Selenoprotein S and the Causal Risk of Hypertension in Pregnancy: A Mendelian Randomization Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Data Sources
2.3. Genetic Instruments
2.4. Statistical Analysis
3. Results
3.1. Genetic Instruments and Statistical Power Assessment
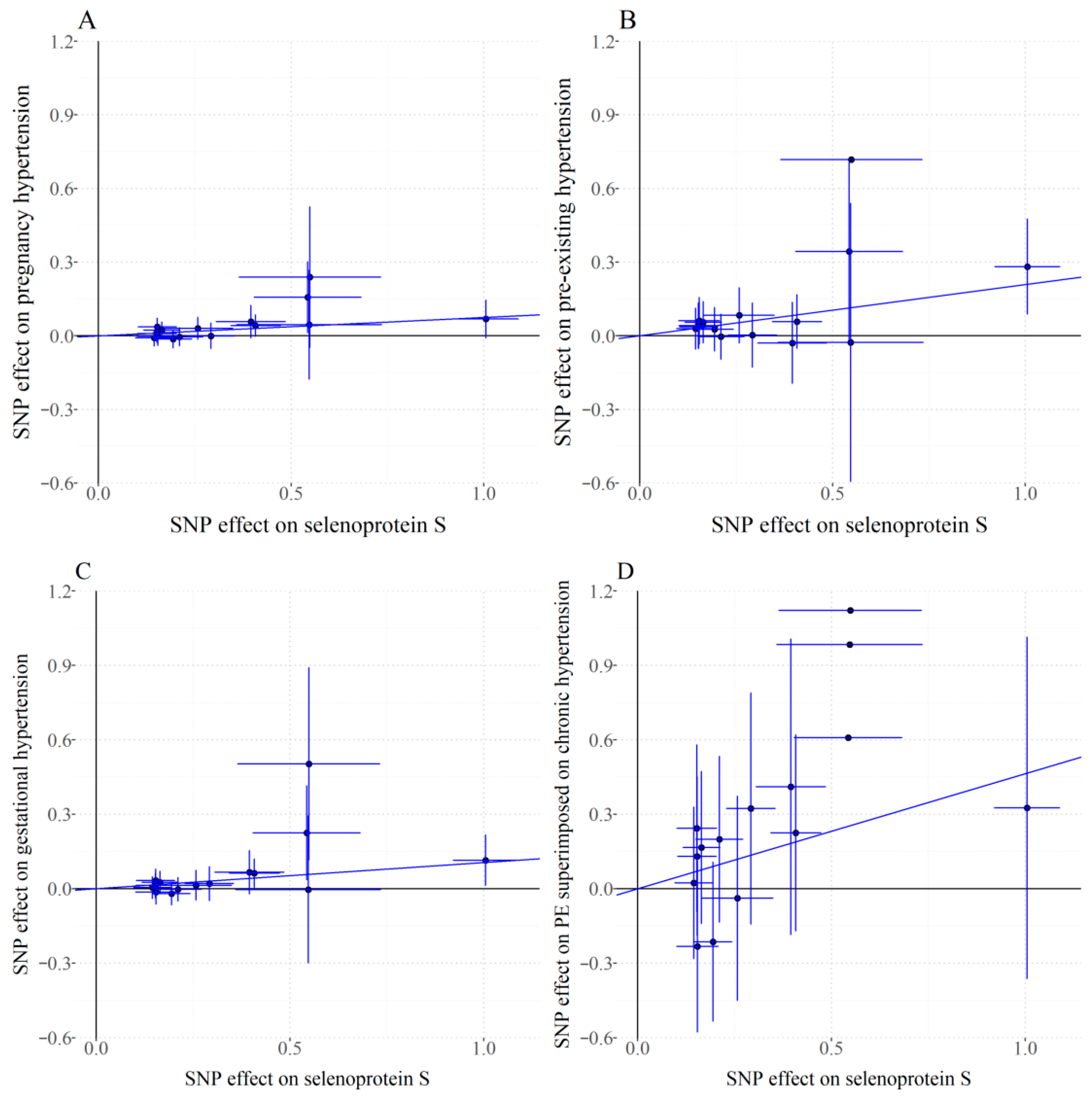
3.2. Primary MR Findings
3.3. Sensitivity Analysis
4. Discussion
4.1. Primary Findings
4.2. Our Findings in the Context of Current Literature
4.3. Strengths and Limitations
4.4. Further Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SELENOS | selenoprotein S |
| PIH | pregnancy induced hypertension |
| MR | Mendelian randomization |
| GWAS | Genome-wide association study |
| IVW | inverse-variance weighted |
| PEH | pre-existing hypertension |
| GH | gestational hypertension |
| PE | Preeclampsia |
| OR | odds ratio |
| CI | confidence interval |
| BP | Blood pressure |
| DIC | disseminated intravascular coagulation |
| ER | endoplasmic reticulum |
| SELENOP | selenoprotein P |
| WHO | World Health Organization |
| MoBa | Norwegian Mother, Father and Child Cohort Study |
| IVs | instrumental variables |
| MR-RAPS | Mendelian Randomization Robust Adjusted Profile Scoring |
| MR-PRESSO | Mendelian Randomization Pleiotropy RESidual Sum and Outlier |
| RCT | randomized controlled trial |
| SNPs | Single-nucleotide polymorphisms |
| LD | linkage disequilibrium |
| ROS | reactive oxygen species |
| sFlt-1 | soluble fms-like tyrosine kinase-1 |
| VEGF | vascular endothelial growth factor |
| ERAD | ER-associated degradation |
| UPR | unfolded protein response |
References
- Cífková, R. Hypertension in Pregnancy: A Diagnostic and Therapeutic Overview. High Blood Press. Cardiovasc. Prev. 2023, 30, 289–303. [Google Scholar] [CrossRef]
- Wu, P.; Green, M.; Myers, J.E. Hypertensive disorders of pregnancy. BMJ 2023, 381, e071653. [Google Scholar] [CrossRef] [PubMed]
- Fishel Bartal, M.; Sibai, B.M. Eclampsia in the 21st century. Am. J. Obstet. Gynecol. 2022, 226, S1237–S1253. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Gupta, A. Hypertensive Disorders of Pregnancy. Cardiol. Clin. 2019, 37, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Kametas, N.A.; Nzelu, D.; Nicolaides, K.H. Chronic hypertension and superimposed preeclampsia: Screening and diagnosis. Am. J. Obstet. Gynecol. 2022, 226, S1182–S1195. [Google Scholar] [CrossRef]
- Suri, J.; Suri, J.C.; Arora, R.; Gupta, M.; Adhikari, T. The Impact of Sleep-Disordered Breathing on Severity of Pregnancy-Induced Hypertension and Feto-Maternal Outcomes. J. Obstet. Gynaecol. India 2019, 69, 111–121. [Google Scholar] [CrossRef]
