Transferable Architecture for Segmenting Maxillary Sinuses on Texture-Enhanced Occipitomental View Radiographs
Abstract
1. Introduction
2. Materials and Methods
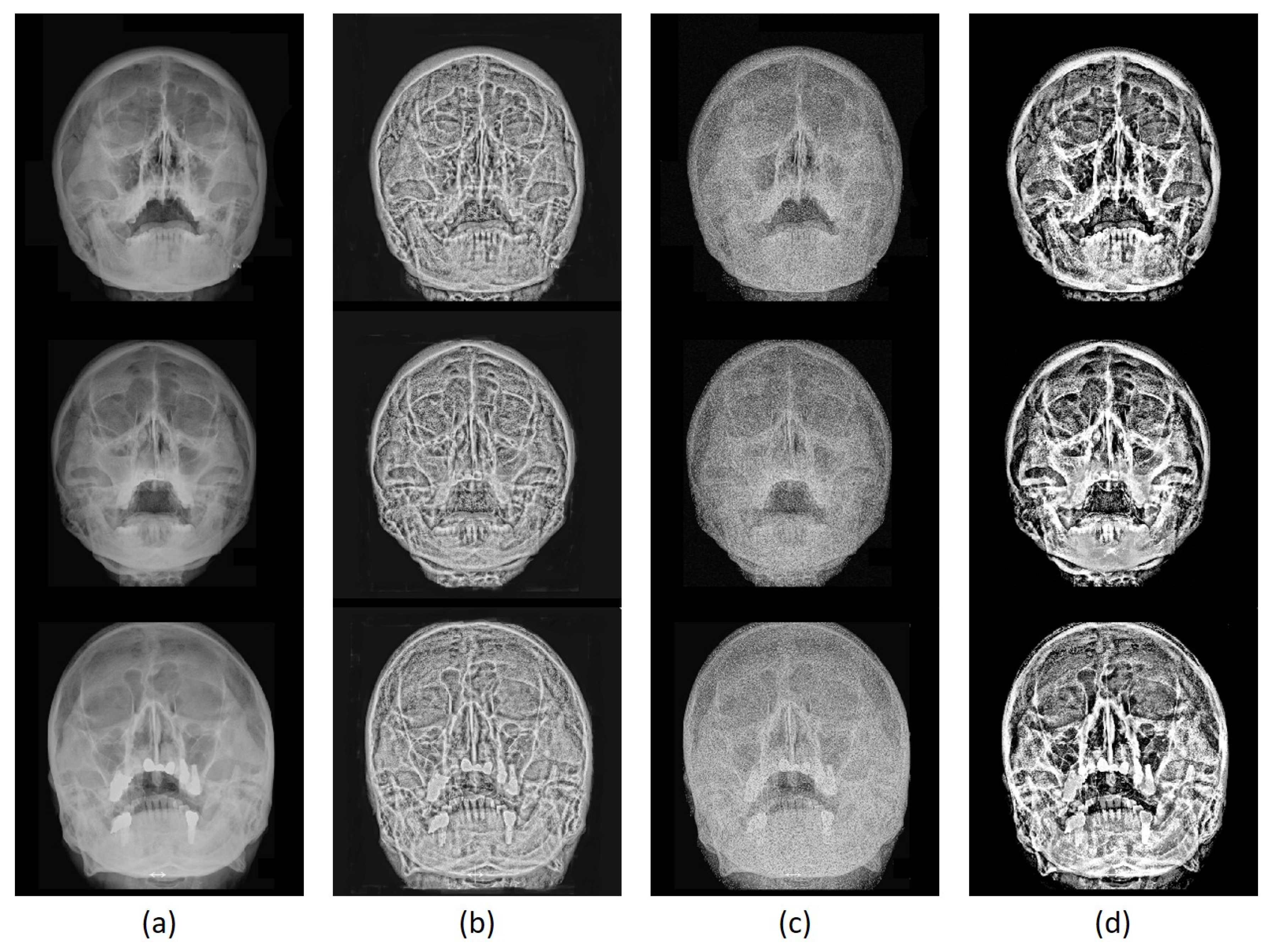
2.1. ToMA for Contrast Enhancement
- if (or ), then is grown row-wise (or column-wise) by one pixel;
- if (or ), then is grown row-wise (or column-wise) by one pixel;
- if (or ), then the expansion of is terminated horizontally (or vertically); and
- if both vertical and horizontal expansions have been terminated, a new segment (i.e., ) is created;
2.2. Transferable Neural Network Architecture
3. Results
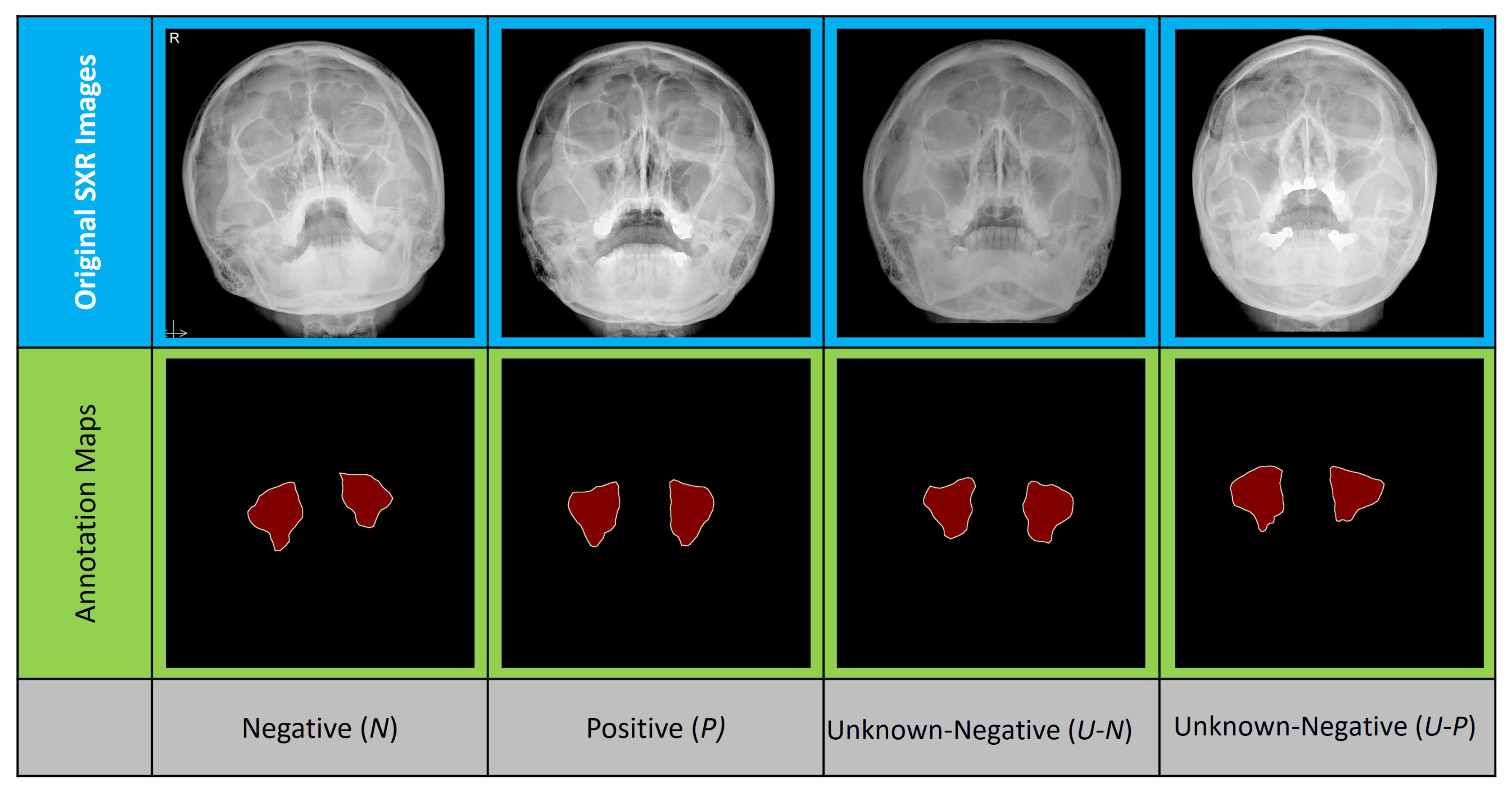
3.1. Dataset Specifications
3.2. Implementation Environment
3.3. Image Enhancement Evaluation
3.3.1. Quantitative Evaluation
3.3.2. Qualitative Evaluation
3.3.3. Complexity Evaluation
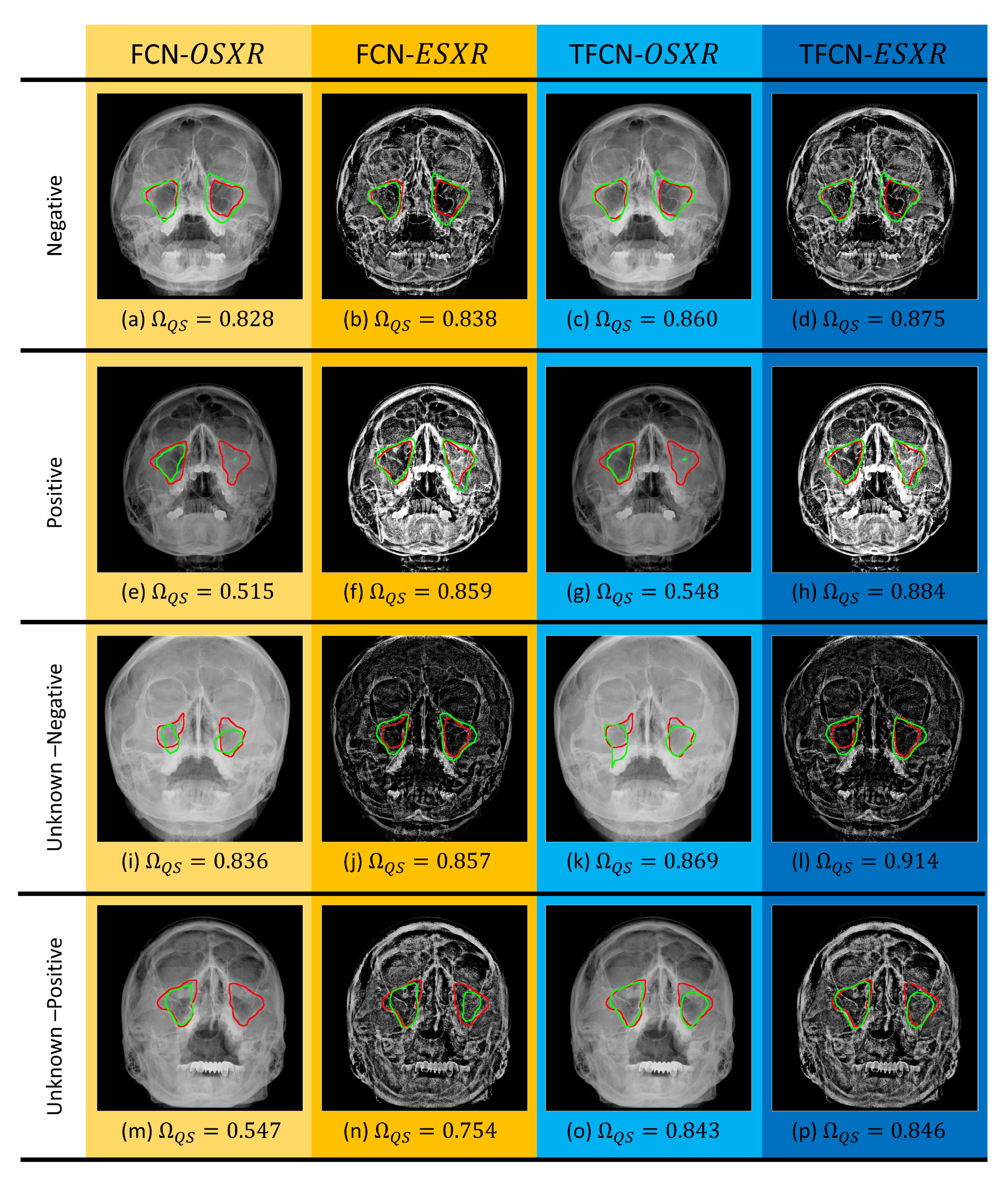
3.4. Image Segmentation Evaluation
Quantitative Evaluation
4. Discussion
4.1. Complexity Evaluation
4.2. Study Limitation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| SXR | Skull Radiography |
| ToMA | Texture-based Morphological Analysis |
| FCN | Transferable Fully Convolutional Network |
| T-FCN | Transferable fully Convolutional Network |
| CT | Computed Tomography |
