Proteins Adsorbed during Intraoperative Hemoadsorption and Their In Vitro Effects on Endothelium
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Spectrum of Blood Proteins Adsorbed on the CytoSorb Polymer Matrix
3.2. Alteration in Endothelial Cells Functionality after Treatment with Eluted Protein Mixture
3.2.1. Cell Viability Assay
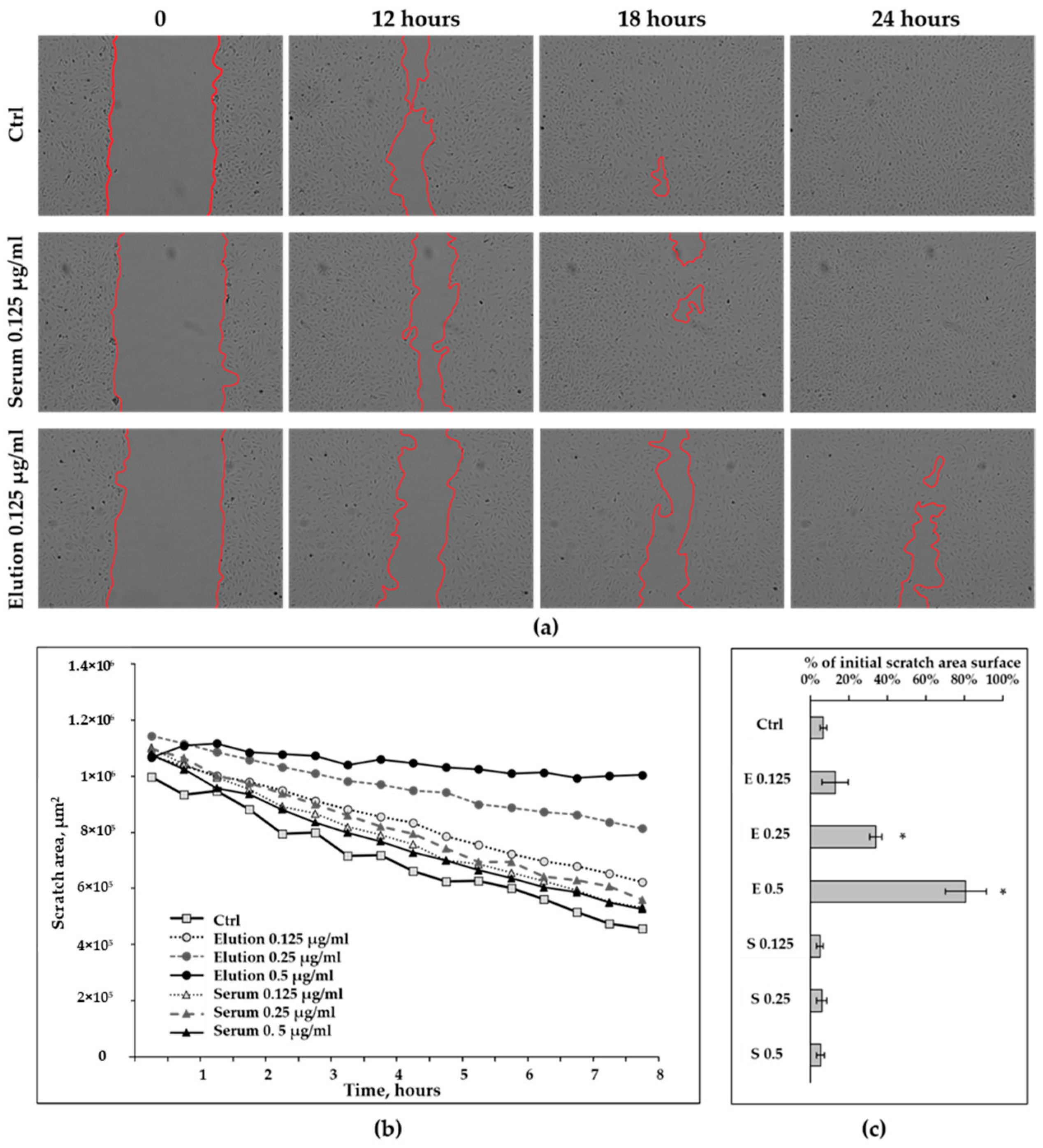
3.2.2. Wound-Healing Assay
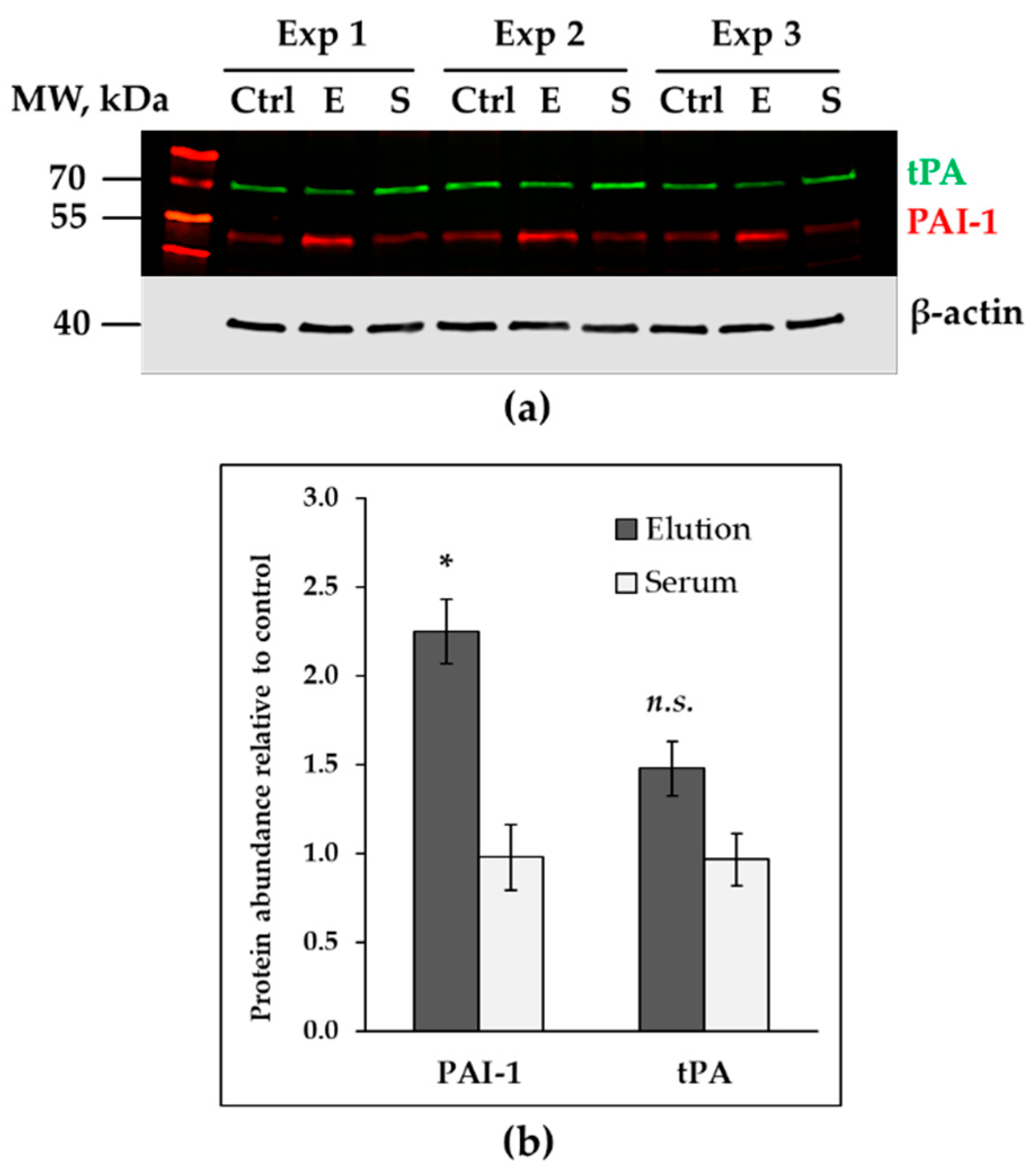
3.3. Up-Regulated Genes in Treated HAECs
3.4. Down-Regulated Genes in Treated Endothelial Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BUB1 | Mitotic checkpoint kinase |
| CPB | Cardiopulmonary bypass |
| CCB | Coomassie colloidal blue |
| ECGM2 | Endothelial cell growth medium 2 |
| ECs | Endothelial cells |
| FCS | Fetal calf serum |
| HAECs | Human aortic endothelial cells |
| KIF20A | Kinesin superfamily 20A |
| PAI-1 | Plasminogen activator inhibitor 1 |
| PBS | Phosphate-buffered saline |
| RIN | RNA integrity number |
| RIPA | Radioimmunoprecipitation assay buffer |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SIRS | Systemic inflammatory response syndrome |
| SEM | Standard error of mean |
| TBS | Tris-buffered saline |
| tPA | Tissue plasminogen activator |
References
- Squiccimarro, E.; Labriola, C.; Malvindi, P.G.; Margari, V.; Guida, P.; Visicchio, G.; Kounakis, G.; Favale, A.; Dambruoso, P.; Mastrototaro, G.; et al. Prevalence and Clinical Impact of Systemic Inflammatory Reaction after Cardiac Surgery. J. Cardiothorac. Vasc. Anesth. 2019, 33, 1682–1690. [Google Scholar] [CrossRef] [PubMed]
- Bronicki, R.A.; Hall, M. Cardiopulmonary Bypass-Induced Inflammatory Response: Pathophysiology and Treatment. Pediatr. Crit. Care Med. 2016, 17 (Suppl. 1), S272–S278. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen Windsant, I.C.; de Wit, N.C.J.; Sertorio, J.T.C.; van Bijnen, A.A.; Ganushchak, Y.M.; Heijmans, J.H.; Tanus-Santos, J.E.; Jacobs, M.J.; Maessen, J.G.; Buurman, W.A. Hemolysis during cardiac surgery is associated with increased intravascular nitric oxide consumption and perioperative kidney and intestinal tissue damage. Front. Physiol. 2014, 5, 340. [Google Scholar] [CrossRef] [PubMed]
