Figure 1.
The basis functions (solid orange) and (broken blue used by MARS).
Figure 1.
The basis functions (solid orange) and (broken blue used by MARS).
Figure 2.
Diagram of a single hidden layer, feedforward neural network.
Figure 2.
Diagram of a single hidden layer, feedforward neural network.
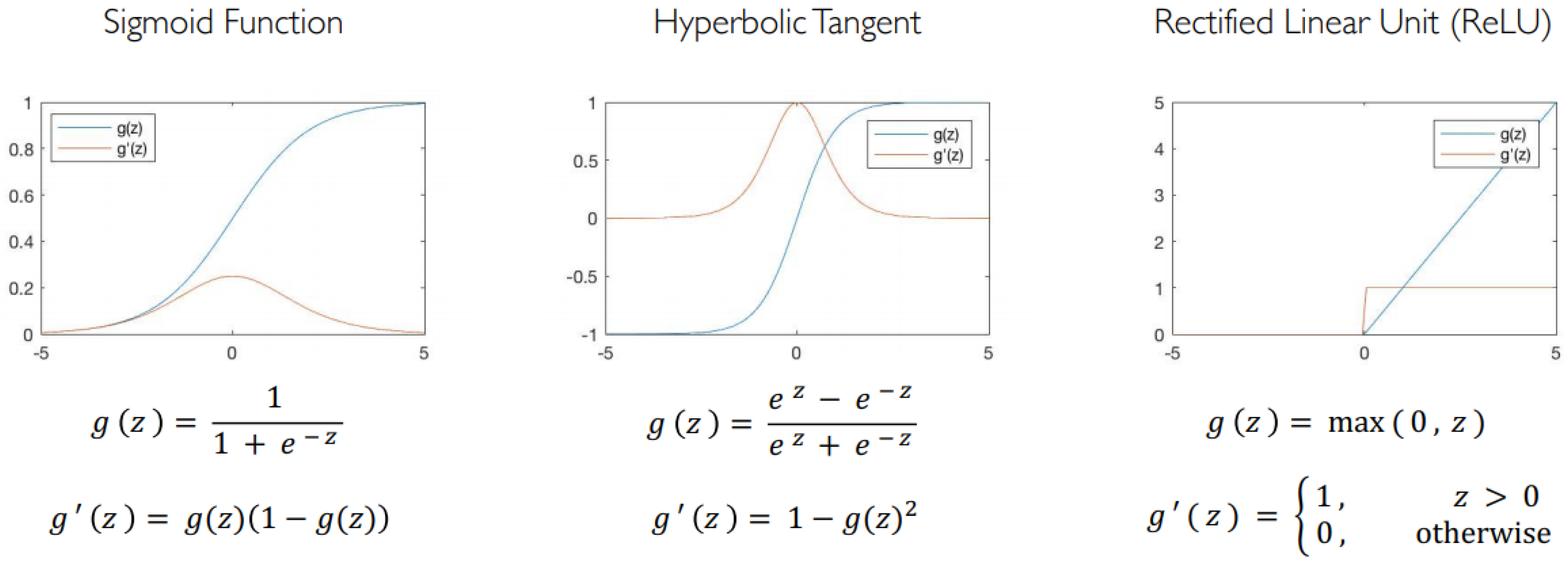
Figure 3.
Common activation functions.
Figure 3.
Common activation functions.
Figure 4.
Scatter plot and correlation between variables by response for the breast cancer data.
Figure 4.
Scatter plot and correlation between variables by response for the breast cancer data.
Figure 5.
Scatter plot and correlation between variables by response in heart disease data.
Figure 5.
Scatter plot and correlation between variables by response in heart disease data.
Figure 6.
Scatter plot and correlation between variables by response in prostate cancer data.
Figure 6.
Scatter plot and correlation between variables by response in prostate cancer data.
Figure 7.
Correlation between variables and PCA for the breast cancer data.
Figure 7.
Correlation between variables and PCA for the breast cancer data.
Figure 8.
Variable PCA for the breast cancer data.
Figure 8.
Variable PCA for the breast cancer data.
Figure 9.
Breast cancer PCA bipot.
Figure 9.
Breast cancer PCA bipot.
Figure 10.
Lasso using 10 fold cross validation for the breast cancer data.
