Fitting Early Phases of the COVID-19 Outbreak: A Comparison of the Performances of Used Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data
2.2. Epidemic Models
2.3. Compartmental Models
2.4. Data-Driven Models
2.5. Model Evaluation
3. Results
3.1. Total Cases Fit
3.2. New Cases Fit
4. Discussion
4.1. Study Limitations
4.2. Future Research Developments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Italian Civil Protection Department; Morettini, M.; Sbrollini, A.; Marcantoni, I.; Burattini, L. COVID-19 in Italy: Dataset of the Italian Civil Protection Department. Data Brief 2020, 30, 105526. [Google Scholar] [CrossRef]
- Du, S.; Wang, J.; Zhang, H.; Cui, W.; Kang, Z.; Yang, T.; Lou, B.; Chi, Y.; Long, H.; Ma, M.; et al. Predicting COVID-19 Using Hybrid AI Model. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Šušteršič, T.; Blagojević, A.; Cvetković, D.; Cvetković, A.; Lorencin, I.; Šegota, S.B.; Milovanović, D.; Baskić, D.; Car, Z.; Filipović, N. Epidemiological Predictive Modeling of COVID-19 Infection: Development, Testing, and Implementation on the Population of the Benelux Union. Front. Public Health 2021, 9, 727274. [Google Scholar] [CrossRef]
- Ramazi, P.; Haratian, A.; Meghdadi, M.; Mari Oriyad, A.; Lewis, M.A.; Maleki, Z.; Vega, R.; Wang, H.; Wishart, D.S.; Greiner, R. Accurate Long-Range Forecasting of COVID-19 Mortality in the USA. Sci. Rep. 2021, 11, 13822. [Google Scholar] [CrossRef]
- Kermack, W.O.; McKendrick, A.G. A Contribution to the Mathematical Theory of Epidemics. Proc. R. Soc. Lond. Ser. A 1927, 115, 700–772. [Google Scholar]
- Rizkalla, C.; Blanco-Silva, F.; Gruver, S. Modeling the Impact of Ebola and Bushmeat Hunting on Western Lowland Gorillas. EcoHealth 2007, 4, 151–155. [Google Scholar] [CrossRef]
- Ng, T.W.; Turinici, G.; Danchin, A. A Double Epidemic Model for the SARS Propagation. BMC Infect. Dis. 2003, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Giordano, G.; Blanchini, F.; Bruno, R.; Colaneri, P.; Di Filippo, A.; Di Matteo, A.; Colaneri, M. Modelling the COVID-19 Epidemic and Implementation of Population-Wide Interventions in Italy. Nat. Med. 2020, 26, 855–860. [Google Scholar] [CrossRef]
- Ferrari, L.; Gerardi, G.; Manzi, G.; Micheletti, A.; Nicolussi, F.; Biganzoli, E.; Salini, S. Modeling Provincial COVID-19 Epidemic Data Using an Adjusted Time-Dependent SIRD Model. Int. J. Environ. Res. Public Health 2021, 18, 6563. [Google Scholar] [CrossRef]
- Antonelli, E.; Piccolomini, E.L.; Zama, F. Switched Forced SEIRDV Compartmental Models to Monitor COVID-19 Spread and Immunization in Italy. Infect. Dis. Model. 2022, 7, 1–15. [Google Scholar] [CrossRef]
- Roda, W.C.; Varughese, M.B.; Han, D.; Li, M.Y. Why Is It Difficult to Accurately Predict the COVID-19 Epidemic? Infect. Dis. Model. 2020, 5, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Li, K.; Jiang, Y.; Guo, X.; Zhao, T. Prediction and analysis of Coronavirus Disease 2019. arXiv 2019, arXiv:2003.05447. [Google Scholar]
- Remuzzi, A.; Remuzzi, G. Health Policy COVID-19 and Italy: What Next? Lancet 2020, 2, 10–13. [Google Scholar] [CrossRef]
- Vattay, G. Predicting the Ultimate Outcome of the COVID-19 Outbreak in Italy. arXiv 2020, arXiv:2003.07912. [Google Scholar]
- Wu, K.; Darcet, D.; Wang, Q.; Sornette, D. Generalized Logistic Growth Modeling of the COVID-19 Outbreak in 29 Provinces in China and in the Rest of the World. Nonlinear Dyn. 2020, 101, 1561–1581. [Google Scholar] [CrossRef] [PubMed]
- Ma, J. Estimating Epidemic Exponential Growth Rate and Basic Reproduction Number. Infect. Dis. Model. 2020, 5, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, M. Epilocal: A Real-Time Tool for Local Epidemic Monitoring. arXiv 2020, arXiv:2003.07928. [Google Scholar] [CrossRef]
- Agosto, A.; Giudici, P. A Poisson Autoregressive Model to Understand COVID-19 Contagion Dynamics. Risks 2020, 8, 77. [Google Scholar] [CrossRef]
- Benvenuto, D.; Giovanetti, M.; Vassallo, L.; Angeletti, S.; Ciccozzi, M. Application of the ARIMA Model on the COVID-2019 Epidemic Dataset. Data Brief 2020, 29, 105340. [Google Scholar] [CrossRef]
- Ding, G.; Li, X.; Jiao, F.; Shen, Y. Brief Analysis of the ARIMA Model on the COVID-19 in Italy. medRxiv 2020. [Google Scholar] [CrossRef]