- Erez, O.; Othman, M.; Rabinovich, A.; Leron, E.; Gotsch, F.; Thachil, J. DIC in Pregnancy—Pathophysiology, Clinical Characteristics, Diagnostic Scores, and Treatments. J. Blood Med. 2022, 13, 21–44. [Google Scholar] [CrossRef]
- Jiao, Y.; Liu, Y.; Li, H.; Song, Z.; Wang, S.; Zhang, J.; Li, J.; Liu, J.; Wang, P.; Chen, Y. Value of proteinuria in evaluating the severity of HELLP and its maternal and neonatal outcomes. BMC Pregnancy Childbirth 2023, 23, 591. [Google Scholar] [CrossRef]
- Sweeney, L.C.; Lundsberg, L.S.; Culhane, J.F.; Partridge, C.; Son, M. Co-existing chronic hypertension and hypertensive disorders of pregnancy and associated adverse pregnancy outcomes. J. Matern.-Fetal Neonatal Med. 2024, 37, 2305675. [Google Scholar] [CrossRef]
- Bucci, T.; Meek, C.L.; Awor, S.; Lip, G.Y.H.; Merriel, A. Five-year risk of all-cause death and cardiovascular events in women with gestational diabetes and hypertensive disorders of pregnancy. Curr. Probl. Cardiol. 2024, 49, 102698. [Google Scholar] [CrossRef]
- Garovic, V.D.; White, W.M.; Vaughan, L.; Saiki, M.; Parashuram, S.; Garcia-Valencia, O.; Weissgerber, T.L.; Milic, N.; Weaver, A.; Mielke, M.M. Incidence and Long-Term Outcomes of Hypertensive Disorders of Pregnancy. J. Am. Coll. Cardiol. 2020, 75, 2323–2334. [Google Scholar] [CrossRef]
- Mojadadi, A.; Au, A.; Salah, W.; Witting, P.; Ahmad, G. Role for Selenium in Metabolic Homeostasis and Human Reproduction. Nutrients 2021, 13, 3256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, X.; Wei, Y. Selenium and Selenoproteins in Health. Biomolecules 2023, 13, 799. [Google Scholar] [CrossRef]
- Kipp, A.P.; Strohm, D.; Brigelius-Flohé, R.; Schomburg, L.; Bechthold, A.; Leschik-Bonnet, E.; Heseker, H. Revised reference values for selenium intake. J. Trace Elem. Med. Biol. 2015, 32, 195–199. [Google Scholar] [CrossRef]
- Genchi, G.; Lauria, G.; Catalano, A.; Sinicropi, M.S.; Carocci, A. Biological Activity of Selenium and Its Impact on Human Health. Int. J. Mol. Sci. 2023, 24, 2633. [Google Scholar] [CrossRef]
- Bai, S.; Zhang, M.; Tang, S.; Li, M.; Wu, R.; Wan, S.; Chen, L.; Wei, X.; Feng, S. Effects and Impact of Selenium on Human Health, A Review. Molecules 2024, 30, 50. [Google Scholar] [CrossRef]
- Hoque, B.; Shi, Z. Association between selenium intake, diabetes and mortality in adults: Findings from National Health and Nutrition Examination Survey (NHANES) 2003–2014. Br. J. Nutr. 2022, 127, 1098–1105. [Google Scholar] [CrossRef]
- Chaudière, J. Biological and Catalytic Properties of Selenoproteins. Int. J. Mol. Sci. 2023, 24, 10109. [Google Scholar] [CrossRef]
