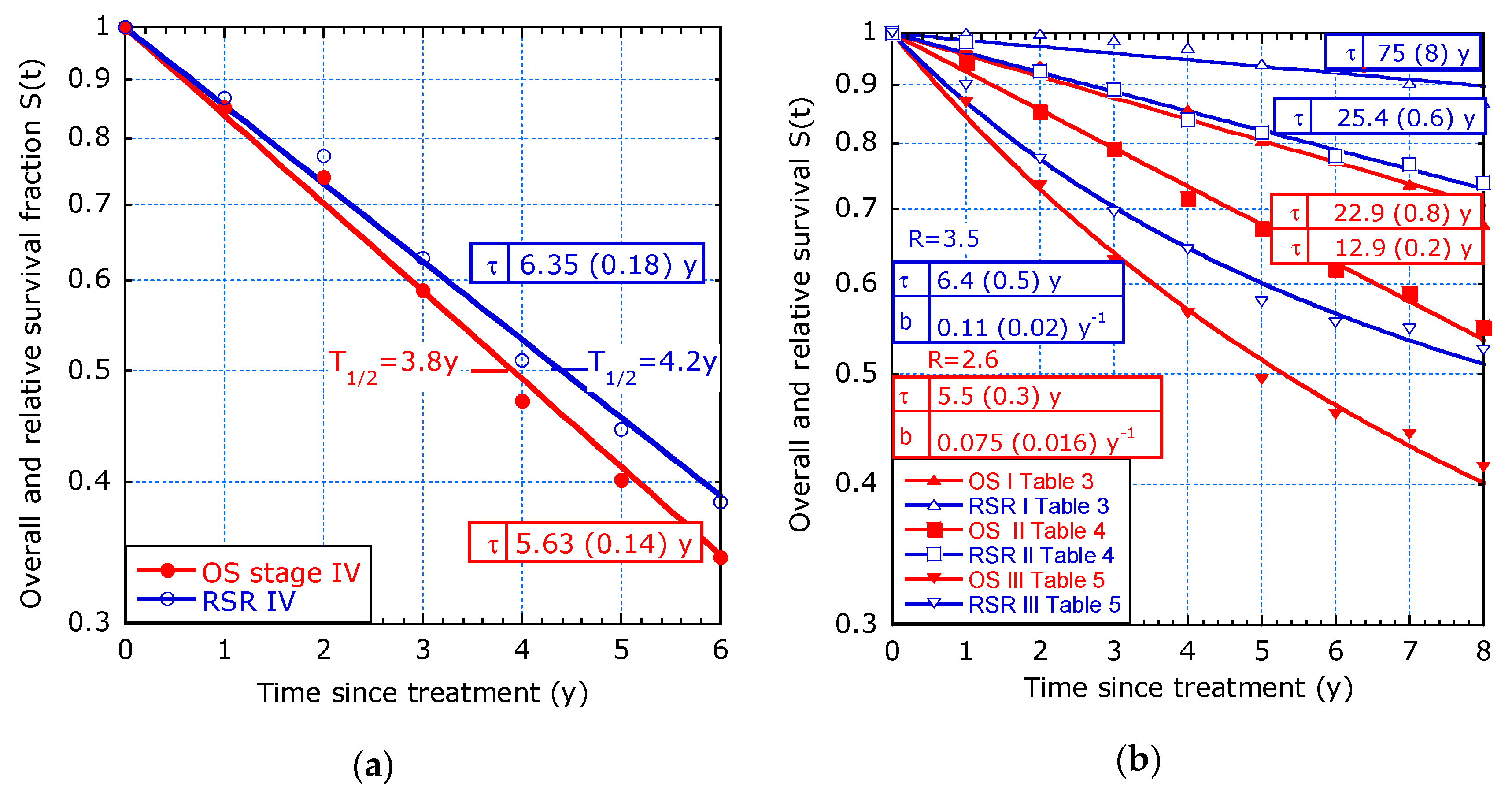
- Study by van Gestel et al. [3] on colorectal cancer (CRC), mainly with tumour stage III.
- 2.