- Chintalapudi, N.; Battineni, G.; Amenta, F. COVID-19 Virus Outbreak Forecasting of Registered and Recovered Cases after Sixty Day Lockdown in Italy: A Data Driven Model Approach. J. Microbiol. Immunol. Infect. 2020, 53, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Christodoulos, C.; Michalakelis, C.; Varoutas, D. Forecasting with Limited Data: Combining ARIMA and Diffusion Models. Technol. Forecast. Soc. Chang. 2010, 77, 558–565. [Google Scholar] [CrossRef]
- Azzolina, D.; Lanera, C.; Comoretto, R.; Francavilla, A.; Rosi, P.; Casotto, V.; Navalesi, P.; Gregori, D. Automatic Forecast of Intensive Care Unit Admissions: The Experience during the COVID-19 Pandemic in Italy. J. Med. Syst. 2023, 47, 84. [Google Scholar] [CrossRef] [PubMed]
- Flaxman, S.; Mishra, S.; Gandy, A.; Unwin, H.J.T.; Coupland, H.; Mellan, T.A.; Berah, T.; Eaton, J.W.; Guzman, P.N.P.; Schmit, N.; et al. Estimating the Number of Infections and the Impact of Non-Pharmaceutical Interventions on COVID-19 in 11 European Countries; Imperial College London: London, UK, 2020. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, W. Government Responses Matter: Predicting COVID-19 Cases in US under an Empirical Bayesian Time Series Framework. medRxiv 2020. [Google Scholar] [CrossRef]
- Yang, Z.; Zeng, Z.; Wang, K.; Wong, S.; Liang, W.; Zanin, M.; Liu, P.; Cao, X.; Gao, Z.; Mai, Z.; et al. Modified SEIR and AI Prediction of the Epidemics Trend of COVID-19 in China under Public Health Interventions. J. Thorac. Dis. 2020, 2019, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ge, Q.; Li, S.; Jin, L.; Xiong, M. Artificial Intelligence Forecasting of COVID-19 in China. arXiv 2020, arXiv:2002.07112. [Google Scholar] [CrossRef]
- Li, Y.; Wang, B.; Peng, R.; Zhou, C.; Zhao, B. Mathematical Modeling and Epidemic Prediction of COVID-19 and Its Significance to Epidemic Prevention and Control Measures. Ann. Infect. Dis. Epidemiol. 2020, 5, 1052. [Google Scholar]
- Kucharski, A.J.; Russell, T.W.; Diamond, C.; Liu, Y.; Edmunds, J.; Funk, S.; Eggo, R.M.; Sun, F.; Jit, M.; Munday, J.D.; et al. Early Dynamics of Transmission and Control of COVID-19: A Mathematical Modelling Study. Lancet Infect. Dis. 2020, 20, 553–558. [Google Scholar] [CrossRef]
- Wen, C.; Wei, J.; Ma, Z.F.; He, M.; Zhao, S.; Ji, J.; He, D. Heterogeneous Epidemic Modelling within an Enclosed Space and Corresponding Bayesian Estimation. Infect. Dis. Model. 2022, 7, 1–24. [Google Scholar] [CrossRef]
- Lin, Q.; Zhao, S.; Gao, D.; Lou, Y.; Yang, S.; Musa, S.S.; Wang, M.H.; Cai, Y.; Wang, W.; Yang, L.; et al. A Conceptual Model for the Coronavirus Disease 2019 (COVID-19) Outbreak in Wuhan, China with Individual Reaction and Governmental Action. Int. J. Infect. Dis. 2020, 93, 211–216. [Google Scholar] [CrossRef]
- Azzolina, D.; Lorenzoni, G.; Silvestri, L.; Prosepe, I.; Berchialla, P.; Gregori, D. Regional Differences in Mortality Rates during the COVID-19 Epidemic in Italy. Disaster Med. Public Health Prep. 2021, 16, 1355–1361. [Google Scholar] [CrossRef]
- Boccia, S.; Ricciardi, W.; Ioannidis, J.P.A. What Other Countries Can Learn from Italy during the COVID-19 Pandemic. JAMA Intern Med. 2020, 180, 927. [Google Scholar] [CrossRef] [PubMed]
- Magnani, C.; Azzolina, D.; Gallo, E.; Ferrante, D.; Gregori, D. How Large Was the Mortality Increase Directly and Indirectly Caused by the COVID-19 Epidemic? An Analysis on All-Causes Mortality Data in Italy. Int. J. Environ. Res. Public Health 2020, 17, 3452. [Google Scholar] [CrossRef]
- Istituto Superiore di Sanità Epidemia COVID-19 Aggiornamento Nazionale. Available online: https://www.epicentro.iss.it/coronavirus/sars-cov-2-dashboard (accessed on 28 June 2020).
- Lanera, C.; Azzolina, D.; Pirotti, F.; Prosepe, I.; Lorenzoni, G.; Berchialla, P.; Gregori, D. A Web-Based Application to Monitor and Inform about the COVID-19 Outbreak in Italy: The {COVID-19ita} Initiative. Healthcare 2022, 10, 473. [Google Scholar] [CrossRef] [PubMed]
- Nesteruk, I. Statistics Based Predictions of Coronavirus 2019-NCoV Spreading in Mainland China. medRxiv 2020. [Google Scholar] [CrossRef]
- Fanelli, D.; Piazza, F. Analysis and Forecast of COVID-19 Spreading in China, Italy and France. Chaos Solitons Fractals 2020, 134, 109761. [Google Scholar] [CrossRef]
- Pangea Formazione Pangea Formazione. Available online: https://www.pangeadds.eu/demos/covid19/ (accessed on 1 June 2023).