Figure 10.
Lasso using 10 fold cross validation for the breast cancer data.
Figure 11.
Ridge using 10 fold cross-validation for the breast cancer data.
Figure 11.
Ridge using 10 fold cross-validation for the breast cancer data.
Figure 12.
Coefficients for our LR-lasso model for the breast cancer data. The dotted vertical line in the plot represents the with misclassification error (MCE) within one standard error of the minimum MCE.
Figure 12.
Coefficients for our LR-lasso model for the breast cancer data. The dotted vertical line in the plot represents the with misclassification error (MCE) within one standard error of the minimum MCE.
Figure 13.
Coefficients for our LR-ridge model for the breast cancer data. The dotted vertical line in the plot represents the with misclassification error (MCE) within one standard error of the minimum MCE.
Figure 13.
Coefficients for our LR-ridge model for the breast cancer data. The dotted vertical line in the plot represents the with misclassification error (MCE) within one standard error of the minimum MCE.
Figure 14.
Variables importance plot for lasso and ridge regression for the breast cancer data.
Figure 14.
Variables importance plot for lasso and ridge regression for the breast cancer data.
Figure 15.
Selecting and the number of components using 10-fold cross-validation for the breast cancer data.
Figure 15.
Selecting and the number of components using 10-fold cross-validation for the breast cancer data.
Figure 16.
Selecting the number of components by CV for the breast cancer data.
Figure 16.
Selecting the number of components by CV for the breast cancer data.
Figure 17.
Selecting the number of components by CV for the breast cancer data.
Figure 17.
Selecting the number of components by CV for the breast cancer data.
Figure 18.
MARS variables importance plot for the breast cancer data.
Figure 18.
MARS variables importance plot for the breast cancer data.
Figure 19.
Selecting the split size.
Figure 19.
Selecting the split size.
Figure 20.
Selecting the tree size for the breast cancer data.
Figure 20.
Selecting the tree size for the breast cancer data.
Figure 21.
RF important variables in predicting breast cancer.
Figure 21.
RF important variables in predicting breast cancer.
Figure 22.
Selecting the tree size for the breast cancer data.
Figure 22.
Selecting the tree size for the breast cancer data.
Figure 23.
GBM important variables for the breast cancer data.
Figure 23.
GBM important variables for the breast cancer data.
Figure 24.
Selecting the cost parameter for the breast cancer data.
Figure 24.
Selecting the cost parameter for the breast cancer data.
Figure 25.
SVM important variables for the breast cancer data.
Figure 25.
SVM important variables for the breast cancer data.
Figure 26.
FFNN important variables for the breast cancer data.
Figure 26.
FFNN important variables for the breast cancer data.
Figure 27.
RF for the breast cancer data.
Figure 27.
RF for the breast cancer data.
Table 1.
Name of variables and description for the heart disease data.
Table 1.
Name of variables and description for the heart disease data.
| Variable | Description |
|---|
| Id | Sample code number |
| I Cl.thickness | Clump Thickness |
| Cell.size | Uniformity of Cell Size |
| Cell.shape | Uniformity of Cell Shape |
| Marg.adhesion | Marginal Adhesion |
| Epith.c.size | Single Epithelial Cell Size |
| Bare.nuclei | Bare Nuclei |
| Bl.cromatin | Bland Chromatin |
| Normal.nucleoli | Normal Nucleoli |
| Mitoses | Mitoses |
| Class | Class |
Table 2.
% of variation explained by each component for the breast cancer data.
Table 2.
% of variation explained by each component for the breast cancer data.
| STATISTICS | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 |
|---|
| Std deviation | 2.43 | 0.88 | 0.73 | 0.68 | 0.62 | 0.55 | 0.54 | 0.51 | 0.30 |
| Prop of Variance | 0.66 | 0.09 | 0.06 | 0.05 | 0.04 | 0.03 | 0.03 | 0.03 | 0.01 |
| Cum Proportion | 0.66 | 0.74 | 0.80 | 0.85 | 0.89 | 0.93 | 0.96 | 0.99 | 1.00 |
Table 3.
Coefficient, prediction accuracy of lasso and ridge LR model for the breast cancer data.
Table 3.