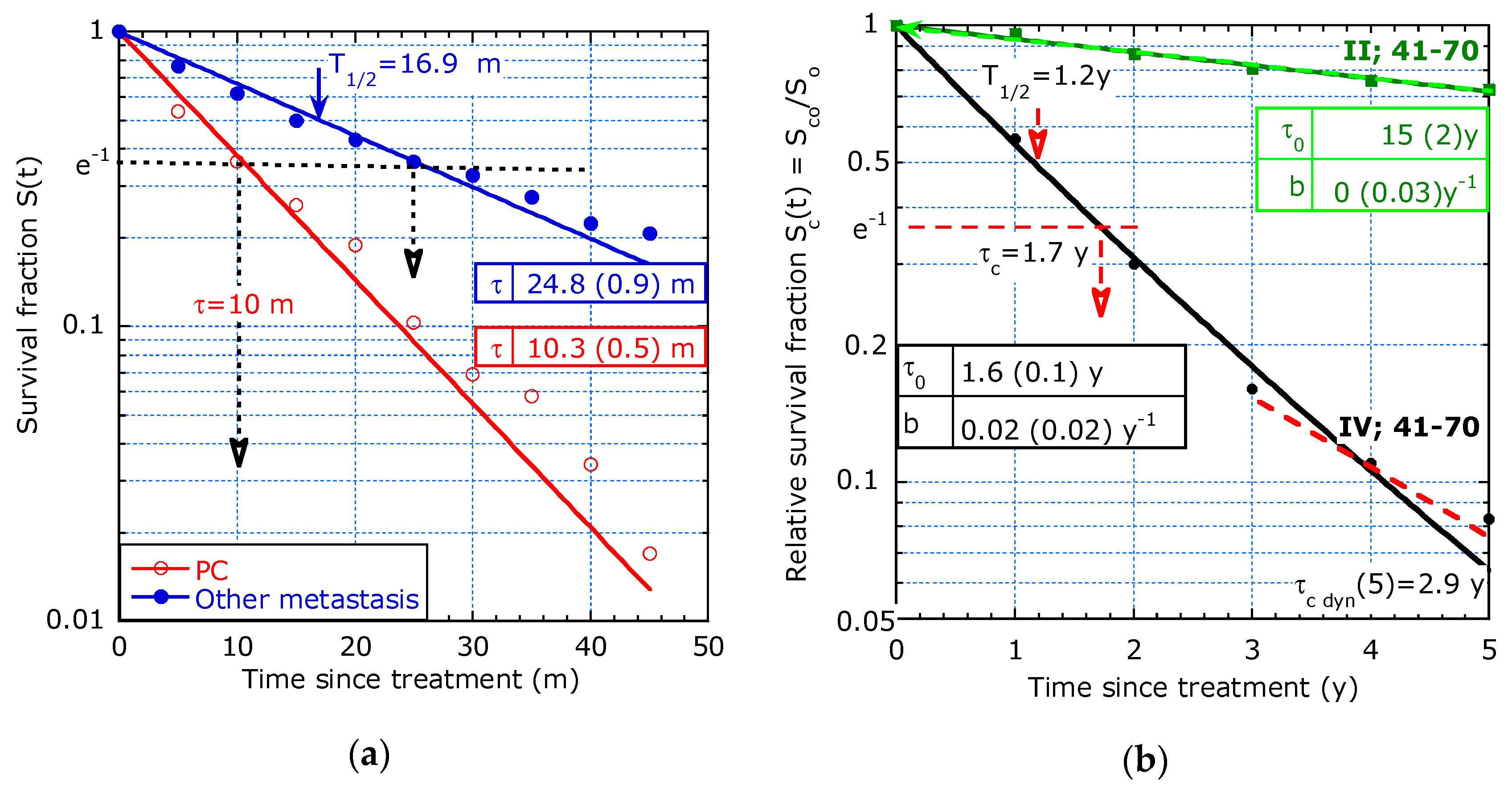
- Study by Orsini et al. [4] on 37,056 rectal cancer patients in the Netherlands between 1989 and 2010 according to the stage and stratified by age (until 40 years and 41–70 years).
- 3.
- Study by Stelzner et al. [5] on CRC selected by stage, with the aim to justify a preference for relative survival as a standard measure for surgical audit and quality control.
- 4.
- Study by Bollschweiler [6] on gastric cancer with patients having different tumour stages after gastrectomy with the aim to discuss the interpretation of KM survival plots.
- 5.
- Study by ‘t Lam-Boer et al. [7] on OS depending on different surgical therapies of 1617 patients between 2004 and 2012 with CRC of stage IV and metastases confined to the liver only. The aim of their study was to investigate changes over time in the utilization of liver resections, as well as possible institutional variations.
- 6.
- Study from Razenberg et al. [8] on 4430 patients with CRC and PC treated with cytoreductive surgery (CRS) and hyperthermic intraperitoneal chemotherapy (HIPEC).
- 7.
- Study by Kuijpers et al. [9] evaluating results of CRS+HIPEC for PC of CRC origin in the Netherlands following a national protocol.
- 8.
- Study by Glehen et al. [10] on PC of digestive or primary origin with mixtures of tumour stages and treatments such as HIPEC and early postoperative intraperitoneal chemotherapy (EPIC).
- 9.
- Study from Gooiker et al. [11] on pancreatic cancer with the aim to analyse the impact of nationwide centralization of pancreatic surgery on long-term survival.
- 10.
- Survival analysis from studies with KM plots hampered by limited data.
- 11.
- 12.
- Study by Chang et al. [16] on advanced pancreatic cancer treated with gemicitabine plus S-1.