- Izadi, F. Generalized Additive Models to Capture the Death Rates in Canada COVID-19. arXiv 2020, arXiv:2008.01030. [Google Scholar]
- Dong, E.; Du, H.; Gardner, L. An Interactive Web-Based Dashboard to Track COVID-19 in Real Time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Brauer, F.; Van Den Driessche, P.; Wu, J. Mathematical Epidemiology; Springer: Berlin, Germany, 2008; pp. 19–79. [Google Scholar]
- Lega, J.; Brown, H.E. Data-Driven Outbreak Forecasting with a Simple Nonlinear Growth Model. Epidemics 2016, 17, 19–26. [Google Scholar] [CrossRef]
- Cantó, B.; Coll, C.; Sánchez, E. Estimation of Parameters in a Structured SIR Model. Adv. Differ. Equ. 2017, 2017, 33. [Google Scholar] [CrossRef]
- Paiva, H.M.; Afonso, R.J.M.; de Oliveira, I.L.; Garcia, G.F. A Data-Driven Model to Describe and Forecast the Dynamics of COVID-19 Transmission. PLoS ONE 2020, 15, e0236386. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, H. A New Metric of Absolute Percentage Error for Intermittent Demand Forecasts. Int. J. Forecast. 2016, 32, 669–679. [Google Scholar] [CrossRef]
- Hyndman, R.J.; Koehler, A.B. Another Look at Measures of Forecast Accuracy. Int. J. Forecast. 2006, 22, 679–688. [Google Scholar] [CrossRef]
- Willmott, C.; Matsuura, K. Advantages of the Mean Absolute Error (MAE) over the Root Mean Square Error (RMSE) in Assessing Average Model Performance. Clim. Res. 2005, 30, 79–82. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2019. [Google Scholar]
- Yadav, S.K.; Akhter, Y. Statistical Modeling for the Prediction of Infectious Disease Dissemination with Special Reference to COVID-19 Spread. Front. Public Health 2021, 9, 645405. [Google Scholar] [CrossRef]
- Challener, D.W.; Dowdy, S.C.; O’Horo, J.C. Analytics and Prediction Modeling During the COVID-19 Pandemic. Mayo Clin. Proc. 2020, 95, S8–S10. [Google Scholar] [CrossRef] [PubMed]
- Poletto, C.; Scarpino, S.V.; Volz, E.M. Applications of Predictive Modelling Early in the COVID-19 Epidemic. Lancet Digit. Health 2020, 2, e498–e499. [Google Scholar] [CrossRef]
- Ma, N.; Ma, W.; Li, Z. Multi-Model Selection and Analysis for COVID-19. Fractal Fract. 2021, 5, 120. [Google Scholar] [CrossRef]
- Brauer, F. Mathematical Epidemiology: Past, Present, and Future. Infect. Dis. Model. 2017, 2, 113–127. [Google Scholar] [CrossRef]
- May, R.M.; Anderson, R.M. Transmission Dynamics of HIV Infection. Nature 1987, 326, 137–142. [Google Scholar] [CrossRef]
