Mathematical Modeling of Japanese Encephalitis under Aquatic Environmental Effects
Abstract
1. Introduction
2. Model Formulation
- we do not consider immigration of infected humans;
- the human population is not constant (we consider a disease-induced death rate, due to fatality, of 25%);
- we assume that the coefficient of transmission of the virus is constant and does not vary with seasons, which is reasonable due to the short course of the disease;
- mosquitoes are assumed to be born susceptible.
3. Mathematical Analysis of the JE Model
4. Numerical Simulations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Python Code for Figure 1–3
References
- Boyer, S.; Peng, B.; Pang, S.; Chevalier, V.; Duong, V.; Gorman, C.; Dussart, P.; Fontenille, D.; Cappelle, J. Dynamics and diversity of mosquito vectors of Japanese encephalitis virus in Kandal province, Cambodia. J. Asia-Pac. Entomol. 2020, 23, 1048–1054. [Google Scholar] [CrossRef]
- Wang, C.J.; Zeng, Z.L.; Zhang, F.S.; Guo, S.G. Clinical features of adult anti-N-methyl-d-aspartate receptor encephalitis after Japanese encephalitis. J. Neurol. Sci. 2020, 417, 117080. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Ni, H.; Beasley, D.; Ekkelenkamp, M.; Cardosa, M.; Barrett, A. Origin and evolution of Japanese encephalitis virus in southeast Asia. J. Virol. 2003, 77, 3091–3098. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, H.S.; Monteiro, M.T.T.; Torres, D.F.M. Seasonality effects on dengue: Basic reproduction number, sensitivity analysis and optimal control. Math. Methods Appl. Sci. 2016, 39, 4671–4679. [Google Scholar] [CrossRef]