| SCT | Sinsus Computed Tomography |
| CAD | computer-aided detection |
| CE | Combined Enhancement Measure |
References
- Hamilos, D.L. Chronic sinusitis. J. Allergy Clin. Immunol. 2000, 106, 213–227. [Google Scholar] [CrossRef]
- Drumond, J.P.N.; Allegro, B.B.; Novo, N.F.; de Miranda, S.L.; Sendyk, W.R. Evaluation of the prevalence of maxillary sinuses abnormalities through spiral computed tomography (CT). Int. Arch. Otorhinol. 2017, 21, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Lazar, R.H.; Younis, R.T.; Parvey, L.S. Comparison of plain radiographic coronal CT and intraoperative findings in children with chronic sinusitis. Otolarnygol.-Head Neck Surg. 1992, 107, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Konen, E.; Faibel, M.; Kleinbaum, Y.; Wolf, M.; Lusky, A.; Hoffman, C.; Eyal, A.; Tadmor, R. The value of the occipitomental (Waters’) view in diagnosis of sinusitis: A comparative study with computed tomography. Clin. Radiol. 2000, 55, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Mettler, F.A.; Huda, W.; Yoshizumi, T.T.; Mahesh, M. Effective doses in radiology and diagnostic nuclear medicine: A catalog. Radiology 2008, 248, 254–259. [Google Scholar] [CrossRef]