- Iranzo, V.; Pérez-González, S. Epidemiological Models and COVID-19: A Comparative View. Hist. Philos. Life Sci. 2021, 43, 104. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.M.; Vela-Pérez, M.; Ferrández, M.R.; Kubik, A.B.; Ivorra, B. Modeling the Impact of SARS-CoV-2 Variants and Vaccines on the Spread of COVID-19. Commun. Nonlinear Sci. Numer. Simul. 2021, 102, 105937. [Google Scholar] [CrossRef] [PubMed]
- Giordano, G.; Colaneri, M.; Di Filippo, A.; Blanchini, F.; Bolzern, P.; De Nicolao, G.; Sacchi, P.; Colaneri, P.; Bruno, R. Modeling Vaccination Rollouts, SARS-CoV-2 Variants and the Requirement for Non-Pharmaceutical Interventions in Italy. Nat. Med. 2021, 27, 993–998. [Google Scholar] [CrossRef]
- Prodanov, D. Analytical Parameter Estimation of the SIR Epidemic Model. Applications to the COVID-19 Pandemic. Entropy 2020, 23, 59. [Google Scholar] [CrossRef] [PubMed]
- Prodanov, D. Asymptotic Analysis of the SIR Model and the Gompertz Distribution. J. Comput. Appl. Math. 2023, 422, 114901. [Google Scholar] [CrossRef]
- Gregori, D.; Azzolina, D.; Lanera, C.; Prosepe, I.; Destro, N.; Lorenzoni, G.; Berchialla, P. A First Estimation of the Impact of Public Health Actions against COVID-19 in Veneto (Italy). J. Epidemiol. Community Health 2020, 74, 858–860. [Google Scholar] [CrossRef] [PubMed]
- Alabdulrazzaq, H.; Alenezi, M.N.; Rawajfih, Y.; Alghannam, B.A.; Al-Hassan, A.A.; Al-Anzi, F.S. On the Accuracy of ARIMA Based Prediction of COVID-19 Spread. Results Phys. 2021, 27, 104509. [Google Scholar] [CrossRef]
- Mingione, M.; Alaimo Di Loro, P.; Farcomeni, A.; Divino, F.; Lovison, G.; Maruotti, A.; Lasinio, G.J. Spatio-Temporal Modelling of COVID-19 Incident Cases Using Richards’ Curve: An Application to the Italian Regions. Spat. Stat. 2021, 49, 100544. [Google Scholar] [CrossRef]
- Ohnishi, A.; Namekawa, Y.; Fukui, T. Universality in COVID-19 Spread in View of the Gompertz Function. Prog. Theor. Exp. Phys. 2020, 2020, 123J01. [Google Scholar] [CrossRef]
- Fokas, A.S.; Dikaios, N.; Kastis, G.A. Mathematical Models and Deep Learning for Predicting the Number of Individuals Reported to Be Infected with SARS-CoV-2. J. R. Soc. Interface 2020, 17, 20200494. [Google Scholar] [CrossRef]
- Thompson, R.N.; Hollingsworth, T.D.; Isham, V.; Arribas-Bel, D.; Ashby, B.; Britton, T.; Challenor, P.; Chappell, L.H.K.; Clapham, H.; Cunniffe, N.J.; et al. Key Questions for Modelling COVID-19 Exit Strategies. Proc. R. Soc. B 2020, 287, 20201405. [Google Scholar] [CrossRef] [PubMed]