- Ndaïrou, F.; Area, I.; Nieto, J.J.; Torres, D.F.M. Mathematical modeling of COVID-19 transmission dynamics with a case study of Wuhan. Chaos Solitons Fractals 2020, 135, 109846. [Google Scholar] [CrossRef] [PubMed]
- Lemos-Paião, A.P.; Silva, C.J.; Torres, D.F.M.; Venturino, E. Optimal control of aquatic diseases: A case study of Yemen’s cholera outbreak. J. Optim. Theory Appl. 2020, 185, 1008–1030. [Google Scholar] [CrossRef]
- Naresh, R.; Pandey, S. Modeling and analysis of the spread of Japanese encephalitis with environmental effects. Appl. Appl. Math. 2009, 4, 155–175. [Google Scholar]
- Agarwal, M.; Verma, V. The impact of media on the spreading and control of Japanese encephalitis. Int. J. Math. Sci. Comput. 2012, 2, 23–31. [Google Scholar]
- Panja, P.; Mondal, S.K.; Chattopadhyay, J. Stability and bifurcation analysis of Japanese encephalitis model with/without effects of some control parameters. Comput. Appl. Math. 2018, 37, 1330–1351. [Google Scholar] [CrossRef]
- Zhao, S.; Lou, Y.; Chiu, A.P.Y.; He, D. Modelling the skip-and-resurgence of Japanese encephalitis epidemics in Hong Kong. J. Theoret. Biol. 2018, 454, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, D. Final size distributions for epidemics. Math. Biosci. 1975, 23, 33–46. [Google Scholar] [CrossRef]
- Purdom, P.W. Environmental Health; Elsevier: New York, NY, USA, 2013. [Google Scholar]