3.2. Merits of Providing Dynamic Lifetime
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kleinbaum, D.G.; Klein, M. Survival Analysis—A Self-Learning Text, 3rd ed.; Springer: New York, NY, USA, 2012. [Google Scholar]
- Kalbfleisch, J.D.; Prentice, R.L. The Statistical Analysis of Failure Time Data, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2002. [Google Scholar]
- Van Gestel, Y.R.B.M.; Thomassen, I.; Lemmens, V.E.P.P.; Pruijt, J.F.M.; van Herk-Sukel, M.P.P.; Rutten, H.J.T.; Creemers, G.J.; de Hingh, I.H.J.T. Metachronous peritoneal carcinomatosis after curative treatment of colorectal cancer. Eur. J. Surg. Oncol. 2014, 40, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Orsini, R.G.; Verhoeven, R.H.A.; Lemmens, V.E.P.P.; van Steenbergen, L.N.; de Hingh, I.H.J.T.; Nieuwenhuijzen, G.A.P.; Rutten, H.J.T. Comparable survival for young rectal cancer patients, despite unfavourable morphology and more advanced-stage disease. Eur. J. Cancer 2015, 51, 1675–1682. [Google Scholar] [CrossRef] [PubMed]
- Stelzner, S.; Hellmich, G.; Koch, R.; Witzigmann, H. Exactitude of relative survival compared with cause-specific survival and competing risk estimations based on a clinical database of patients with colorectal carcinoma. Dis. Colon Rectum 2009, 52, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Bollschweiler, E. Benefits and limitations of Kaplan-Meier calculations of survival chance in cancer surgery. Langenbeck’s Arch. Surg. 2003, 388, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Al Ali, C.; Verhoeven, R.H.A.; Roumen, R.M.H.; Lemmens, V.E.P.P.; Rijken, A.M.; De Wilt, J.H.W. Large variation in the utilization of liver resections in stage IV colorectal cancer patients with metastases confined to the liver. Eur. J. Surg. Oncol. 2015, 41, 1217–1225. [Google Scholar]
- Razenberg, L.G.E.M.; van Gestel, Y.R.B.M.; Creemers, G.J.; Verwaal, V.J.; Lemmens, V.E.P.P.; de Hingh, I.H.J.T. Trends in cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for the treatment of synchronous peritoneal carcinomatosis of colorectal origin in the Netherlands. Eur. J. Surg. Oncol. 2015, 41, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Kuijpers, A.M.J.; Mirck, B.; Aalbers, A.G.J.; Nienhuijs, S.W.; de Hingh, I.H.J.T.; Wiezer, M.J.; van Ramshorst, B.; van Ginkel, R.J.; Havenga, K.; Bremers, A.J.; et al. Cytoreduction and HIPEC in The Netherlands: Nationwide Long-term Outcome Following the Dutch Protocol. Ann. Surg. Oncol. 2013, 20, 4224–4230. [Google Scholar] [CrossRef] [PubMed]
- Glehen, O.; Gilly, F.N.; Boutitie, F.; Bereder, J.M.; Quenet, F.; Sideris, L.; Mansvelt, B.; Lorimier, G.; Msika, S.; Elias, D. Toward Curative treatment of peritoneal carcinomatosis from nonovarian origin by cytoreductive surgery combined with perioperative intraperitoneal chemotherapy a multi-Institutional study of 1290 patients. Cancer 2010, 116, 5608–5618. [Google Scholar] [CrossRef] [PubMed]
- Gooiker, G.A.; Lemmens, V.E.P.P.; Besselink, M.G.; Busch, O.R.; Bonsing, B.A.; Molenaar, I.Q.; Tollenaar, R.A.E.M.; de Hingh, I.H.J.T.; Wouters, M.W.J.M. Impact of centralization of pancreatic cancer surgery on resection rates and survival. Br. J. Surg. 2014, 101, 1000–1005. [Google Scholar] [CrossRef] [PubMed]
- Van Oudheusden, T.R.; Braam, H.J.; Nienhuijs, S.W.; Wiezer, M.J.; Van Ramshorst, B.; Luyer, M.D.; Lemmens, V.E.P.P.; de Hingh, I.H.J.T. Cytoreduction and hyperthermic intraperitoneal chemotherapy: A feasible and effective option for colorectal cancer patients after emergency surgery in the presence of peritoneal carcinomatosis. Ann. Surg. Oncol. 2014, 21, 2621–2626. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.K.; Yun, S.S.; Kim, H.J.; Lee, D.S. The risk factors of early recurrence after hepatectomy in hepatocel-lular carcinoma. Ann. Surg. Treat. Res. 2014, 86, 283–288. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rades, D.; Hansen, H.C.; Janssen, S.; Schild, S.E. Comparison of diagnosis-specific survival scores for patients with small-cell lung cancer irradiated for brain metastases. Cancers 2019, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, N.P.M.; Bos, A.C.R.K.; Lemmens, V.E.P.P.; Tanis, P.J.; Hugen, N.; Nagtegaal, I.D.; de Wilt, J.H.W.; Verhoeven, R.H.A. An overview of 25 years of incidence, treatment and outcome of colorectal cancer patients. Int. J. Cancer 2018, 143, 2758–2766. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.F.; Huang, P.W.; Chen, J.S.; Chen, Y.Y.; Lu, C.H.; Chang, P.H.; Hung, Y.S.; Chou, W.C. Prognostic Factors for Advanced Pancreatic Cancer Treated with Gemcitabine Plus S-1: Retrospective Analysis and Development of a Prognostic Model. Cancers 2019, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Nafteux, P.R.; Lerut, A.M.; Moons, J.; Holscher, A.H.; Bollschweiler, E.; Henegouwen, M.I.; Lagarde, S.M.; van Lanschot, J.J.; Messager, M.; Mariette, C.; et al. International multicenter study on the impact of extracapsular lymph node involvement in primary surgery adenocarcinoma of the esophagus on overall survival and staging systems. Ann. Surg. 2015, 262, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Synergy Software. Available online: www.synergy.com (accessed on 27 May 2019).