- Duntas, L.H. Selenium and at-risk pregnancy: Challenges and controversies. Thyroid Res. 2020, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Mesdaghinia, E.; Shahin, F.; Ghaderi, A.; Shahin, D.; Shariat, M.; Banafshe, H. The Effect of Selenium Supplementation on Clinical Outcomes, Metabolic Profiles, and Pulsatility Index of the Uterine Artery in High-Risk Mothers in Terms of Preeclampsia Screening with Quadruple Test: A Randomized, Double-Blind, Placebo-Controlled Clinical Trial: Selenium and preeclampsia. Biol. Trace Elem. Res. 2023, 201, 567–576. [Google Scholar] [PubMed]
- Silva, I.; Bracchi, I.; Keating, E. The association between selenium levels and hypertensive disorders of pregnancy: A systematic review of the literature. Br. J. Nutr. 2023, 130, 651–665. [Google Scholar] [CrossRef]
- Atamer, Y.; Koçyigit, Y.; Yokus, B.; Atamer, A.; Erden, A.C. Lipid peroxidation, antioxidant defense, status of trace metals and leptin levels in preeclampsia. Eur. J. Obstet. Gynecol. Reprod. Biol. 2005, 119, 60–66. [Google Scholar] [CrossRef]
- Moses, E.K.; Johnson, M.P.; Tømmerdal, L.; Forsmo, S.; Curran, J.E.; Abraham, L.J.; Charlesworth, J.C.; Brennecke, S.P.; Blangero, J.; Austgulen, R. Genetic association of preeclampsia to the inflammatory response gene SEPS1. Am. J. Obstet. Gynecol. 2008, 198, 336.e1–336.e5. [Google Scholar] [CrossRef] [PubMed]
- Holmquist, E.; Brantsæter, A.L.; Meltzer, H.M.; Jacobsson, B.; Barman, M.; Sengpiel, V. Maternal selenium intake and selenium status during pregnancy in relation to preeclampsia and pregnancy-induced hypertension in a large Norwegian Pregnancy Cohort Study. Sci. Total Environ. 2021, 798, 149271. [Google Scholar] [CrossRef] [PubMed]
- Davey Smith, G.; Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Walia, G.K.; Sachdeva, M.P. ‘Mendelian randomization’: An approach for exploring causal relations in epidemiology. Public Health 2017, 145, 113–119. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian Randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef]
- Burgess, S.; Bowden, J.; Fall, T.; Ingelsson, E.; Thompson, S.G. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology 2017, 28, 30–42. [Google Scholar] [CrossRef]
- Burgess, S.; Small, D.S.; Thompson, S.G. A review of instrumental variable estimators for Mendelian randomization. Stat. Methods Med. Res. 2017, 26, 2333–2355. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef]
- Sun, B.B.; Maranville, J.C.; Peters, J.E.; Stacey, D.; Staley, J.R.; Blackshaw, J.; Burgess, S.; Jiang, T.; Paige, E.; Surendran, P.; et al. Genomic atlas of the human plasma proteome. Nature 2018, 558, 73–79. [Google Scholar] [CrossRef]