- Hussein, A.O.; Ahmed, B.H.; Omer, M.A.A.; Manafal, M.F.M.; Elhaj, A.B. Assessment of clinical x-ray and CT in diagnosis of paranasal sinus diseases. Int. J. Sci. Res. 2014, 3, 7–11. [Google Scholar]
- Sundaram, M.; Ramar, K.; Arumugam, N.; Prabin, G. Histogram based contrast enhancement for mammogram images. In Proceedings of the International Conference of Signal Processing, Communication and Computer Network Technology, Thuckafay, India, 21–22 July 2011; pp. 842–846. [Google Scholar]
- Ibrahim, H.; Hoo, S.C. Local contrast enhancement utilizing bidirectional switching equalization of separated and clipped subhistograms. Math. Probl. Eng. 2014. [Google Scholar] [CrossRef]
- Zhou, Y.; Shi, C.; Lai, B.; Jimenez, G. Contrast enhancement of medical images using a new version of the World Cup Optimization algorithm. Quant. Imag. Med. Surgery 2019, 9, 1528–1547. [Google Scholar] [CrossRef]
- Rundo, L.; Tangherloni, A.; Nobile, M.S.; Militello, C.; Besozzi, D.; Mauri, G.; Cazzaniga, P. MedGA: A novel evolutionary method for image enhancement in medical imaging systems. Expert Syst. Appl. 2019, 119, 387–399. [Google Scholar] [CrossRef]
- Muslim, H.S.; Khan, S.A.; Hussain, S.; Jamal, A.A.; Qasim, H.S. A knowledge-based image enhancement and denoising approach. Comput. Math. Organ. Theory 2019, 25, 108–121. [Google Scholar] [CrossRef]