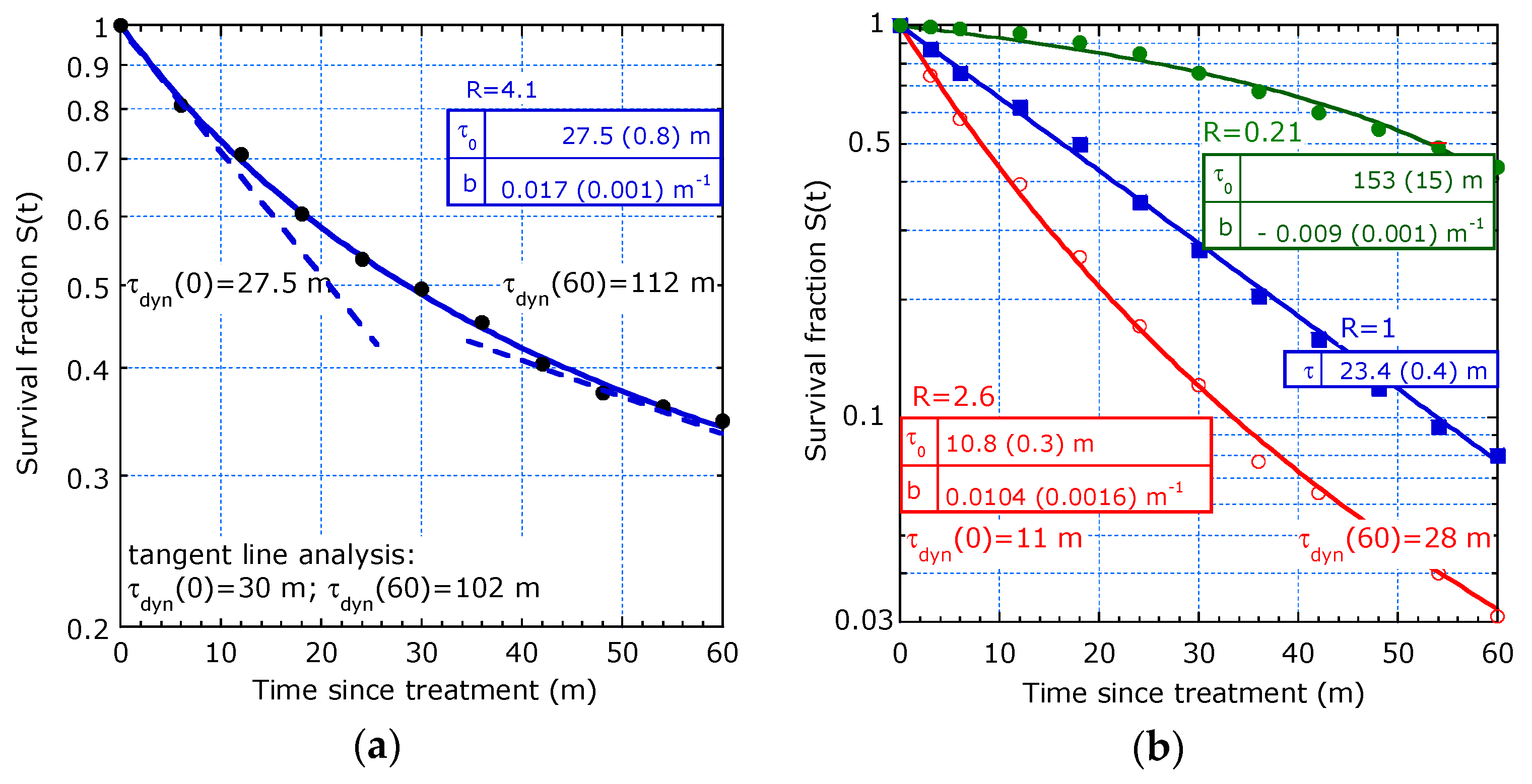
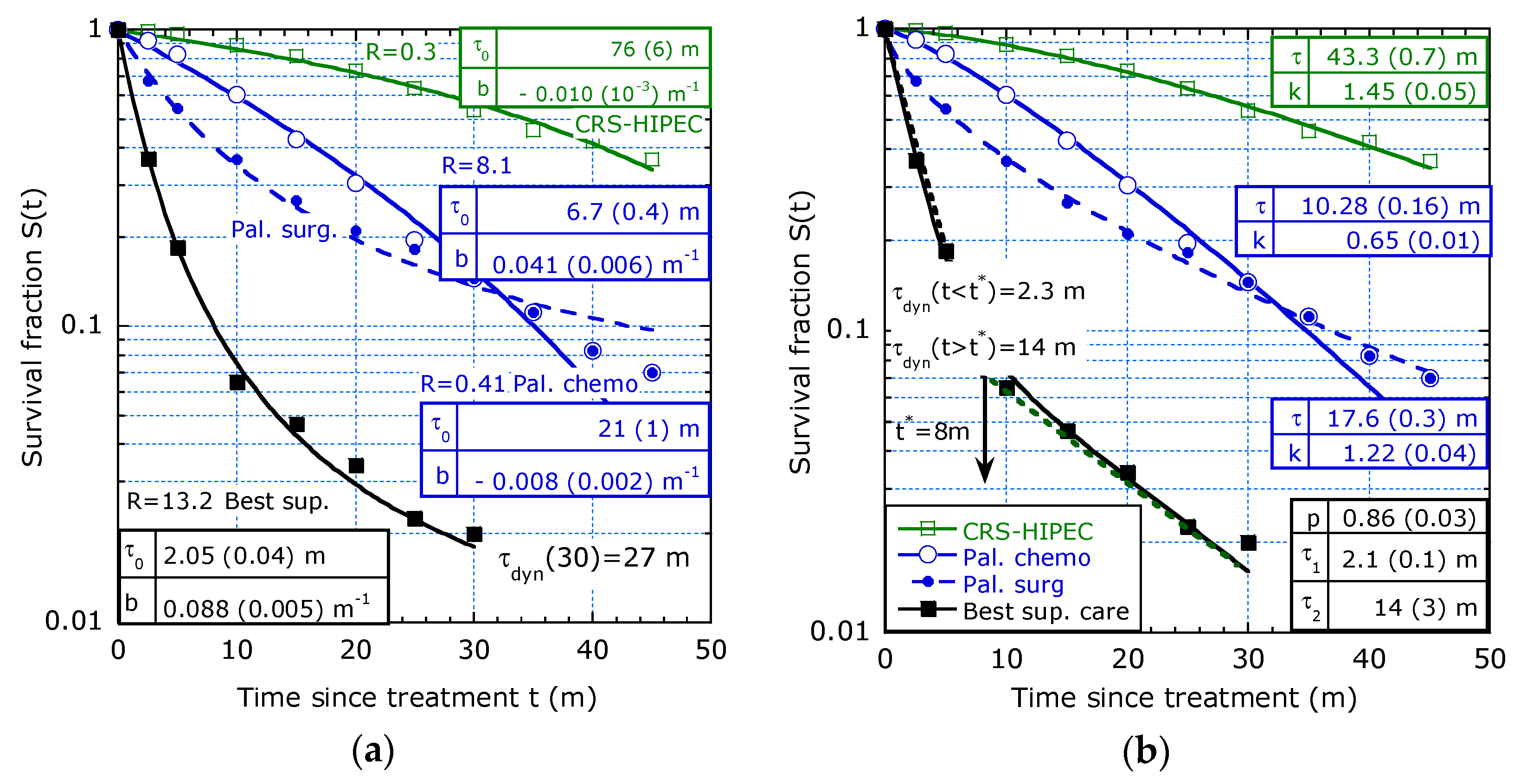
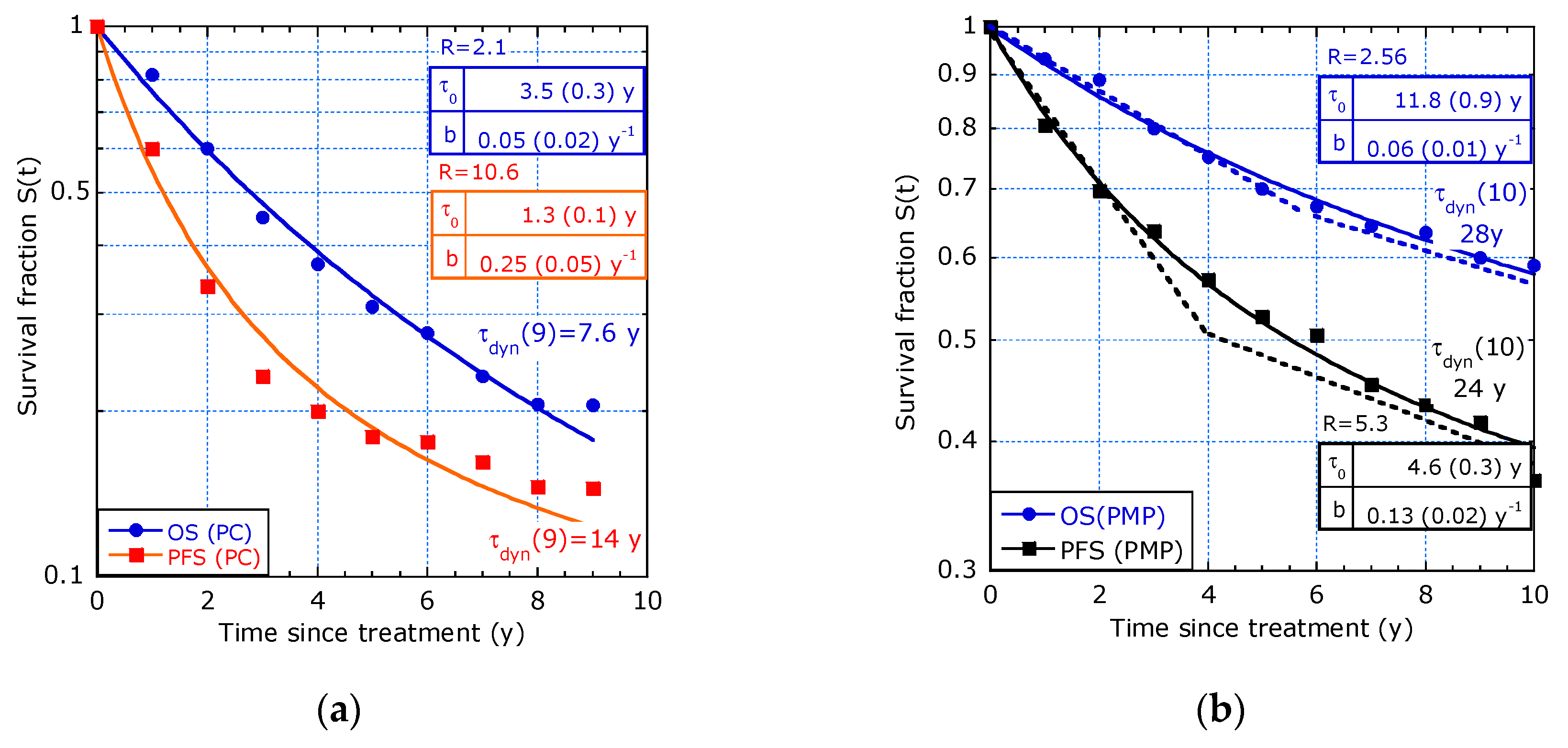
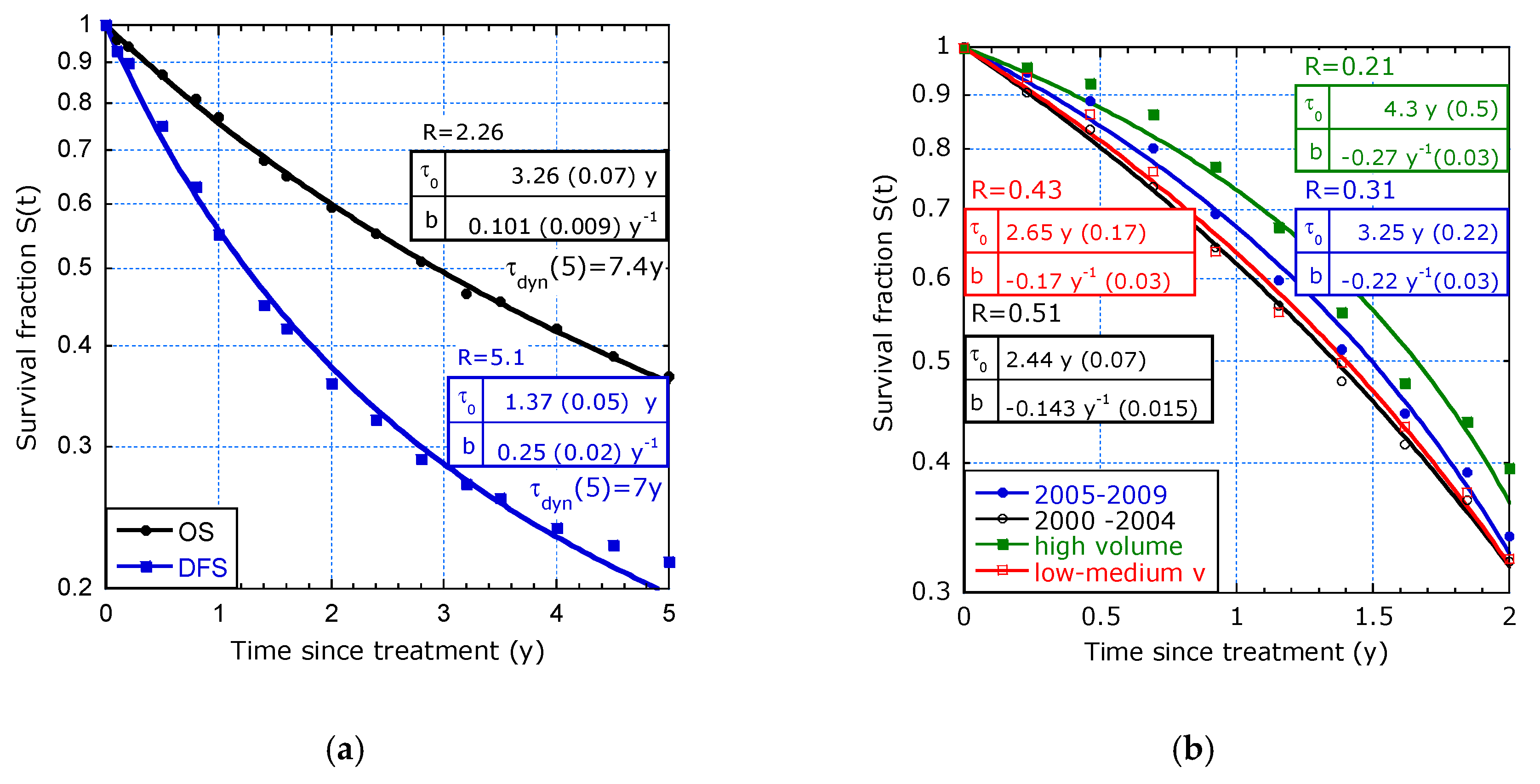
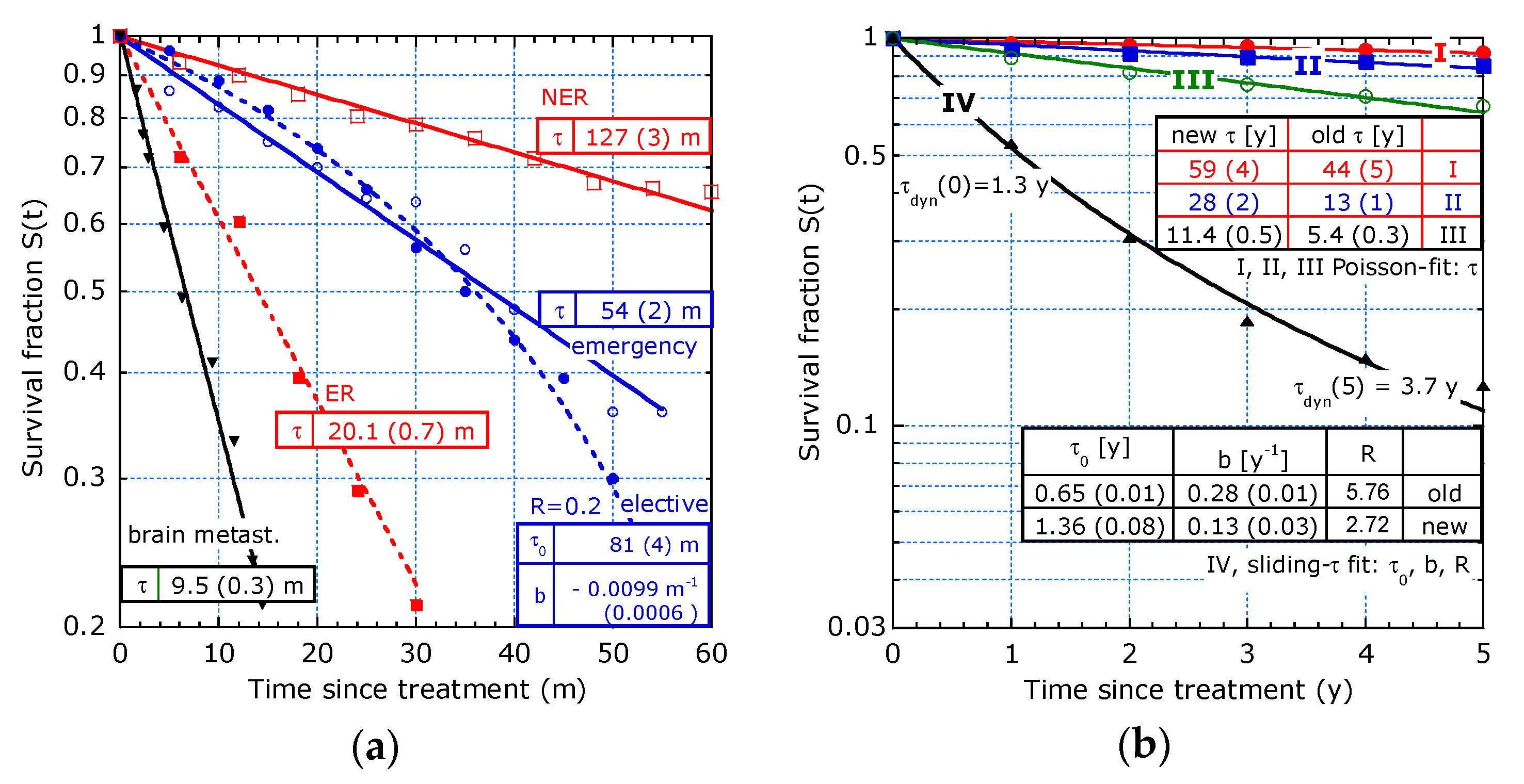
| Reference and tmax | T1/2 [y] | τdyn(0) [y] 1 | b [y−1] | R (ΔR/R) 2 | |
|---|---|---|---|---|---|
| [9] 9 y, PC: | OS | 2.6 | 3.5 (0.3) | 0.05 (0.02) | 2.1 (25%) |
| PFS | 1.2 | 1.3 (0.1) | 0.25 (0.05) | 10.5 (27%) | |
| [9] 10 y, PMP: | OS | >10 | 11.8 (0.9) | 0.06 (0.01) | 2.56 (12%) |