- Lewnard, J.A.; Lo, N.C. Scientific and Ethical Basis for Social-Distancing Interventions against COVID-19. Lancet Infect. Dis. 2020, 20, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Padoan, A.; Sciacovelli, L.; Basso, D.; Negrini, D.; Zuin, S.; Cosma, C.; Faggian, D.; Matricardi, P.; Plebani, M. IgA-Ab Response to Spike Glycoprotein of SARS-CoV-2 in Patients with COVID-19: A Longitudinal Study. Clin. Chim. Acta 2020, 507, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Amendola, A.; Bianchi, S.; Gori, M.; Colzani, D.; Canuti, M.; Borghi, E.; Raviglione, M.C.; Zuccotti, G.V.; Tanzi, E. Evidence of SARS-CoV-2 RNA in an Oropharyngeal Swab Specimen, Milan, Italy, Early December 2019. Emerg. Infect Dis. 2021, 27, 648–650. [Google Scholar] [CrossRef] [PubMed]
- Galvani, A.P.; Parpia, A.S.; Pandey, A.; Sah, P.; Colón, K.; Friedman, G.; Campbell, T.; Kahn, J.G.; Singer, B.H.; Fitzpatrick, M.C. Universal Healthcare as Pandemic Preparedness: The Lives and Costs That Could Have Been Saved during the COVID-19 Pandemic. Proc. Natl. Acad. Sci. USA 2022, 119, e2200536119. [Google Scholar] [CrossRef] [PubMed]
- Alamo, T.; Reina, D.G.; Millán Gata, P.; Preciado, V.M.; Giordano, G. Data-Driven Methods for Present and Future Pandemics: Monitoring, Modelling and Managing. Annu. Rev. Control 2021, 52, 448–464. [Google Scholar] [CrossRef] [PubMed]
- Rohrich, R.J.; Hamilton, K.L.; Avashia, Y.; Savetsky, I. The COVID-19 Pandemic: Changing Lives and Lessons Learned. Plast. Reconstr. Surg. Glob. Open 2020, 8, e2854. [Google Scholar] [CrossRef] [PubMed]
- Chowell, G.; Hincapie-Palacio, D.; Ospina, J.; Pell, B.; Tariq, A.; Dahal, S.; Moghadas, S.; Smirnova, A.; Simonsen, L.; Viboud, C. Using Phenomenological Models to Characterize Transmissibility and Forecast Patterns and Final Burden of Zika Epidemics. PLoS Curr. 2016, 8. [Google Scholar] [CrossRef]
- Flaxman, S.; Mishra, S.; Gandy, A.; Unwin, H.J.T.; Mellan, T.A.; Coupland, H.; Whittaker, C.; Zhu, H.; Berah, T.; Eaton, J.W.; et al. Estimating the Effects of Non-Pharmaceutical Interventions on COVID-19 in Europe. Nature 2020, 584, 257–261. [Google Scholar] [CrossRef]
- Malthus, T.; Gilbert, G. An Essay on the Principle of Population; Oxford University Press: Oxford, UK, 2008; 172p. [Google Scholar]