- Giacinto, O.; Satriano, U.; Nenna, A.; Spadaccio, C.; Lusini, M.; Mastroianni, C.; Nappi, F.; Chello, M. Inflammatory Response and Endothelial Dysfunction Following Cardiopulmonary Bypass: Pathophysiology and Pharmacological Targets. Recent Pat. Inflamm. Allergy Drug Discov. 2019, 13, 158–173. [Google Scholar] [CrossRef]
- Brettner, F.; Chappell, D.; Schwartz, L.; Lukasz, A.; Kümpers, P.; Becker, B.F.; Reichart, B.; Rehm, M.; Bruegger, D. Vascular Endothelial Dysfunction during Cardiac Surgery: On-Pump versus Off-Pump Coronary Surgery. Eur. Surg. Res. 2017, 58, 354–368. [Google Scholar] [CrossRef]
- Koning, N.J.; Overmars, M.A.; van den Brom, C.E.; van Bezu, J.; Simon, L.E.; Vonk, A.B.; Girbes, A.R.; van Nieuw Amerongen, G.P.; Boer, C. Endothelial hyperpermeability after cardiac surgery with cardiopulmonary bypass as assessed using an in vitro bioassay for endothelial barrier function. Br. J. Anaesth. 2016, 116, 223–232. [Google Scholar] [CrossRef]
- Sturtzel, C. Endothelial Cells. Adv. Exp. Med. Biol. 2017, 1003, 71–91. [Google Scholar] [CrossRef]
- Träger, K.; Skrabal, C.; Fischer, G.; Datzmann, T.; Schroeder, J.; Fritzler, D.; Hartmann, J.; Liebold, A.; Reinelt, H. Hemoadsorption Treatment of Patients with Acute Infective Endocarditis during Surgery with Cardiopulmonary Bypass—A Case Series. Int. J. Artif. Organs 2017, 40, 240–249. [Google Scholar] [CrossRef]
- Bernardi, M.H.; Rinoesl, H.; Dragosits, K.; Ristl, R.; Hoffelner, F.; Opfermann, P.; Lamm, C.; Preißing, F.; Wiedemann, D.; Hiesmayr, M.J.; et al. Effect of hemoadsorption during cardiopulmonary bypass surgery—A blinded, randomized, controlled pilot study using a novel adsorbent. Crit. Care 2016, 20, 96. [Google Scholar] [CrossRef]
- Gleason, T.G.; Argenziano, M.; Bavaria, J.E.; Kane, L.C.; Coselli, J.S.; Engelman, R.M.; Tanaka, K.A.; Awad, A.; Sekela, M.E.; Zwischenberger, J.B. Hemoadsorption to Reduce Plasma-Free Hemoglobin during Cardiac Surgery: Results of REFRESH I Pilot Study. Semin. Thorac. Cardiovasc. Surg. 2019, 31, 783–793. [Google Scholar] [CrossRef]
- Poli, E.C.; Alberio, L.; Bauer-Doerries, A.; Marcucci, C.; Roumy, A.; Kirsch, M.; De Stefano, E.; Liaudet, L.; Schneider, A.G. Cytokine clearance with CytoSorb® during cardiac surgery: A pilot randomized controlled trial. Crit. Care 2019, 23, 108. [Google Scholar] [CrossRef] [PubMed]