- Good, B.H. Linkage disequilibrium between rare mutations. Genetics 2022, 220, iyac004. [Google Scholar] [CrossRef]
- Pierce, B.L.; Burgess, S. Efficient design for Mendelian randomization studies: Subsample and 2-sample instrumental variable estimators. Am. J. Epidemiol. 2013, 178, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Ong, J.S.; MacGregor, S. Implementing MR-PRESSO and GCTA-GSMR for pleiotropy assessment in Mendelian randomization studies from a practitioner’s perspective. Genet. Epidemiol. 2019, 43, 609–616. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Staff, A.C.; Fjeldstad, H.E.; Fosheim, I.K.; Moe, K.; Turowski, G.; Johnsen, G.M.; Alnaes-Katjavivi, P.; Sugulle, M. Failure of physiological transformation and spiral artery atherosis: Their roles in preeclampsia. Am. J. Obstet. Gynecol. 2022, 226, S895–S906. [Google Scholar] [CrossRef]
- Nonn, O.; Fischer, C.; Geisberger, S.; El-Heliebi, A.; Kroneis, T.; Forstner, D.; Desoye, G.; Staff, A.C.; Sugulle, M.; Dechend, R.; et al. Maternal Angiotensin Increases Placental Leptin in Early Gestation via an Alternative Renin-Angiotensin System Pathway: Suggesting a Link to Preeclampsia. Hypertension 2021, 77, 1723–1736. [Google Scholar] [CrossRef] [PubMed]
- Mistry, H.D.; Wilson, V.; Ramsay, M.M.; Symonds, M.E.; Broughton Pipkin, F. Reduced selenium concentrations and glutathione peroxidase activity in preeclamptic pregnancies. Hypertension 2008, 52, 881–888. [Google Scholar] [CrossRef]
- Jung, E.; Romero, R.; Yeo, L.; Gomez-Lopez, N.; Chaemsaithong, P.; Jaovisidha, A.; Gotsch, F.; Erez, O. The etiology of preeclampsia. Am. J. Obstet. Gynecol. 2022, 226, S844–S866. [Google Scholar] [CrossRef]
- Ghelichkhani, F.; Gonzalez, F.A.; Kapitonova, M.A.; Schaefer-Ramadan, S.; Liu, J.; Cheng, R.; Rozovsky, S. Selenoprotein S: A versatile disordered protein. Arch. Biochem. Biophys. 2022, 731, 109427. [Google Scholar] [CrossRef]
- Tang, W.K.; Zhang, T.; Ye, Y.; Xia, D. Structural basis for nucleotide-modulated p97 association with the ER membrane. Cell Discov. 2017, 3, 17045. [Google Scholar] [CrossRef]
- Rayman, M.P.; Searle, E.; Kelly, L.; Johnsen, S.; Bodman-Smith, K.; Bath, S.C.; Mao, J.; Redman, C.W. Effect of selenium on markers of risk of pre-eclampsia in UK pregnant women: A randomised, controlled pilot trial. Br. J. Nutr. 2014, 112, 99–111. [Google Scholar] [CrossRef]
- Dahlen, C.R.; Reynolds, L.P.; Caton, J.S. Selenium supplementation and pregnancy outcomes. Front. Nutr. 2022, 9, 1011850. [Google Scholar] [CrossRef]