- Nguyen, N.Q.; Lee, S.Q. Robust boundary segmentation in medical images using a consecutive deep encoder-decoder network. IEEE Access 2019, 7, 33795–33808. [Google Scholar] [CrossRef]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.O.; Villena-Martinez, V.; Garcia-Rodriguez, J. A review on deep learning techniques applied to semantic segmentation. arXiv 2017, arXiv:1704.06857. [Google Scholar]
- Weng, Y.; Zhou, T.; Li, Y.; Qiu, X. NAS-Unet: Neural architecture search for medical image segmentation. IEEE Access 2019, 7, 44247–44257. [Google Scholar] [CrossRef]
- Zhang, G.; Dong, S.; Xu, H.; Zhang, H.; Wu, Y.; Zhang, Y.; Xi, X.; Yin, Y. Correction learning for medical image segmentation. IEEE Access 2019, 7, 143597–143607. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. CoRR 2014, 2014, 1–10. [Google Scholar]
- Zhao, W.; Zhang, h.; Yan, Y.; Fu, Y.; Wang, H. A Semantic Segmentation Algorithm Using FCN with Combination of BSLIC. Appl. Sci. 2018, 8, 500. [Google Scholar] [CrossRef]
- Yasrab, R. ECRU: An Encoder-Decoder Based Convolution Neural Network (CNN) for Road-Scene Understanding. J. Imaging 2018, 4, 116. [Google Scholar] [CrossRef]