- Haidari, Z.; Wendt, D.; Thielmann, M.; Mackowiak, M.; Neuhäuser, M.; Jakob, H.; Ruhparwar, A.; El-Gabry, M. Intraoperative Hemoadsorption in Patients With Native Mitral Valve Infective Endocarditis. Ann. Thorac. Surg. 2020, 110, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Ankawi, G.; Xie, Y.; Yang, B.; Xie, Y.; Xie, P.; Ronco, C. What Have We Learned about the Use of Cytosorb Adsorption Columns? Blood Purif. 2019, 48, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Henn, M.C.; Moon, M.R. Hemoadsorption Is Safe during Cardiac Surgery—But Does It Improve Outcomes? Semin. Thorac. Cardiovasc. Surg. 2019, 31, 794–795. [Google Scholar] [CrossRef]
- Diab, M.; Lehmann, T.; Bothe, W.; Akhyari, P.; Platzer, S.; Wendt, D.; Deppe, A.C.; Strauch, J.; Hagel, S.; Gunther, A.; et al. Cytokine Hemoadsorption During Cardiac Surgery Versus Standard Surgical Care for Infective Endocarditis (REMOVE): Results From a Multicenter Randomized Controlled Trial. Circulation 2022, 145, 959–968. [Google Scholar] [CrossRef]
- Poli, E.C.; Rimmelé, T.; Schneider, A.G. Hemoadsorption with CytoSorb®. Intensive Care Med. 2019, 45, 236–239. [Google Scholar] [CrossRef]
- Singh, A.; Mehta, Y.; Trehan, N. Bilirubin Removal Using CytoSorb Filter in a Cardiac Surgical Patient. J. Cardiothorac. Vasc. Anesth. 2019, 33, 881–883. [Google Scholar] [CrossRef]
- Piwowarczyk, P.; Kutnik, P.; Potręć-Studzińska, B.; Sysiak-Sławecka, J.; Rypulak, E.; Borys, M.; Czczuwar, M. Hemoadsorption in isolated conjugated hyperbilirubinemia after extracorporeal membrane oxygenation support. Cholestasis of sepsis: A case report and review of the literature on differential causes of jaundice in ICU patient. Int. J. Artif. Organs 2019, 42, 263–268. [Google Scholar] [CrossRef]
- Calabrò, M.G.; Febres, D.; Recca, G.; Lembo, R.; Fominskiy, E.; Scandroglio, A.M.; Zangrillo, A.; Pappalardo, F. Blood Purification with CytoSorb in Critically Ill Patients: Single-Center Preliminary Experience. Artif. Organs 2019, 43, 189–194. [Google Scholar] [CrossRef]
- Wiegele, M.; Krenn, C.G. Cytosorb™ in a patient with Legionella pneumonia-associated rhabdomyolysis: A case report. Asaio J. 2015, 61, e14–e16. [Google Scholar] [CrossRef]