- Freitas, R.G.; Nogueira, R.J.; Antonio, M.A.; Barros-Filho Ade, A.; Hessel, G. Selenium deficiency and the effects of supplementation on preterm infants. Rev. Paul. Pediatr. 2014, 32, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P.; Wijnen, H.; Vader, H.; Kooistra, L.; Pop, V. Maternal selenium status during early gestation and risk for preterm birth. CMAJ 2011, 183, 549–555. [Google Scholar]
- Mirone, M.; Giannetta, E.; Isidori, A.M. Selenium and reproductive function. A systematic review. J. Endocrinol. Investig. 2013, 36, 28–36. [Google Scholar]
- Ma’ayeh, M.; Costantine, M.M. Prevention of preeclampsia. Semin. Fetal Neonatal Med. 2020, 25, 101123. [Google Scholar] [CrossRef]
- Rolnik, D.L.; Wright, D.; Poon, L.C.; O’Gorman, N.; Syngelaki, A.; de Paco Matallana, C.; Akolekar, R.; Cicero, S.; Janga, D.; Singh, M.; et al. Aspirin versus Placebo in Pregnancies at High Risk for Preterm Preeclampsia. N. Engl. J. Med. 2017, 377, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Vanderlelie, J.J.; Perkins, A.V.; Redman, C.W.; Ahmadi, K.R.; Rayman, M.P. Genetic polymorphisms that affect selenium status and response to selenium supplementation in United Kingdom pregnant women. Am. J. Clin. Nutr. 2016, 103, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Perry, A.; Stephanou, A.; Rayman, M.P. Dietary factors that affect the risk of pre-eclampsia. BMJ Nutr. Prev. Health 2022, 5, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Kelishadi, M.R.; Ashtary-Larky, D.; Davoodi, S.H.; Clark, C.C.T.; Asbaghi, O. The effects of selenium supplementation on blood lipids and blood pressure in adults: A systematic review and dose-response meta-analysis of randomized control trials. J. Trace Elem. Med. Biol. 2022, 74, 127046. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P.; Bath, S.C.; Westaway, J.; Williams, P.; Mao, J.; Vanderlelie, J.J.; Perkins, A.V.; Redman, C.W. Selenium status in U.K. pregnant women and its relationship with hypertensive conditions of pregnancy. Br. J. Nutr. 2015, 113, 249–258. [Google Scholar] [CrossRef] [PubMed]


| SNP | Chr | Position (Build 37) | Nearest Gene | EA | OA | β | SE | p Value | F Statistic |
|---|---|---|---|---|---|---|---|---|---|
| rs117261169 | 19 | 45491032 | CLPTM1 | T | C | −0.55 | 0.1 | 1.20 × 108 | 37.43 |
| rs2965169 | 19 | 45251156 | BCL3 | C | A | −0.15 | 0.03 | 2.04 × 109 | 37.06 |
| rs151330717 | 19 | 45196964 | CEACAM16 | A | G | −0.55 | 0.09 | 5.13 × 109 | 39.63 |
| rs405509 | 19 | 45408836 | APOE | G | T | −0.17 | 0.02 | 1.45 × 1011 | 45.36 |
| rs28399657 | 19 | 45318351 | BCAM | G | A | −0.54 | 0.07 | 1.78 × 1014 | 60.4 |