- Jia, Y.; Shelhamer, E.; Donahue, J.; Karayev, S.; Long, J.; Girshick, R.; Guadarrama, S.; Darrell, T. Caffe: Convolution architecture for fast feature embedding. CoRR 2014, 2014, 675–678. [Google Scholar]
- Alp’s Image Segmentation Tool. 2017. Available online: shorturl.at/fjopM (accessed on 30 September 2017).
- Wei, Q.; Dunbrack, R.L. The role of balanced training and testing datasets for binary classifiers in bioinformatics. PLoS ONE 2013, 8, e67863. [Google Scholar]
- Lado, M.J.; Tahoces, P.G.; Mendez, A.J.; Souto, M.; Vidal, J.J. A wavelet-based algorithm for detection clustered microcalcifications in digital mammograms. Med. Phys. 1999, 26, 1294–1305. [Google Scholar] [CrossRef]
- Singh, S.; Bovis, K. An evaluation of contrast enhancement techniques for mammographic breast masses. IEEE Trans. Inf. Technol. Biomed. 2005, 9, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Morrow, W.M.; Paranjape, R.B.; Rangayyan, R.M.; Desautels, J.E.L. Region-based contrast enhancement of mammograms. IEEE Trans. Med. Imaging 1992, 11, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Radau, P.; Lu, Y.; Connelly, K.; Paul, G.; Dick, A.J.; Wright, G.A. Evaluation framework for algorithms segmenting short axis cardiac MRI. MIDAS J. 2009, 49. Available online: http://hdlhandlenet/10380/3070 (accessed on 9 May 2020).