- Braun, I.; Deppe, A.C.; Weber, C.; Mihaylova, M.; Paunel-Görgülü, A.; Schlachtenberger, G.; Gerfer, S.; Djordjevic, I.; Choi, Y.H.; Wahlers, T. Limitation of Circulating cfDNA Under the Use of a Cytokine Elimination Adsorber (CytoSorb) in Cardiac Surgery. Thorac. Cardiovasc. Surg. 2018, 66 (Suppl. 1), S1–S110. [Google Scholar] [CrossRef]
- Hassan, K.; Kannmacher, J.; Wohlmuth, P.; Budde, U.; Schmoeckel, M.; Geidel, S. Cytosorb Adsorption During Emergency Cardiac Operations in Patients at High Risk of Bleeding. Ann. Thorac. Surg. 2019, 108, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Wachter, K.; Navarrete Santos, A.; Grosskopf, A.; Baldensperger, T.; Glomb, M.A.; Szabo, G.; Simm, A. AGE-Rich Bread Crust Extract Boosts Oxidative Stress Interception via Stimulation of the NRF2 Pathway. Nutrients 2021, 13, 3874. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Nunes-Nesi, A.; Kruger, P.; Nagel, A.; Hannemann, J.; Giorgi, F.M.; Childs, L.; Osorio, S.; Walther, D.; Selbig, J.; et al. Robin: An intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol. 2010, 153, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Glaí, M.; MöLler, B.; Zirkel, A.; WäChter, K.; HüTtelmaier, S.; Posch, S. Cell migration analysis: Segmenting scratch assay images with level sets and support vector machines. Pattern Recogn. 2012, 45, 3154–3165. [Google Scholar] [CrossRef]
- Herwig, R.; Hardt, C.; Lienhard, M.; Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 2016, 11, 1889–1907. [Google Scholar] [CrossRef]
- Harm, S.; Schildböck, C.; Hartmann, J. Cytokine Removal in Extracorporeal Blood Purification: An in vitro Study. Blood Purif. 2020, 49, 33–43. [Google Scholar] [CrossRef]
- David, S.; Thamm, K.; Schmidt, B.M.W.; Falk, C.S.; Kielstein, J.T. Effect of extracorporeal cytokine removal on vascular barrier function in a septic shock patient. J. Intensive Care 2017, 5, 12. [Google Scholar] [CrossRef]
- Denzinger, M.; Staendker, L.; Ehlers, K.; Schneider, J.M.; Schulz, T.; Hein, T.; Wiese, S.; Roecker, A.; Gross, R.; Münch, J.; et al. Bioassay for Endothelial Damage Mediators Retrieved by Hemoadsorption. Sci. Rep. 2019, 9, 14522. [Google Scholar] [CrossRef]
