1. Introduction
Spectroscopy techniques, such as infrared and Raman spectroscopy, are increasingly being used to measure and analyze the physical and chemical properties of materials. There are two types of analysis methods related to this technique. The first is to identify the constituents of a given spectrum, and the second is to identify the spectrum itself by comparing it directly to other known spectra in the database [
1,
2]. The second type of analysis is addressed in this study.
Spectral identification methods can be divided into two categories: classification methods based on machine learning (ML) and algorithms based on the similarity evaluation [
3]. The first methods show good classification performance through an optimal learning model by training a given database with a ML-based algorithm. Conventionally, k-nearest neighbor (KNN) [
4], random forest (RF) [
5] and artificial neural network (ANN) [
6] methods have been proposed, and various 1D-convolutional neural network(CNN) models based on deep learning have recently been proposed [
7,
8]. Good identification performance is expected from these methods if a sufficient number of samples in each spectrum is obtained.
However, most existing Raman libraries provide one sample for each type, such that ML methods require significant time to build up sufficient samples of the target material. The other methods are more suitable for utilizing existing Raman libraries. Representative methods include correlation search [
9] and cosine similarity, the Hit-quality index (HQI) [
10], and the Euclidean distance (ED) search [
11,
12]. These methods are intuitive and have often been used for identifying different types of Raman spectra. In recent years, methods have been proposed that improve identification performance in various applications along with the moving window technique [
13,
14].
The spectral database is growing exponentially, and therefore, searching for similar spectra is significantly more demanding. Further, larger databases are being built, as existing databases can be merged and reused along with technologies that complement the characteristics of measurement equipment [
9,
15], making high-speed search an essential and demanding task. A highly viable, fast search method is particularly important in embedded systems with limited computing power, such as handheld spectrometer systems [
16,
17,
18]. These portable Raman spectrometers are often used at accident sites, crime scenes or terrorist threat sites due to their advantages such as portability and maneuverability [
19]. In particular, applications that detect hazardous substances such as explosives and poisons require fast and accurate solutions [
20].
A more suitable identification method for the above fast search applications is the similarity evaluation methods. The first methods require significant calculations in the learning process depending on the volume of the database and the number of samples of the data. To introduce the ML methods to Raman systems with limited computing power, such as handheld Raman spectroscopy, hardware technologies such as the field-programmable gate array (FPGA) must be incorporated [
21,
22]. These methods are currently showing remarkable achievements owing to the breakthroughs in hardware. Meanwhile, because the identification method based on similarity evaluation can be applied even if there is only one sample representing the data type, there is no difficulty in applying it to the existing Raman library. Furthermore, there is no separate learning process, and it has the advantage of simply configuring an identification system.
The simplest and most commonly applied comparison method for spectral identification is to calculate and compare the ED between a given spectrum and the spectrum in the database, i.e., the reference spectra. This method has a structure very similar to the HQI method and determines the spectrum with the closest distance to the input spectrum as the identity of the input material.
Several fast search methods have been proposed in the context of vector quantization (VQ). However, the Raman spectrum is generally higher dimensional than the image covered by VQ, and hence, an appropriate method is required to solve this problem. Therefore, to introduce the major algorithms of VQ into the Raman identification system, it is necessary to analyze the mathematical modelling methods and key characteristics of each algorithm.
The conventional fast ED comparison methods can be classified into two groups. The methods of the first group do not solve the nearest neighbor problem itself; however, these methods find approximately the same solutions in terms of the mean squared error. These methods generally rely on the use of data structures such as K-dimensional trees and other types of structures that facilitate the fast search of reference data [
23,
24]. These methods are very fast, but they are not considered in this study because an exact solution, and not an approximate one, is required.
Conversely, the methods provided in the second group deal with exact solutions to the nearest neighbor search problem. These typically include the partial distance search (PDS) and projection-based search algorithms. A very simple but effective approach is the PDS method reported by Bei and Gray [
25]. In this method, when the cumulative sum of the EDs between the reference data and the input signal is larger than the distance of the current closest candidate, the distance calculation is terminated to reduce computational costs [
26,
27]. This method does not require memory overhead; however, the reduction in computational cost is limited.