Coefficient, prediction accuracy of lasso and ridge LR model for the breast cancer data.
| Features | LR-Ridge Coefficient | LR-Lasso Coefficient |
|---|
| Cl.thickness | 0.1712177 | 0.23606 |
| Cell.size | 0.1253754 | 0.07934 |
| Cell.shape | 0.1434834 | 0.18897 |
| Marg.adhesion | 0.1140987 | 0.05437 |
| Epith.c.size | 0.1267267 | NA |
| Bare.nuclei | 0.1421901 | 0.20966 |
| Bl.cromatin | 0.1804671 | 0.2299 |
| Normal.nucleoli | 0.1115392 | 0.08242 |
| Mitoses | 0.1274586 | NA |
| Pred. Accuracy | 0.9756 | 0.9659 |
Table 4.
Coefficient of principal components for the breast cancer data.
Table 4.
Coefficient of principal components for the breast cancer data.
| | Estimate | Std. Error | z Value | p-Value |
|---|
| (Intercept) | −1.263 | 0.3278 | −3.853 | 0.00011 |
| PC1 | −2.2708 | 0.2597 | −8.746 | 0.00000 |
| PC2 | 0.1489 | 0.387 | 0.385 | 0.07302 |
| PC3 | 0.7754 | 0.3923 | 1.977 | 0.04809 |
Table 5.
Selecting the optimal model for FFNN using misclassification error for the breast cancer data.
Table 5.
Selecting the optimal model for FFNN using misclassification error for the breast cancer data.
| Actva. | Epochs | Hidden | IDR | L1 | L2 | L.Rate | Miss.Err |
|---|
| Maxout | 100 | [9,3,2] | 0.050 | 8e-05 | 4e-05 | 0.020 | 0.006 |
| Maxout | 100 | [5,5,2] | 0.000 | 9e-05 | 8e-05 | 0.010 | 0.015 |
| Maxout | 100 | [9,2] | 0.000 | 2e-05 | 1e-02 | 0.010 | 0.015 |
| Maxout | 50 | [9,3,2] | 0.000 | 3e-05 | 5e-05 | 0.020 | 0.015 |
| Maxout | 100 | [9,2] | 0.000 | 9e-05 | 2e-02 | 0.020 | 0.015 |
Table 6.
All models applied to the breast cancer data.
Table 6.
All models applied to the breast cancer data.
| Model | Sensitivity | Specificity | Accuracy (95% CI) |
|---|
| LR-Ridge | 0.9724 | 0.9452 | 0.9656 (0.9440, 0.9920) |
| LR-Lasso | 0.9848 | 0.9315 | 0.9659 (0.9309, 0.9862) |
| LR-Enet | 0.9848 | 0.9452 | 0.9707 (0.9374, 0.9892) |
| LR-PC | 0.9848 | 0.9452 | 0.9707 (0.9374, 0.9892) |
| LR-PLS | 0.9773 | 0.9315 | 0.9610 (0.9246, 0.9830) |
| MARS | 0.9589 | 0.9621 | 0.9610 (0.9246, 0.9830) |
| SVM | 0.9848 | 0.9589 | 0.9756 (0.9440, 0.9920) |
| RF | 0.9773 | 0.9726 | 0.9756 (0.9440, 0.9920) |
| GBM | 0.9773 | 0.9589 | 0.9707 (0.9374, 0.9892) |
| FFNN | 0.9473 | 0.9920 | 0.97561 (0.9309, 0.9862) |
Table 7.
% variation explained by each component for the heart disease data.
Table 7.
% variation explained by each component for the heart disease data.
| | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 |
|---|
| Std Dev | 1.76 | 1.27 | 1.12 | 1.05 | 1.00 | 0.93 | 0.92 | 0.88 | 0.83 |
| Prop. var | 0.24 | 0.12 | 0.10 | 0.09 | 0.08 | 0.07 | 0.06 | 0.06 | 0.05 |
| Cum. Prop. | 0.24 | 0.36 | 0.46 | 0.54 | 0.62 | 0.69 | 0.75 | 0.81 | 0.86 |
Table 8.
% variation explained by each components for the prostate cancer data.
Table 8.