- Wisgrill, L.; Lamm, C.; Hell, L.; Thaler, J.; Berger, A.; Weiss, R.; Weber, V.; Rinoesl, H.; Hiesmayr, M.J.; Spittler, A.; et al. Influence of hemoadsorption during cardiopulmonary bypass on blood vesicle count and function. J. Transl. Med. 2020, 18, 202. [Google Scholar] [CrossRef]
- Rimmelé, T.; Kaynar, A.M.; McLaughlin, J.N.; Bishop, J.V.; Fedorchak, M.V.; Chuasuwan, A.; Peng, Z.; Singbartl, K.; Frederick, D.R.; Zhu, L.; et al. Leukocyte capture and modulation of cell-mediated immunity during human sepsis: An ex vivo study. Crit. Care 2013, 17, R59. [Google Scholar] [CrossRef] [PubMed]
- Bierhansl, L.; Conradi, L.C.; Treps, L.; Dewerchin, M.; Carmeliet, P. Central Role of Metabolism in Endothelial Cell Function and Vascular Disease. Physiol. 2017, 32, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Hitzel, J.; Lee, E.; Zhang, Y.; Bibli, S.I.; Li, X.; Zukunft, S.; Pflüger, B.; Hu, J.; Schürmann, C.; Vasconez, A.E.; et al. Oxidized phospholipids regulate amino acid metabolism through MTHFD2 to facilitate nucleotide release in endothelial cells. Nat. Commun. 2018, 9, 2292. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, X.; Carmeliet, P. Hallmarks of Endothelial Cell Metabolism in Health and Disease. Cell Metab. 2019, 30, 414–433. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Lomelino, C.L.; Andring, J.T.; McKenna, R.; Kilberg, M.S. Asparagine synthetase: Function, structure, and role in disease. J. Biol. Chem. 2017, 292, 19952–19958. [Google Scholar] [CrossRef]
- Klebig, C.; Korinth, D.; Meraldi, P. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 2009, 185, 841–858. [Google Scholar] [CrossRef]
- Chen, C.-T.; Hehnly, H.; Doxsey, S.J. Orchestrating vesicle transport, ESCRTs and kinase surveillance during abscission. Nat. Rev. Mol. Cell Biol. 2012, 13, 483–488. [Google Scholar] [CrossRef]
- van Hinsbergh, V.W. Endothelium--role in regulation of coagulation and inflammation. Semin. Immunopathol. 2012, 34, 93–106. [Google Scholar] [CrossRef]
- Chandler, W.L.; Velan, T. Secretion of tissue plasminogen activator and plasminogen activator inhibitor 1 during cardiopulmonary bypass. Thromb. Res. 2003, 112, 185–192. [Google Scholar] [CrossRef]