Projection methods without transformation of input data such as the equal-average nearest neighbor search (ENNS) and its variants [
28,
29,
30] reduce unnecessary searches by using the mean of the input data. These methods provide a significant reduction in computational time compared to the full search method. However, they have their own weakness in that the performance gain is not significant unless the mean of the data is not distinctly different. To overcome the weakness of these methods, the mean pyramid search (MPS) method [
31,
32] was proposed. This method could avoid the weakness by using a local segment mean. It generally performs better than the ENNS.
Projection methods have likewise been proposed to transform the input data using singular value decomposition (SVD), discrete wavelet transform (DWT), and Karhunen–Loeve Transform (KLT) also known as principal component transformation (PCT). One of the most important properties of these transformations is that most of the data energy can be stored using very few coefficients [
33,
34]. Based on these advantages, improved search methods combined with existing fast algorithms have been proposed, providing faster search capabilities.
Apart from data transformation, our method adopted a cluster structure to reduce the search area. Spectra from the database to be compared were pre-clustered into hierarchical cluster groups based on similarity. In the proposed method, the spectra were sequentially tested in the order of cluster distance, starting with the reference spectrum belonging to the nearest clusters, which helped to quickly find the nearest spectrum. The search process was further accelerated using a pilot search with fewer dimensions of the transformed data to find the closest candidates as quickly as possible.
The remainder of this paper is organized as follows. In
Section 2, the main methods in the existing VQ field were reviewed and compared. In
Section 3, a novel fast search algorithm is described. To confirm the applicability of our method, the simulation results of the proposed method are compared with those of existing methods in
Section 4. Finally, a brief conclusion is presented.
4. Experimental Section
A total of 40 chemicals and 12 explosives were prepared using ≥ 99% concentration standard from Sigma-Aldrich (St. Louis, MO, USA) and the materials were measured using three Raman instruments. They merged with a commercial Raman library (Thermo Fisher Scientific) of 14,033 spectra to form a Raman database of 14,085 spectra. The Raman database consists of one template for each material.
Table 1 shows the detailed specifications of the four Raman spectroscopy systems used to measure the spectrum.
All spectra were adjusted to have a resolution of 201–3500 cm
by resampling and were preprocessed with additive noise reduction and background noise removal [
38,
39].
Figure 3 shows an example of Raman spectra after preprocessing. The types of chemicals are acetonitrile, benzene, cyclohexane, and toluene.
To analyze the performance of the algorithm, 2817 types of the Raman spectrum, which is 20% of the database, was searched from all 14,085 types of the Raman spectrum. Similar to the real spectrum, noise of approximately 15, 20, and 25 dB was added to the input spectrum. The factor influencing the identification performance of the proposed method is the noise of the input spectrum used in the experiment. The identification performance of the spectrum acquired under harsh noise conditions is expected to be relatively low, and the well-removed spectrum can be expected to have good identification performance. Therefore, it is important to introduce suitable noise reduction methods and find optimal parameters.
However, the VQ method has no effect on the identification performance of the full search method, which is the reference identification algorithm. Therefore, to focus on the aim of this study, the main content of this paper is limited to the VQ issue.