- More, J.J. The Levenberg-Marquardt Algorithm: Implementation and Theory; Lecture Notes in Mathematics 630, Numerical Analysis; Watson, G.A., Ed.; Springer: Berlin/Heidelberg, Geramny, 1978; pp. 105–116. [Google Scholar]
- Colman, A. A Dictionary of Psychology, 3rd ed.; Oxford University Press: Oxford, UK, 2009; 882p. [Google Scholar]
- Goldberger, A. Econometric Theory; Wiley: New York, NY, USA, 1980; 399p. [Google Scholar]
- Richards, F.J. A flexible growth function for empirical use. J. Exp. Bot. 1959, 10, 290–300. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef] [PubMed]
- Finney, D.J. Bioassay and the Practice of Statistical Inference. Int. Stat. Rev. 1979, 47, 1–12. [Google Scholar] [CrossRef]
- Ritz, C.; Streibig, J. Bioassay Analysis Using R. J. Stat. Softw. 2005, 12, 1–22. [Google Scholar] [CrossRef]
- Kniss, A.R.; Vassios, J.D.; Nissen, S.J.; Ritz, C. Nonlinear Regression Analysis of Herbicide Absorption Studies. Weed Sci. 2011, 59, 601–610. [Google Scholar] [CrossRef]
- Seber, G.A.F.; Wild, C.J. Nonlinear Regression; Wiley & Sons: New York, NY, USA, 1989. [Google Scholar]
- Hastie, T.; Tibshirani, R. Generalized Additive Models; Chapman&Hall/CRC: Boca Raton, FL, USA, 1986; 335p. [Google Scholar]
- Simon, W. Thin plate regression splines. J. R. Stat. Soc. Ser. B Stat. Methodol. 2003, 65, 95–114. [Google Scholar]
- Cameron, A.C.; Trivedi, P.K. Regression Analysis of Count Data; Cambridge University Press: Cambridge, UK; New York, NY, USA, 1998; 411p. [Google Scholar]
- Asteriou, D.; Hall, S. Applied Econometrics, 2nd ed.; Palgrave Macmillan: Basingstoke, UK; New York, NY, USA, 2011; 499p. [Google Scholar]
- Wit, E.; Heuvel, E.v.d.; Romeijn, J.W. ‘All models are wrong...’: An introduction to model uncertainty. Stat. Neerl. 2012, 66, 217–336. [Google Scholar] [CrossRef]
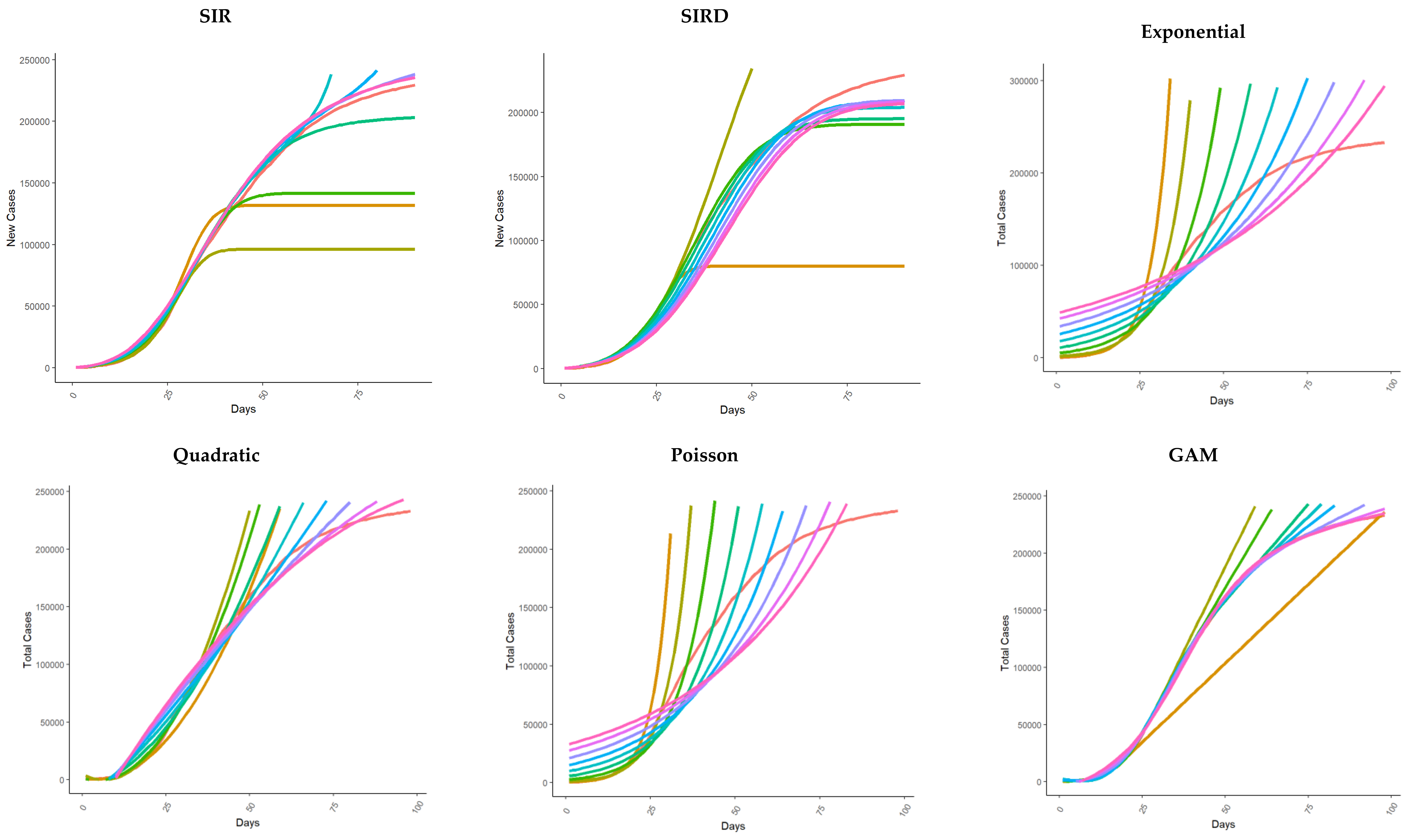
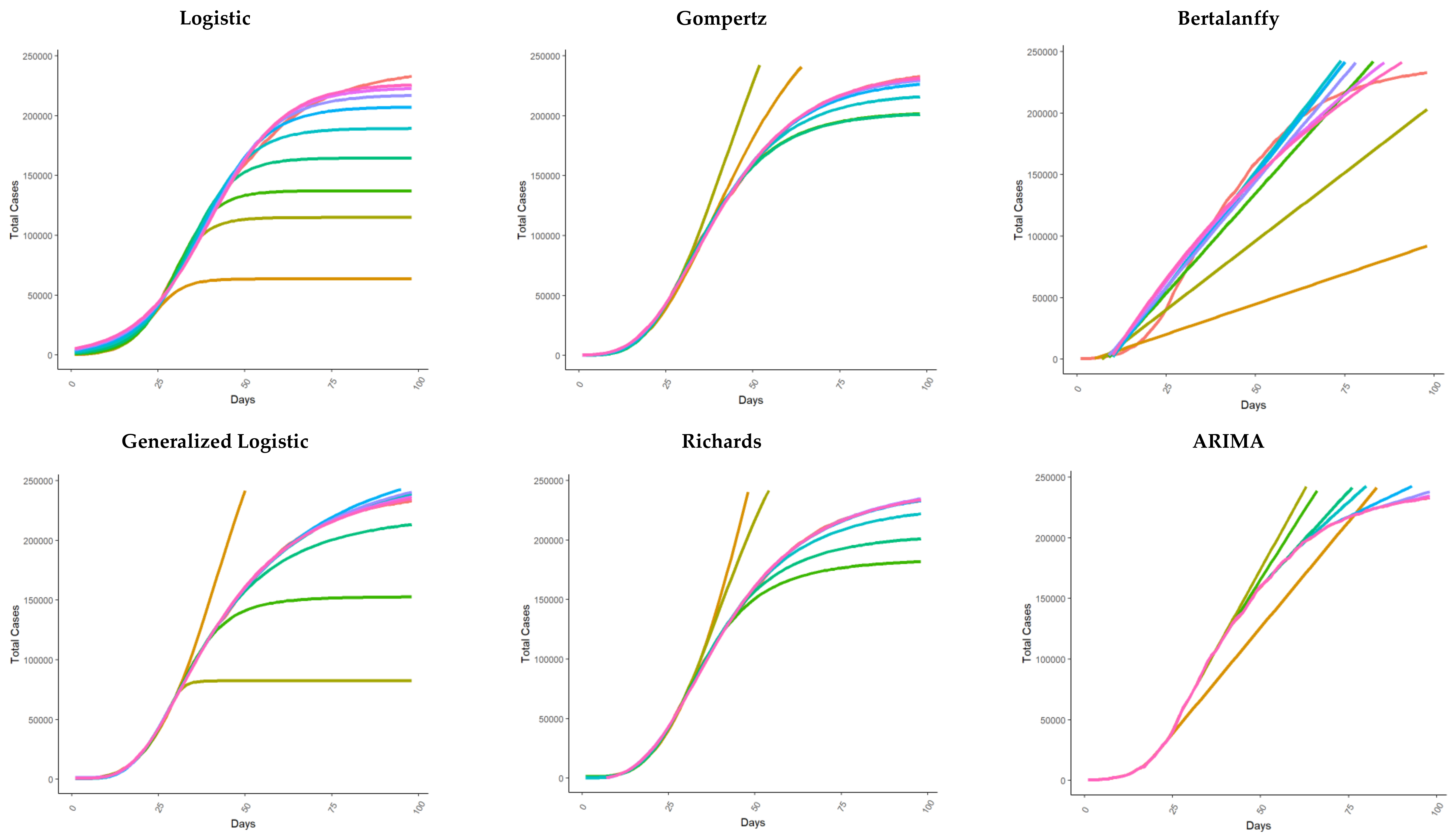
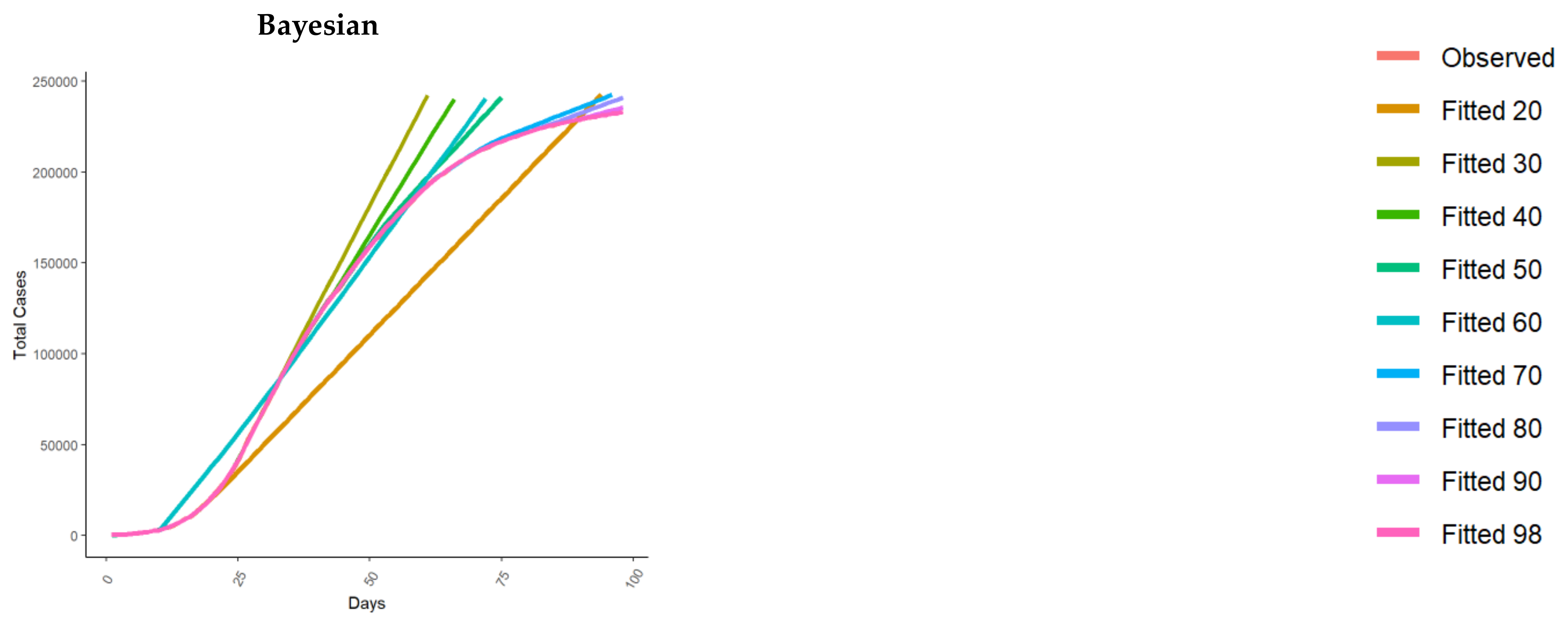


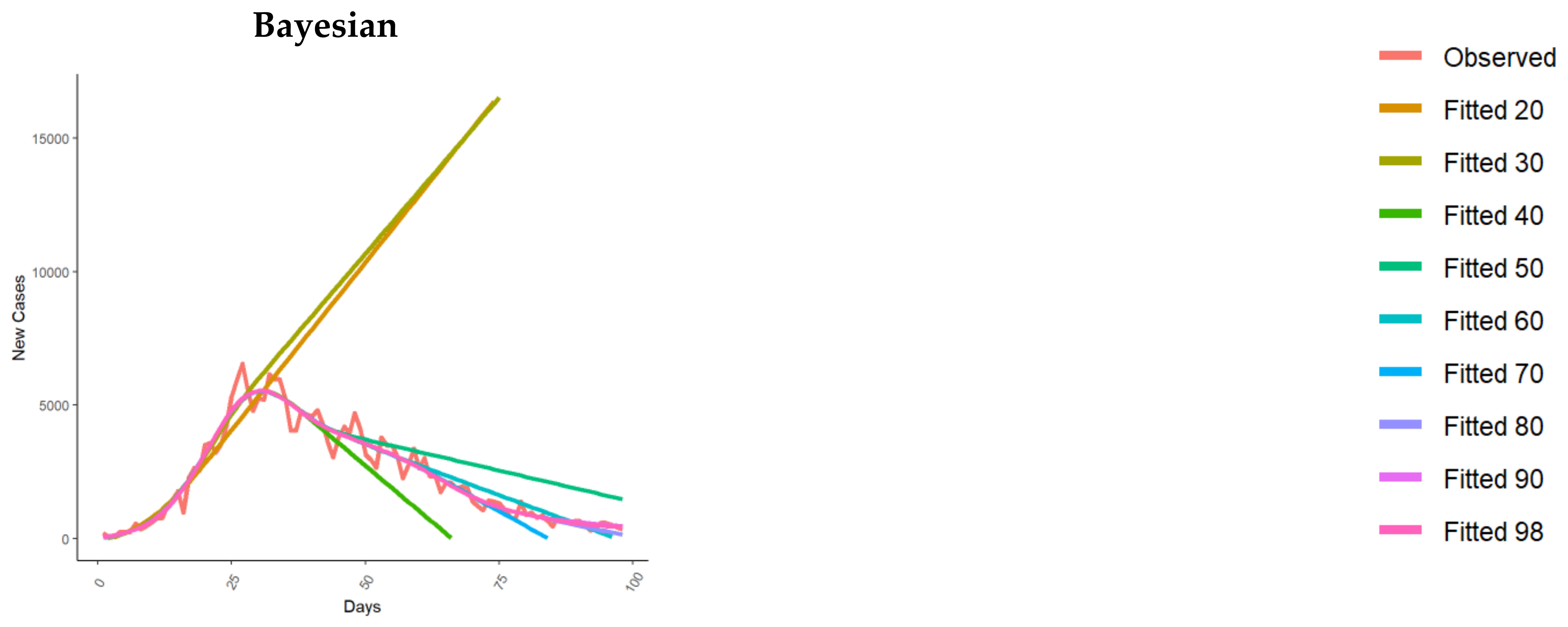
| Model | Reference(s) | |
|---|---|---|
| SIR | Nesteruk 2020 [37] | Nesteruk et al. predicted the numbers of infected, susceptible, and removed persons versus time. They applied a SIR model to predict the characteristics of the epidemic caused by SARS-CoV-2 in mainland China. |
| SIRD | Fanelli and Piazza 2020 [38] | Fanelli et al. analyzed the temporal dynamics of the coronavirus disease 2019 outbreak in China, Italy, and France in the time window of 22 January–15 March 2020, via a SIRD model to provide officials with realistic estimates for the time and magnitude of the epidemic peak. |