| rs28399637 | 19 | 45324138 | BCAM | A | G | 0.15 | 0.03 | 5.62 × 109 | 33.87 |
| rs11668327 | 19 | 45398633 | TOMM40 | C | G | −0.29 | 0.03 | 8.32 × 1020 | 84.7 |
| rs429358 | 19 | 45411941 | APOE | C | T | 0.41 | 0.03 | 5.50 × 1035 | 148.84 |
| rs6859 | 19 | 45382034 | NECTIN2 | G | A | −0.15 | 0.03 | 5.01 × 109 | 34.1 |
| rs7412 | 19 | 45412079 | APOE | T | C | −1.01 | 0.04 | 1.78 × 10120 | 559.13 |
| rs4803759 | 19 | 45327459 | BCAM | C | T | 0.15 | 0.03 | 1.35 × 108 | 33.26 |
| rs62117161 | 19 | 45233385 | BCL3 | G | A | −0.4 | 0.05 | 5.62 × 1018 | 77.13 |
| rs60049679 | 19 | 45429708 | APOC1 | C | G | 0.26 | 0.05 | 3.31 × 108 | 32.35 |
| rs4263041 | 19 | 45438643 | APOC4 | G | A | −0.21 | 0.03 | 6.92 × 1012 | 64.1 |
| rs7343130 | 19 | 45331103 | BCAM | G | A | 0.19 | 0.03 | 5.25 × 1015 | 62.52 |
| Outcome | NO. SNPs | Weighted Median | MR-RAPS | MR-PRESSO | MR-Egger | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | ||
| Pregnancy hypertension | 15 | 1.076 (1.014, 1.143) | 0.016 | 1.078 (1.031, 1.128) | 9.18 × 104 | 1.078 (1.024, 1.134) | 0.004 | 1.109 (1.023, 1.202) | 0.012 |
| Pre-existing hypertension | 15 | 1.301 (1.127, 1.501) | 3.28 × 104 | 1.234 (1.104, 1.379) | 2.18 × 104 | 1.232 (1.096, 1.384) | 4.70 × 10−4 | 1.265 (1.035, 1.546) | 0.022 |
| Gestational hypertension | 15 | 1.120 (1.036, 1.210) | 0.004 | 1.112 (1.049, 1.180) | 3.87 × 104 | 1.111 (1.034, 1.193) | 0.004 | 1.182 (1.062, 1.315) | 0.002 |
| PE superimposed on chronic hypertension | 15 | 1.606 (0.953, 2.707) | 0.075 | 1.598 (1.076, 2.372) | 0.02 | 1.590 (1.069, 2.363) | 0.022 | 1.773 (0.869, 3.615) | 0.115 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, M.; Lv, W.; He, Y.; Liu, W.; Gao, Y. Selenoprotein S and the Causal Risk of Hypertension in Pregnancy: A Mendelian Randomization Study. Healthcare 2025, 13, 2383. https://doi.org/10.3390/healthcare13182383
Cai M, Lv W, He Y, Liu W, Gao Y. Selenoprotein S and the Causal Risk of Hypertension in Pregnancy: A Mendelian Randomization Study. Healthcare. 2025; 13(18):2383. https://doi.org/10.3390/healthcare13182383
Chicago/Turabian StyleCai, Mengqi, Wenrui Lv, Yan He, Weili Liu, and Yuzhen Gao. 2025. "Selenoprotein S and the Causal Risk of Hypertension in Pregnancy: A Mendelian Randomization Study" Healthcare 13, no. 18: 2383. https://doi.org/10.3390/healthcare13182383
APA StyleCai, M., Lv, W., He, Y., Liu, W., & Gao, Y. (2025). Selenoprotein S and the Causal Risk of Hypertension in Pregnancy: A Mendelian Randomization Study. Healthcare, 13(18), 2383. https://doi.org/10.3390/healthcare13182383