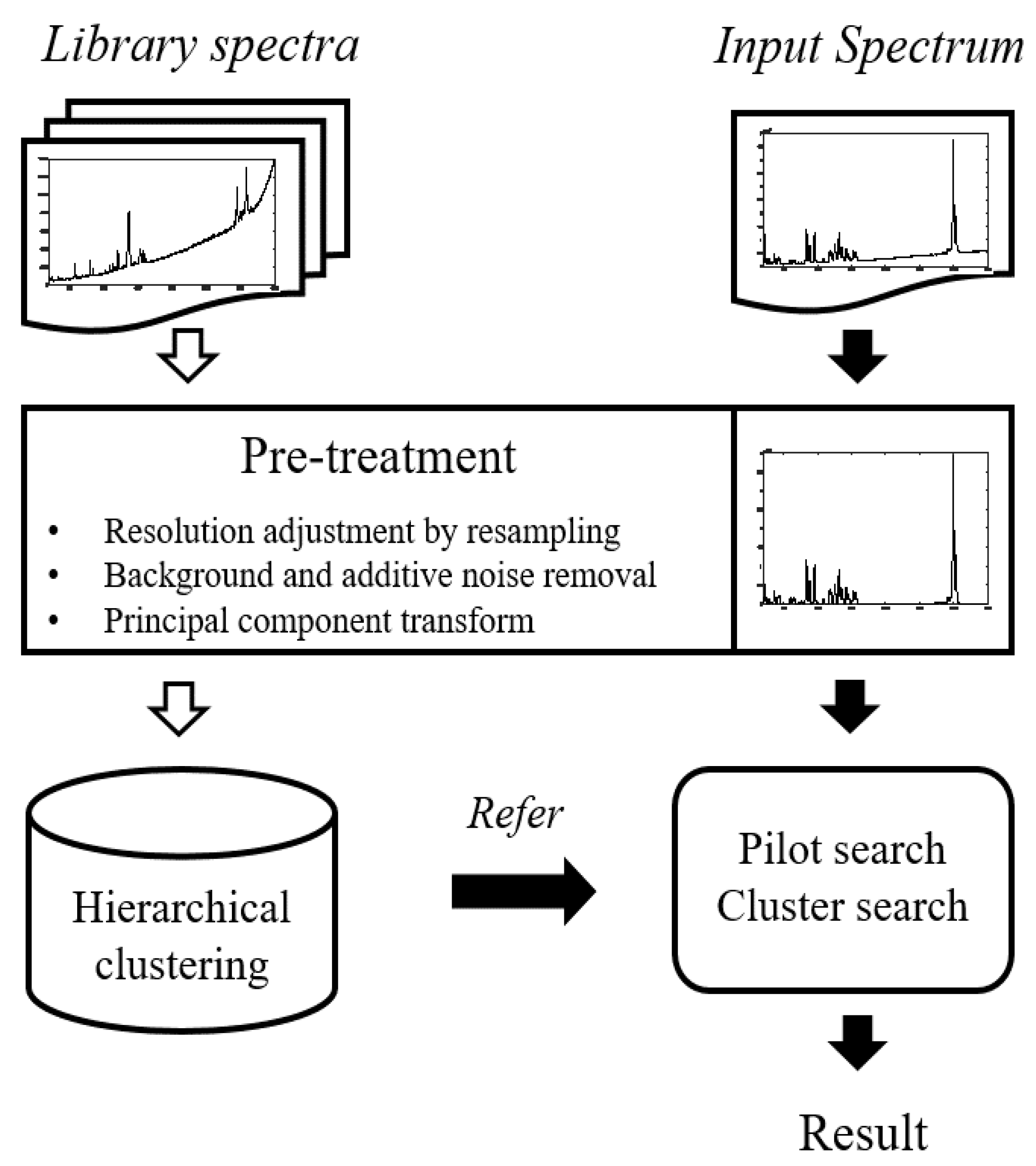
Figure 4 depicts a flowchart of the proposed method including preprocessing. Black arrows on the right indicate real-time processes, while white arrows indicate previously calculated processes.
5. Results and Discussion
There are several ways to evaluate the computational complexity of an algorithm. Among them, the execution speed depends on various aspects of the CPU, such as the instruction mix, pipeline structure, cache memory, and the number of cores, thus rendering it difficult to find an explicit relationship between the search speed and execution time. Therefore, in this study, the number of necessary additions and multiplications was chosen as a criterion for evaluating the search speed.
In general, the fast search technique in VQ presumes the same identification performance as the full search technique, which uses the entire dimension of the input data. Therefore, all experimental results were compared focusing only on the computational complexity, under the same identification performance conditions as the full search.
Preliminary experiments were conducted to determine the appropriate number of clusters and PCs.