| PFS | 6 | 4.6 (0.3) | 0.13 (0.02) | 5.3 (17%) | |
| [10] 5 y: | OS | 2.9 | 3.26 (0.07) | 0.101 (0.009 | 2.2 (6%) |
| [10] 5 y: | DFS | 1.2 | 1.37 (0.05) | 0.25 (0.02) | 5 (9%) |
| [4] 5 y: | stage II | >5 | 15 (2) | 0 | 1 (-) |
| [4] 5 y: | stage IV | 1.2 | 1.6 (0.1) | 0 | 1 (-) |
| Diagnosis Period | 1989–1994 | 1995–2000 | 2001–2010 | |||
|---|---|---|---|---|---|---|
| Age Group (y) | ≤40 | 41–70 | ≤40 | 41–70 | ≤40 | 41–70 |
| short-term τ0,1 (0.5y) (y) | 5.3 | 5.7 | 6.6 | 6.6 | 7.8 | 8.6 |
| τ1,3 (2y) (y) | 6.8 | 7.35 | 6.8 | 8.52 | 9.32 | 10.8 |
| long-term τ3,5 (4y) (y) | 11.4 | 11.8 | 15.0 | 12.6 | 14.8 | 15.4 |
| Clinical Data | Median | τ0,1 (0.5y) | τ1,3 (2y) | τ3,5 (4y) |
|---|---|---|---|---|
| female | 75 | 109 | 83 | 123 |
| male | 59 | 75 | 66 | 140 |
| age < 60 y | 108 | 107 | 72 | 161 |
| age 60–75 y | 61 | 73 | 72 | 138 |
| 1990–2000 period | 43 | 58 | 58 | 110 |
| 2001–2013 period | 96 | 100 | 77 | 167 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vandamme, L.K.J.; Wouters, P.A.A.F.; Slooter, G.D.; de Hingh, I.H.J.T. Cancer Survival Data Representation for Improved Parametric and Dynamic Lifetime Analysis. Healthcare 2019, 7, 123. https://doi.org/10.3390/healthcare7040123
Vandamme LKJ, Wouters PAAF, Slooter GD, de Hingh IHJT. Cancer Survival Data Representation for Improved Parametric and Dynamic Lifetime Analysis. Healthcare. 2019; 7(4):123. https://doi.org/10.3390/healthcare7040123
Chicago/Turabian StyleVandamme, Lode K.J., Peter A.A.F. Wouters, Gerrit D. Slooter, and Ignace H.J.T. de Hingh. 2019. "Cancer Survival Data Representation for Improved Parametric and Dynamic Lifetime Analysis" Healthcare 7, no. 4: 123. https://doi.org/10.3390/healthcare7040123
APA StyleVandamme, L. K. J., Wouters, P. A. A. F., Slooter, G. D., & de Hingh, I. H. J. T. (2019). Cancer Survival Data Representation for Improved Parametric and Dynamic Lifetime Analysis. Healthcare, 7(4), 123. https://doi.org/10.3390/healthcare7040123