| Exponential | Remuzzi and Remuzzi 2020 [13] | Remuzzi et al. observed that in Italy, the number of patients infected since 21 February 2020 closely follows an exponential trend. Assuming the absence of contrast policies and, therefore, if the initial trend remains unchanged, the authors used the data observed up to March 8 to predict the trend of the epidemic curve in the following weeks. |
| Quadratic Regression | https://www.pangeadds.eu/demos/CoVid19/ (accessed on 1 June 2023) [39] | In [39], the authors applied to the Italian data up to 16 March 2020 a generalization of the exponential model, in which the rate of growth may decrease (or increase) linearly over time. Furthermore, this model is also an approximation for the logistic trend in the proximity of the non-exponential regime. |
| Logistic regression, Generalized logistic regression, and Richards | Vattay 2020 [14] Wu et al. 2020 [15] | In [14,15], the authors applied logistic regression models to monitor the effectiveness of measures taken by governments in the early phase of the epidemic in Italy and China, respectively, that were able to break the initial exponential trend. Wu et al. [15], using the Chinese experience up to 10 March, analyzed the calibration results also for Japan, South Korea, Iran, Italy, and Europe to make future scenario projections based on the results from different models. In Wu [15], logistic regression, generalized logistic regression, and Richards regression were applied. |
| Bertalanffy and Gompertz | Jia 2019 [12] | In [12], the authors adopted three kinds of mathematical models, i.e., the logistic model, the Bertalanffy model, and the Gompertz model, to analyze the situation of COVID-19 in China. First, the authors used 2003 SARS data to verify the three models to predict the trend of the epidemic, and then they used the three models to fit and analyze the epidemic trend of COVID-19 in Wuhan and non-Hubei areas in China. They predicted the total number of people expected to be infected, the total death toll, and the end time of the epidemic. |
| Generalized additive model (GAM) | Izadi and Farzali (2020) [40] | In [40], the authors applied a GAM model to capture the trend of the death rate and predict the occurrence of the peak in Canada. |