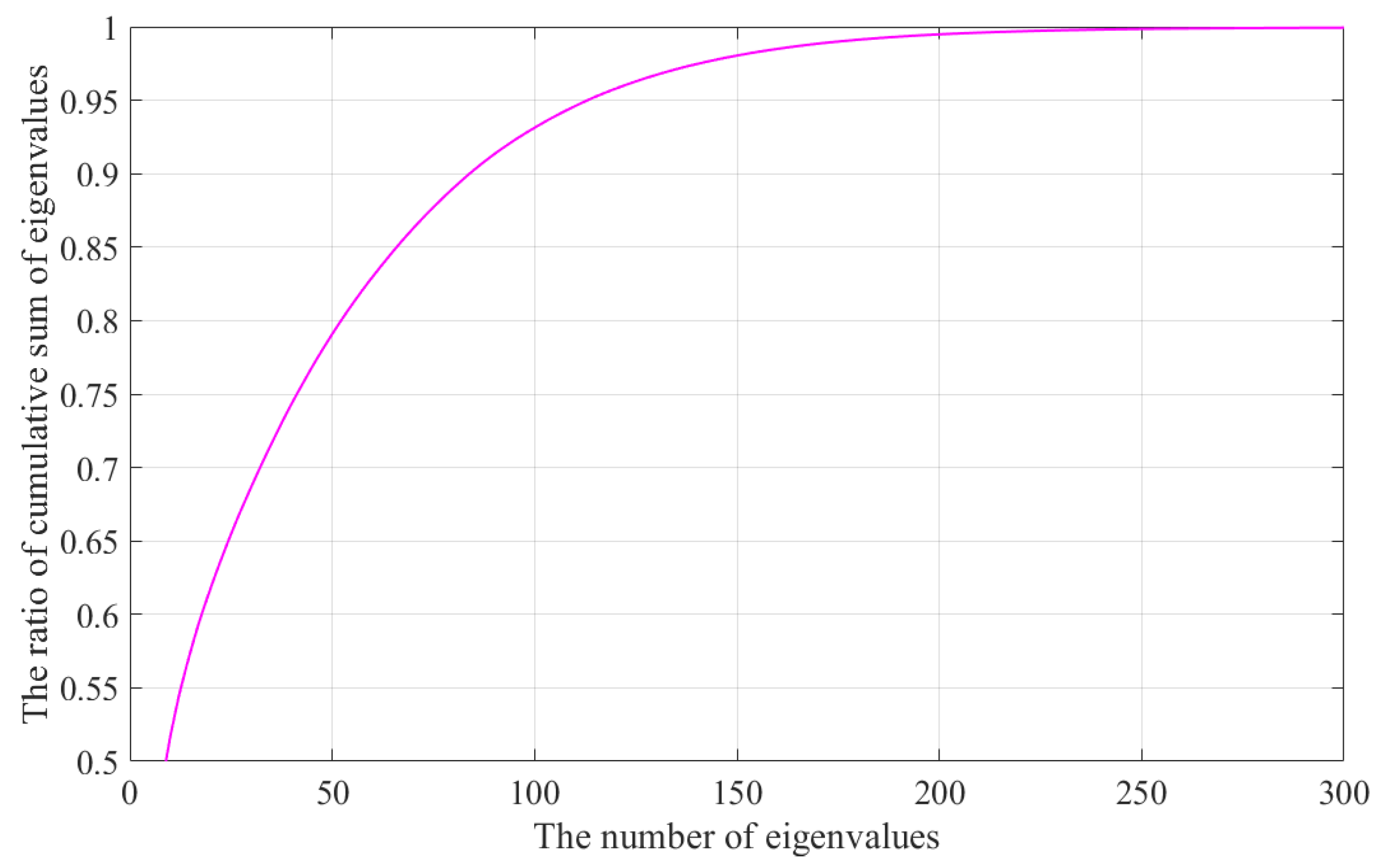
Table 2 shows the results of the PTC + PDS method according to the number of PCs. This method showed the least computational complexity when 150 PCs were used. In
Section 2, it was discussed that 250 PCs contain almost all the information from the 3300 raw data points. However, this does not indicate that it is the best parameter. Based on this result, we determined the optimal number of clusters.
Table 3 shows the number of multiplications and additions according to the number of clusters in CS using 150 PCs.