% variation explained by each components for the prostate cancer data.
| | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 |
|---|
| Std. Dev. | 1.87 | 1.28 | 0.93 | 0.78 | 0.68 | 0.65 | 0.57 | 0.40 |
| Prop. Var. | 0.44 | 0.21 | 0.11 | 0.08 | 0.06 | 0.05 | 0.04 | 0.02 |
| Cum. Prop. | 0.44 | 0.64 | 0.75 | 0.83 | 0.89 | 0.94 | 0.98 | 1.00 |
Table 9.
Comparing predictive performance of the 10 models applied to the heart disease data.
Table 9.
Comparing predictive performance of the 10 models applied to the heart disease data.
| Model | Sensitivity | Specificity | Accuracy (95% CI) |
|---|
| LR-Ridge | 0.9565 | 0.7358 | 0.8384 (0.7509, 0.9047) |
| LR-Lasso | 0.9783 | 0.6792 | 0.8182 (0.7280, 0.8885) |
| LR-Enet | 0.9565 | 0.7358 | 0.8384 (0.7509, 0.9047) |
| LR-PC | 0.9565 | 0.7358 | 0.8384 (0.7509, 0.9047) |
| LR-PLS | 0.9565 | 0.7358 | 0.8384 (0.7509, 0.9047) |
| MARS | 0.6038 | 0.8913 | 0.7374 (0.6393, 0.8207) |
| SVM | 0.9565 | 0.7358 | 0.8384 (0.7509, 0.9047) |
| RF | 0.9130 | 0.6981 | 0.7980 (0.7054, 0.872) |
| GBM | 0.9348 | 0.7358 | 0.8283 (0.7394, 0.8967) |
| FFNN | 0.8000 | 0.8700 | 0.8383 (0.7475, 0.8817) |
Table 10.
Comparing predictive performance of the 10 models applied to the prostate cancer data.
Table 10.
Comparing predictive performance of the 10 models applied to the prostate cancer data.
| Model | Sensitivity | Specificity | Accuracy (95% CI) |
|---|
| LR-Ridge | 0.7500 | 0.9048 | 0.8485 (0.681, 0.9489) |
| LR-Lasso | 1.0000 | 1.0000 | 1.0000 (0.8942, 1.0000) |
| LR-Enet | 1.0000 | 1.0000 | 1.0000 (0.8942, 1.0000) |
| LR-PC | 0.9167 | 1.0000 | 0.9697 (0.8424, 0.9992) |
| LR-PLS | 0.8333 | 0.8095 | 0.8182 (0.6454, 0.9302) |
| MARS | 1.0000 | 1.0000 | 1.0000 (0.8942, 1.0000) |
| SVM | 0.9167 | 0.7619 | 0.8182 (0.6454, 0.9302) |
| RF | 1.0000 | 1.0000 | 1.0000 (0.8942, 1.0000) |
| GBM | 1.0000 | 1.0000 | 1.0000 (0.8942, 1.0000) |
| FFNN | 0.9091 | 0.9091 | 0.9091 (0.7567, 0.9808) |
Table 11.
Performance assessment of the 10 models for the breast cancer data.
Table 11.
Performance assessment of the 10 models for the breast cancer data.
| Model | Specificity | Sens/Recall | Precision | ROC-AUC | PR-AUC |
|---|
| LR-Ridge | 0.9452 | 0.9724 | 0.9857 | 0.9977 | 0.9825 |
| LR-Lasso | 0.9315 | 0.9848 | 0.9714 | 0.9978 | 0.9825 |
| LR-Enet | 0.9452 | 0.9848 | 0.9718 | 0.9978 | 0.9823 |
| LR-PC | 0.9452 | 0.9848 | 0.9718 | 0.9970 | 0.9816 |
| LR-PLS | 0.9315 | 0.9773 | 0.9855 | 0.9949 | 0.9836 |
| RF | 0.9726 | 0.9773 | 0.9595 | 0.9984 | 0.8126 |
| GBM | 0.9589 | 0.9773 | 0.9589 | 0.9958 | 0.9798 |
| MARS | 0.9621 | 0.9589 | 0.9333 | 0.9901 | 0.9723 |
| SVM | 0.9589 | 0.9848 | 0.9351 | 0.9883 | 0.9546 |
| FFNN | 0.9920 | 0.9473 | 0.9730 | 0.9982 | 0.9832 |