| Poisson generalized linear | Bonetti 2020 [17] | In [17], Bonetti developed a Poisson generalized linear model with a logarithmic link function and polynomial regression on time to report day-by-day estimates of the growth rate of the Italian infected subjects. |
| ARIMA | Benvenuto et al. 2020 [19] | In [19], the authors proposed the ARIMA model, a simple econometric approach, to predict the trend of the prevalence and incidence of COVID-19. The authors used data up to February 10, extracted from an online interactive dashboard hosted by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University, Baltimore, USA, created to illustrate the number of confirmed COVID-19 cases, deaths, and recoveries for all affected countries [41]. |
| Empirical Bayesian time series | Liu and Guo 2020 [25] | In [25], Liu and Guo proposed an empirical Bayesian time series framework to predict US cases using data from different countries as a prior reference, using the Johns Hopkins University CSSE data [41]. More in detail, Liu and Guo used the idea of internal time, i.e., the virus spread to different countries at different times, with trajectories different in calendar time but similar in internal time. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sciannameo, V.; Azzolina, D.; Lanera, C.; Acar, A.Ş.; Corciulo, M.A.; Comoretto, R.I.; Berchialla, P.; Gregori, D. Fitting Early Phases of the COVID-19 Outbreak: A Comparison of the Performances of Used Models. Healthcare 2023, 11, 2363. https://doi.org/10.3390/healthcare11162363
Sciannameo V, Azzolina D, Lanera C, Acar AŞ, Corciulo MA, Comoretto RI, Berchialla P, Gregori D. Fitting Early Phases of the COVID-19 Outbreak: A Comparison of the Performances of Used Models. Healthcare. 2023; 11(16):2363. https://doi.org/10.3390/healthcare11162363
Chicago/Turabian StyleSciannameo, Veronica, Danila Azzolina, Corrado Lanera, Aslihan Şentürk Acar, Maria Assunta Corciulo, Rosanna Irene Comoretto, Paola Berchialla, and Dario Gregori. 2023. "Fitting Early Phases of the COVID-19 Outbreak: A Comparison of the Performances of Used Models" Healthcare 11, no. 16: 2363. https://doi.org/10.3390/healthcare11162363
APA StyleSciannameo, V., Azzolina, D., Lanera, C., Acar, A. Ş., Corciulo, M. A., Comoretto, R. I., Berchialla, P., & Gregori, D. (2023). Fitting Early Phases of the COVID-19 Outbreak: A Comparison of the Performances of Used Models. Healthcare, 11(16), 2363. https://doi.org/10.3390/healthcare11162363