- Fleming, G.A.; Billings, F.T.t.; Klein, T.M.; Bichell, D.P.; Christian, K.G.; Pretorius, M. Angiotensin-converting enzyme inhibition alters the inflammatory and fibrinolytic response to cardiopulmonary bypass in children. Pediatr. Crit. Care Med. 2011, 12, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Billings, F.T., IV; Balaguer, J.M.; Yu, C.; Wright, P.; Petracek, M.R.; Byrne, J.G.; Brown, N.J.; Pretorius, M. Comparative effects of angiotensin receptor blockade and ACE inhibition on the fibrinolytic and inflammatory responses to cardiopulmonary bypass. Clin. Pharmacol. Ther. 2012, 91, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Tofler, G.H.; Massaro, J.; O’Donnell, C.J.; Wilson, P.W.F.; Vasan, R.S.; Sutherland, P.A.; Meigs, J.B.; Levy, D.; D’Agostino, R.B., Sr. Plasminogen activator inhibitor and the risk of cardiovascular disease: The Framingham Heart Study. Thromb. Res. 2016, 140, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Ozolina, A.; Strike, E.; Jaunalksne, I.; Krumina, A.; Bjertnaes, L.J.; Vanags, I. PAI-1 and t-PA/PAI-1 complex potential markers of fibrinolytic bleeding after cardiac surgery employing cardiopulmonary bypass. BMC Anesthesiol. 2012, 12, 27. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piskovatska, V.; Navarrete Santos, A.; Kalies, K.; Korca, E.; Stiller, M.; Szabó, G.; Simm, A.; Wächter, K. Proteins Adsorbed during Intraoperative Hemoadsorption and Their In Vitro Effects on Endothelium. Healthcare 2023, 11, 310. https://doi.org/10.3390/healthcare11030310
Piskovatska V, Navarrete Santos A, Kalies K, Korca E, Stiller M, Szabó G, Simm A, Wächter K. Proteins Adsorbed during Intraoperative Hemoadsorption and Their In Vitro Effects on Endothelium. Healthcare. 2023; 11(3):310. https://doi.org/10.3390/healthcare11030310
Chicago/Turabian StylePiskovatska, Veronika, Alexander Navarrete Santos, Katrin Kalies, Edina Korca, Markus Stiller, Gábor Szabó, Andreas Simm, and Kristin Wächter. 2023. "Proteins Adsorbed during Intraoperative Hemoadsorption and Their In Vitro Effects on Endothelium" Healthcare 11, no. 3: 310. https://doi.org/10.3390/healthcare11030310
APA StylePiskovatska, V., Navarrete Santos, A., Kalies, K., Korca, E., Stiller, M., Szabó, G., Simm, A., & Wächter, K. (2023). Proteins Adsorbed during Intraoperative Hemoadsorption and Their In Vitro Effects on Endothelium. Healthcare, 11(3), 310. https://doi.org/10.3390/healthcare11030310