| Layer | c | k | |||
|---|---|---|---|---|---|
| 11 | 3 | 96 | 4 | 100 | |
| 3 | 0 | 0 | 2 | 0 | |
| 5 | 48 | 256 | 4 | 100 | |
| 3 | 0 | 0 | 2 | 0 | |
| 3 | 256 | 384 | 4 | 100 | |
| 3 | 192 | 384 | 4 | 100 | |
| 3 | 192 | 256 | 4 | 100 | |
| 3 | 0 | 0 | 2 | 0 | |
| 6 | 256 | 4096 | 4 | 100 | |
| 1 | 4096 | 4096 | 4 | 100 | |
| 1 | 4096 | 2 | 4 | 100 | |
| 63 | 1 | 2 | 4 | 100 |
| Diagnosis | Subtotals | |||
|---|---|---|---|---|
| N | 45 | 7 | 10 | 62 |
| P | 45 | 7 | 11 | 63 |
| 50 | 15 | 11 | 76 | |
| 7 | 4 | 2 | 13 | |
| Totals | 147 | 33 | 34 | 214 |
| Metric | HM-CLAHE [7] | LCE-BSESCS [8] | Proposed ToMA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | P | U | N | P | U | N | P | U | |
| 0.39 | 0.40 | 0.39 | 0.48 | 0.49 | 0.47 | 0.65 | 0.66 | 0.63 | |
| 1.73 | 1.73 | 1.73 | 0.83 | 0.77 | 0.80 | 0.17 | 0.12 | 0.12 | |
| 2.78 | 1.64 | 1.07 | 0.85 | 0.84 | 0.85 | 54.32 | 36.18 | 28.31 | |
| Fold Type | Diagnosis | Data Amount | True Negative/True Positive |
|---|---|---|---|
| U | True Negative | 64 | 0.84 |
| False Positive | 12 | ||
| True Positive | 11 | ||
| False Negative | 2 |
| Method | Time Cost (seconds) | ||
|---|---|---|---|
| Negative Fold | Positive Fold | Unknown Fold | |
| HM-CLAHE [7] | 154.14 | 152.06 | 151.55 |
| LCE-BSESCS [8] | 107.74 | 93.63 | 102.46 |
| Proposed Method | 26.46 | 26.19 | 26.35 |
| Metric | FCN [16] | T-FCN | ||
|---|---|---|---|---|
| 0.628 | 0.699 | 0.717 | 0.756 | |
| 0.755 | 0.819 | 0.829 | 0.857 | |
| 306.8 | 95.03 | 132.5 | 33.69 | |
| Consumption | FCN [16] | T-FCN | ||
|---|---|---|---|---|
| Learning | 541 | 283 | 7 () | 7 () |
| (mins) | 522 () | 284 () | ||
| Inference | 1.67 | 1.58 | 1.56 | 1.53 |
| (secs/image) | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chondro, P.; Haq, Q.M.u.; Ruan, S.-J.; Li, L.P.-H. Transferable Architecture for Segmenting Maxillary Sinuses on Texture-Enhanced Occipitomental View Radiographs. Mathematics 2020, 8, 768. https://doi.org/10.3390/math8050768
Chondro P, Haq QMu, Ruan S-J, Li LP-H. Transferable Architecture for Segmenting Maxillary Sinuses on Texture-Enhanced Occipitomental View Radiographs. Mathematics. 2020; 8(5):768. https://doi.org/10.3390/math8050768
Chicago/Turabian StyleChondro, Peter, Qazi Mazhar ul Haq, Shanq-Jang Ruan, and Lieber Po-Hung Li. 2020. "Transferable Architecture for Segmenting Maxillary Sinuses on Texture-Enhanced Occipitomental View Radiographs" Mathematics 8, no. 5: 768. https://doi.org/10.3390/math8050768
APA StyleChondro, P., Haq, Q. M. u., Ruan, S.-J., & Li, L. P.-H. (2020). Transferable Architecture for Segmenting Maxillary Sinuses on Texture-Enhanced Occipitomental View Radiographs. Mathematics, 8(5), 768. https://doi.org/10.3390/math8050768