According to
Table 3, the computational complexity decreases as the number of clusters increases. However, once the number of clusters exceeds 80, the computational complexity increases again. Therefore, the number of clusters was set at 80 in this study.
Subsequently, the number of clusters was fixed at 80, and the computational complexity, cluster skip, and element skip according to the number of PCs were analyzed. The results are presented in
Table 4.
Cluster skip refers to the exclusion of all cluster members from the candidates. In contrast, element skip implies excluding some data in the cluster from the candidates. As the number of PCs increases, the number of cluster skips increases, as information regarding the spectrum that can be used in Equation (
10) increases. Therefore, more clusters can be excluded than when the number of PCs is small. However, the overall computational complexity must be considered, as converting more PCs requires additional computation. According to
Table 4, the best results are obtained using 80 PCs. For all PCs considered in the experiment, the sum of the number of cluster and element skips approximates the number of total clusters 80. Hence, it can be confirmed that the cluster structure effectively excludes spectra that cannot be candidates.
Finally, the pilot search was applied to further speed up the search.
Table 5 shows the experimental results obtained while investigating the effect of the number of PCs used in the pilot search. The results show typical trade-off characteristics. As the number of PCs for the pilot search increases, the computational amount of the pilot search decreases, while the computational amount of CS increases. Due to these characteristics, the total number of calculations is similar. This means that the pilot test reliably helps improve the performance. In the following experiments, we chose 40 as the number of PCs for the pilot search.
To analyze the performance of each algorithm, including the proposed method, 2817 Raman spectra were assessed, and the results are listed in
Table 6. PDS significantly reduces the number of additions. In contrast, MPS is more effective in reducing the number of multiplications. This is because MPS relies on the segmental mean, which reduces the need for addition rather than multiplication. The overall performance depends on the characteristics of the data. If the data are not distributed around the mean, the benefit decreases. This is the reason for introducing the CS.
The combination of MPS1D and PDS reduces the overall computation complexity compared to the case of using only MPS1D. The MPS1D_sort method sorts the reference spectra according to the mean in advance. If the difference between the mean is more than K times the minimum distance, further search is not needed as in ENNS, which speeds up the search. This method showed a performance improvement of approximately compared to the MPS1D + PDS.
Subsequently, PCT with PDS shows a reduction of approximately in computational complexity compared to MPS1D_Sort + PDS. The proposed method, CS, showed an improvement of approximately when using the same number of PCs as PCT + PDS, and nearly when using the optimal number of PCs. From these results, it was confirmed that pre-determining the cluster structure of the database is effective for fast search. These properties are not just found in CS. MPS1D, a method of determining the search structure in advance through database analysis, also showed faster search speed compared to the method without structure. Determining an appropriate search structure is a critical issue as this structure is created in advance and does not affect the real-time search.
Finally, the introduction of the pilot search reduced the computational amount by approximately
compared to CS (80 PCs). This result corresponds to a computational complexity equivalent to
that of the full search. The overall average results of the major algorithms are shown in
Figure 5 for convenience.
6. Conclusions
In this paper, we proposed a novel search method that speeds up the search for the identification of Raman spectra. The principal component transformation was introduced along with the cluster structure. The reduced number of data dimensions reduces the computational complexity of distance calculations, and the cluster structure combines the well-known trigonometric inequality with PDS to exclude numerous spectra that cannot be the candidate from the search. Finally, the pilot search was applied to further speed up the search. Optimal parameters of the proposed method were investigated and determined experimentally.
Moreover, various algorithms in the VQ field were modified and introduced into structures suitable for 1D signals and compared with the proposed method. From the results of the experiments, it was found that the proposed method significantly surpassed the existing method in terms of the required number of additions and multiplications.
The proposed method is particularly suitable for systems with relatively limited computing power, such as portable Raman spectroscopy and the compact Raman spectrometer. In addition, applications such as hazardous substance detection require fast and accurate detection, and the search time is particularly long for the large database of 14085 considered in the paper, so the proposed technique can be used appropriately. We expect that the proposed method and its variants will be a promising alternative to the spectral search problems in the above-mentioned applications.