- Ghosh, M.; Chandra, P.; Sinha, P.; Shukla, J.B. Modelling the spread of carrier-dependent infectious diseases with environmental effect. Appl. Math. Comput. 2004, 152, 385–402. [Google Scholar] [CrossRef]
- Ghosh, M.; Chandra, P.; Sinha, P.; Shukla, J.B. Modelling the spread of bacterial infectious disease with environmental effect in a logistically growing human population. Nonlinear Anal. Real World Appl. 2006, 7, 341–363. [Google Scholar] [CrossRef]
- Berge, T.; Bowong, S.; Lubuma, J.M.S. Global stability of a two-patch cholera model with fast and slow transmissions. Math. Comput. Simul. 2017, 133, 142–164. [Google Scholar] [CrossRef]
- Berge, T.; Lubuma, J.M.S.; Moremedi, G.M.; Morris, N.; Kondera-Shava, R. A simple mathematical model for Ebola in Africa. J. Biol. Dyn. 2017, 11, 42–74. [Google Scholar] [CrossRef] [PubMed]
- Codeço, C.T. Endemic and epidemic dynamic of cholera: The role of the aquatic reservoir. BMC Infect. Dis. 2001, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- van den Driessche, P.; Watmough, J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 2002, 180, 29–48. [Google Scholar] [CrossRef]
- Strogatz, S.H. Nonlinear Dynamics and Chaos: With Applications to Physics, Biology, Chemistry, and Engineering; Addison-Wesley Pub.: Reading, MA, USA, 1994. [Google Scholar]
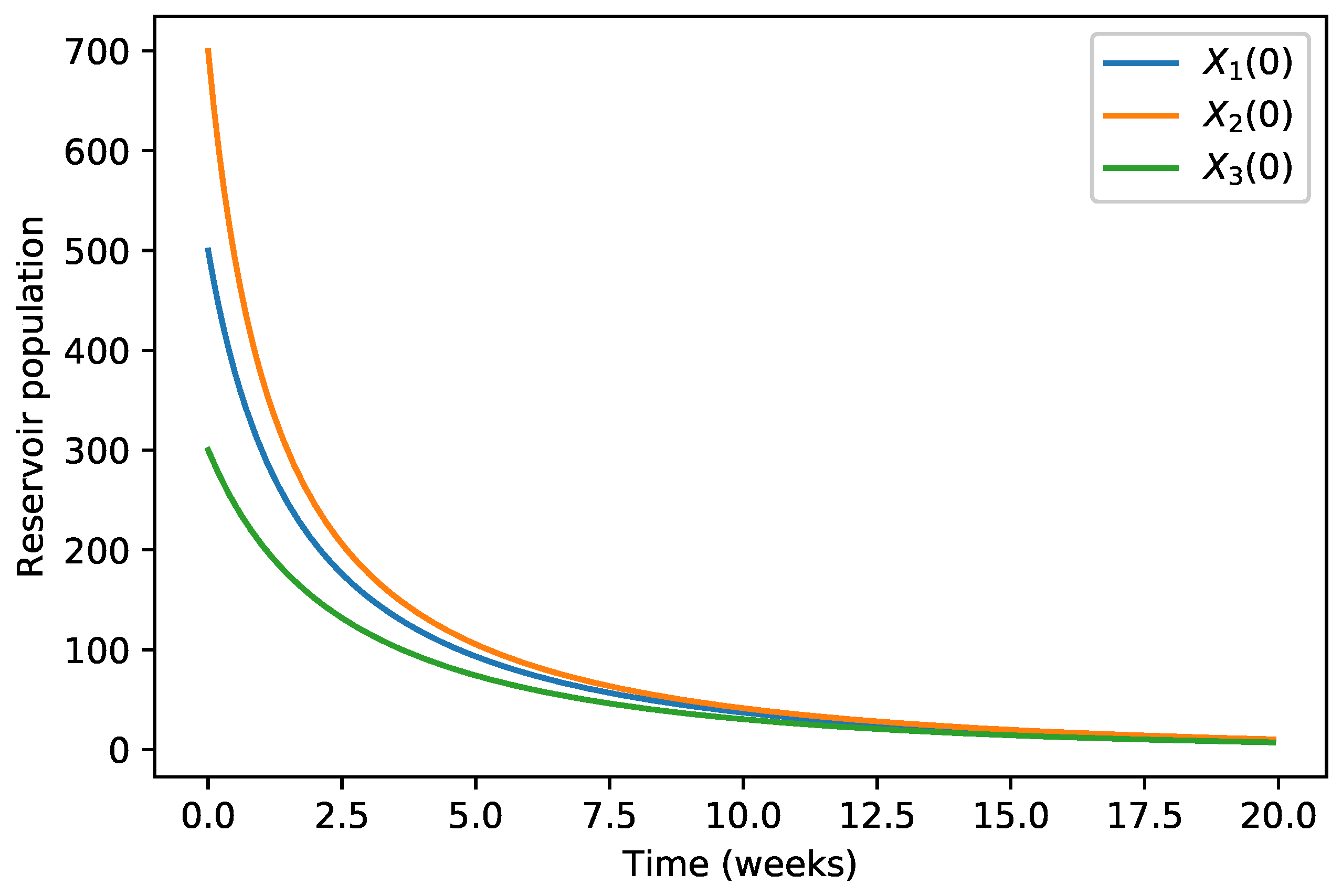

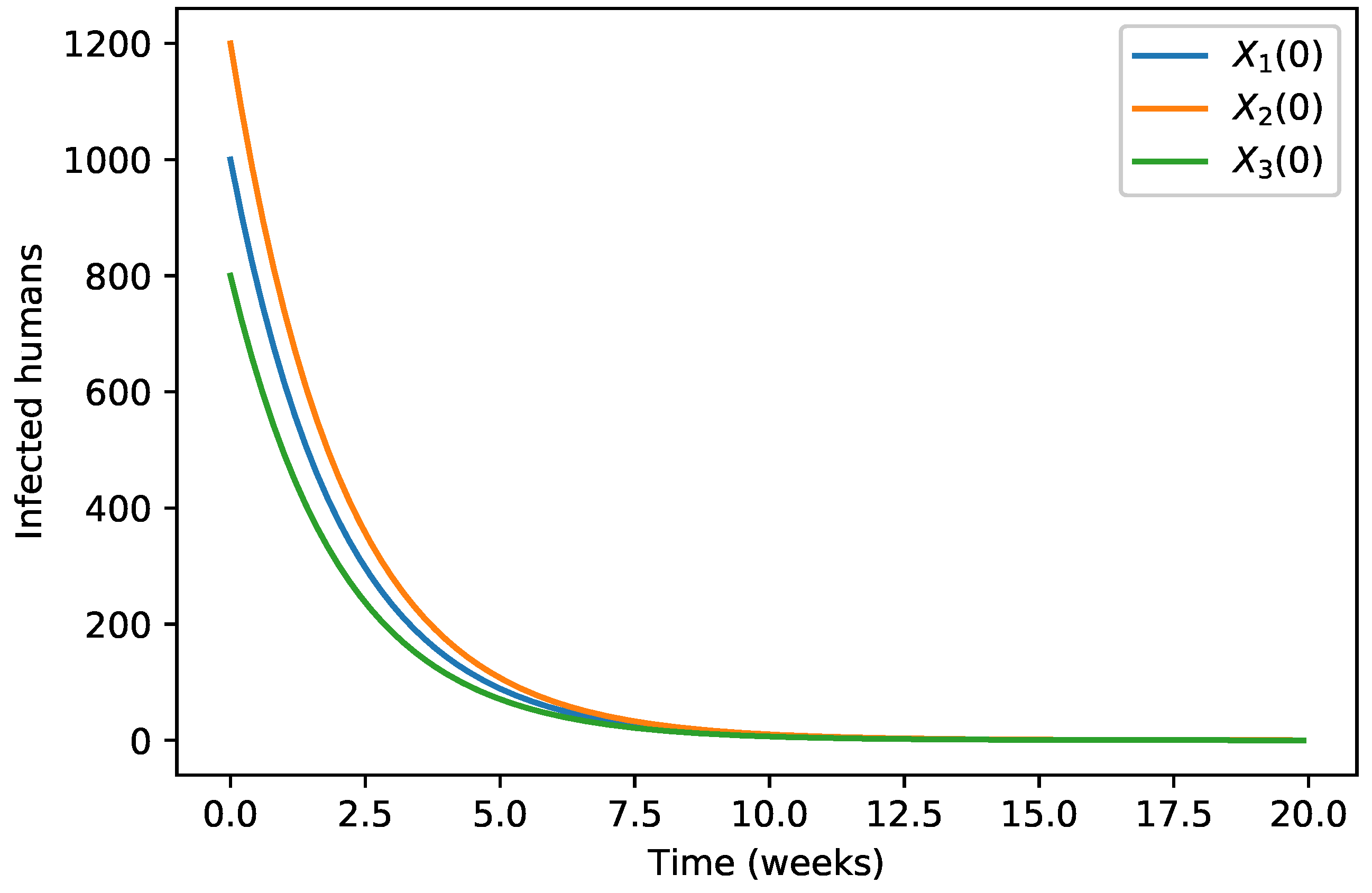

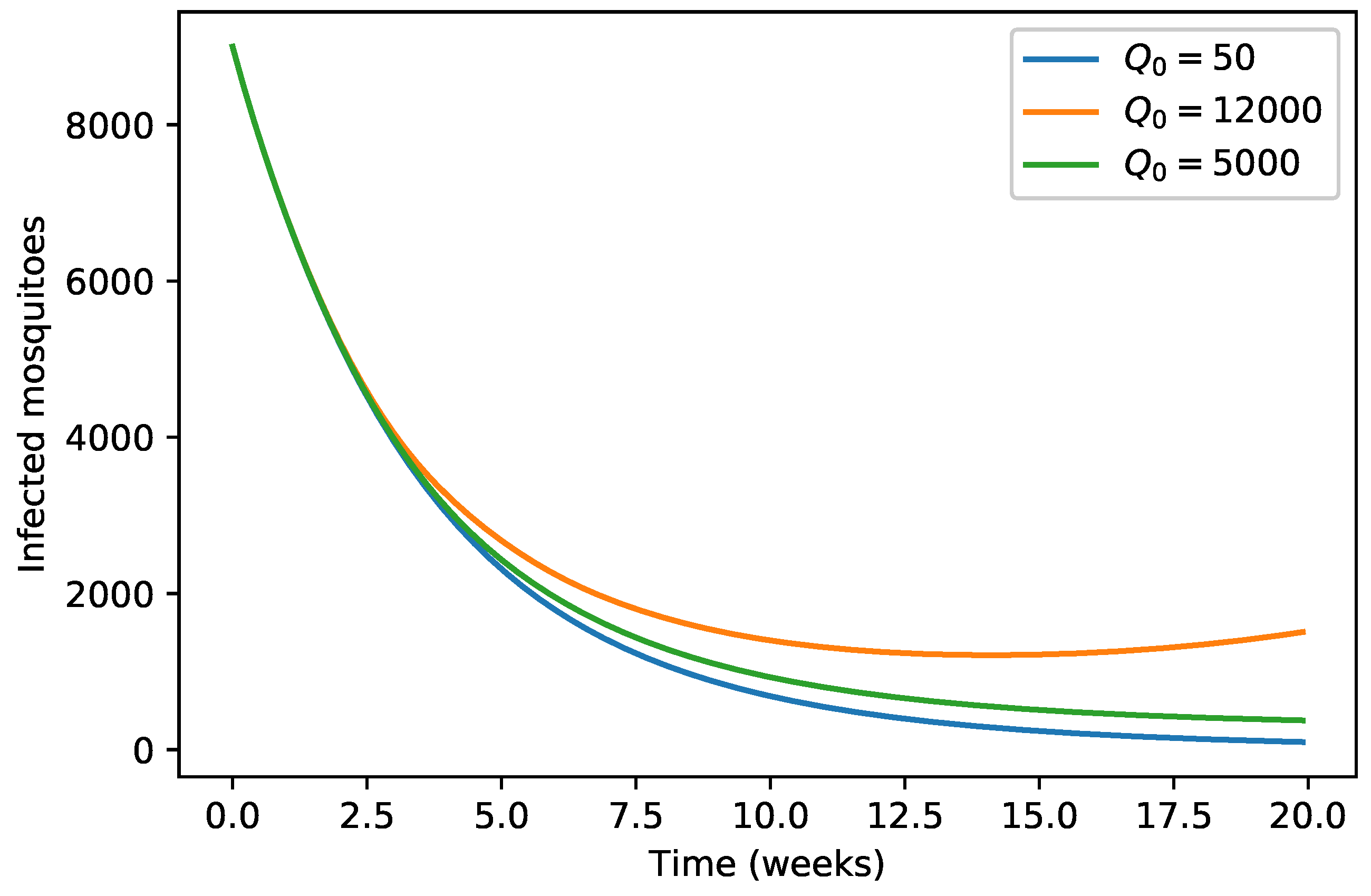
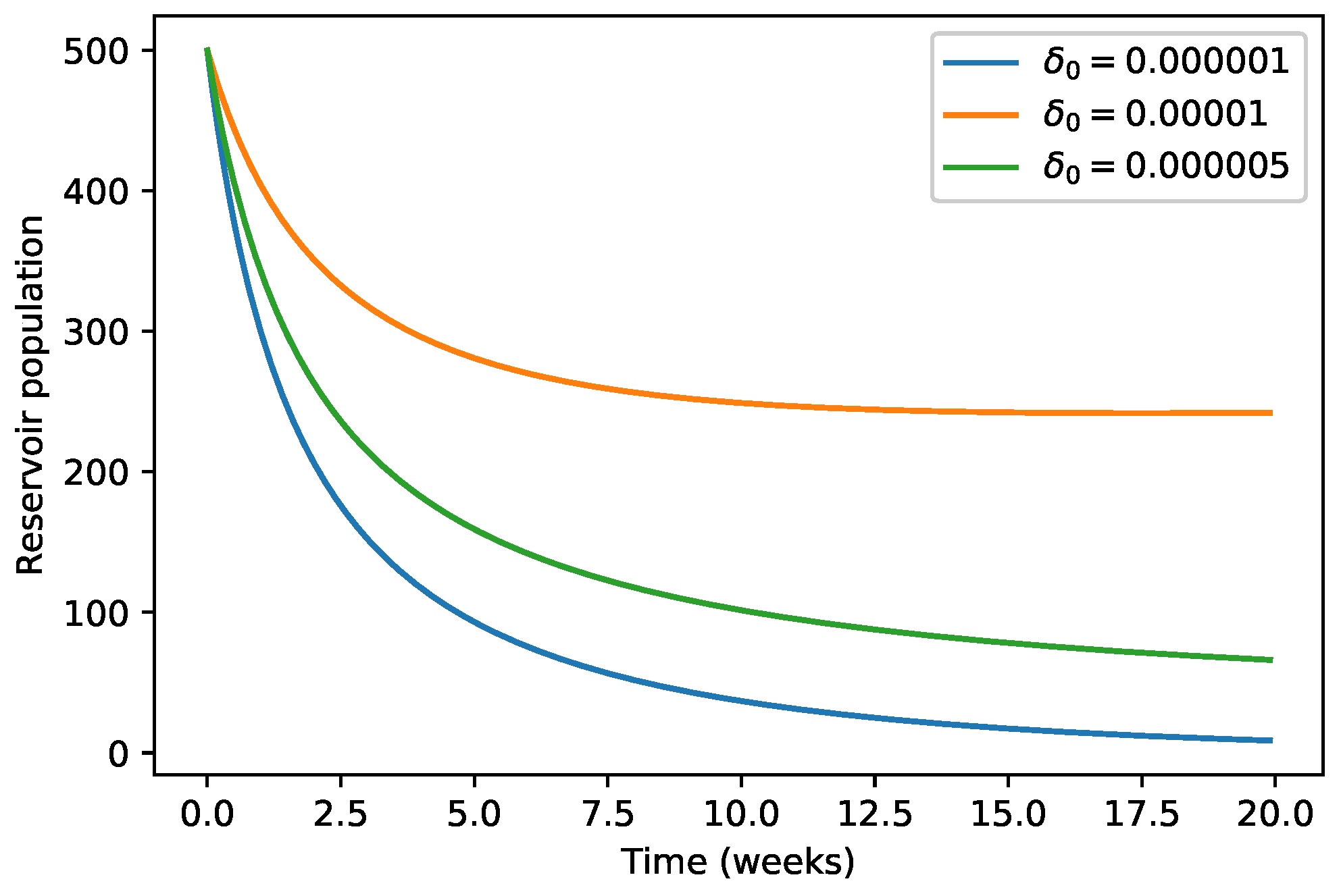
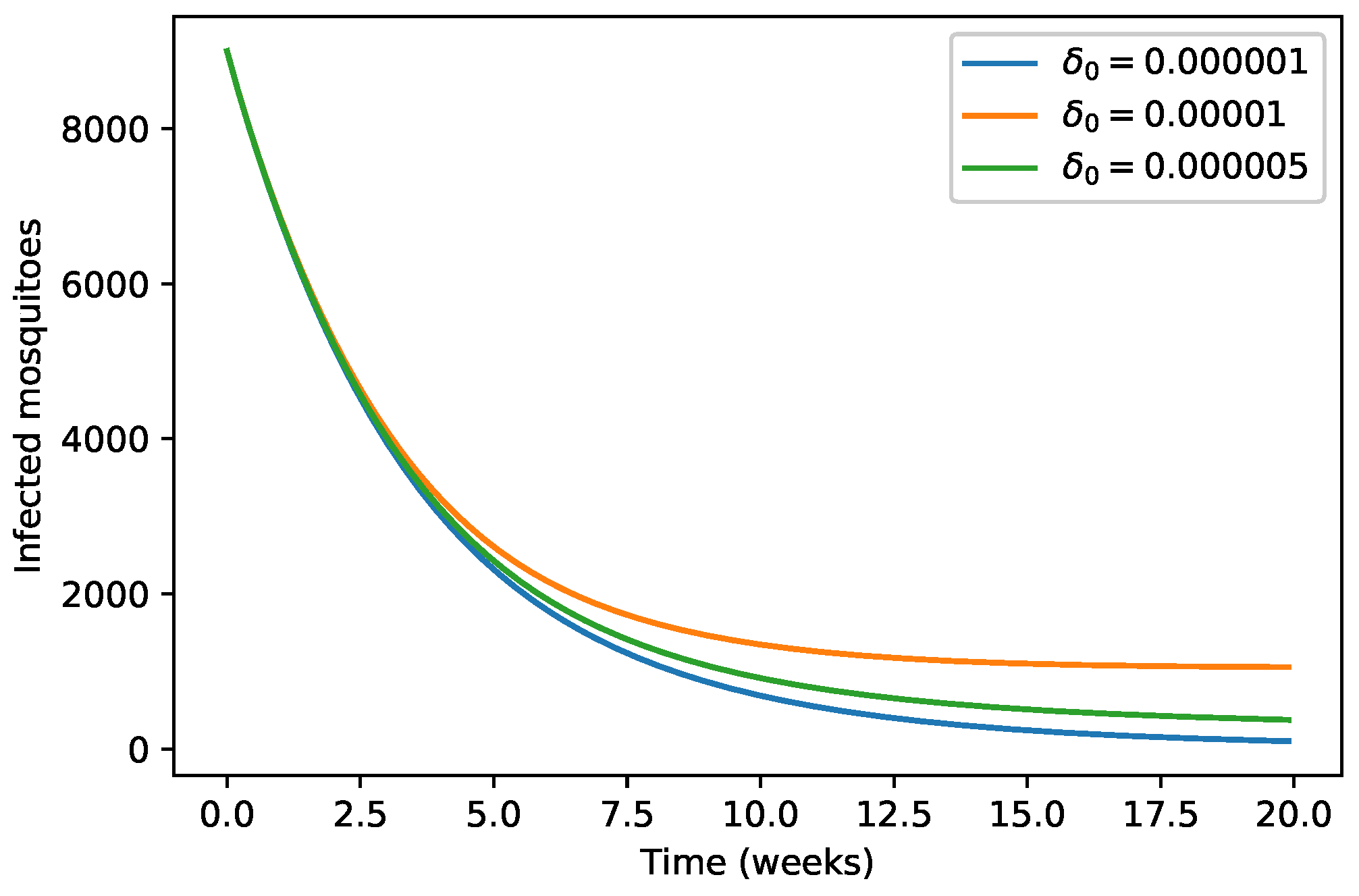
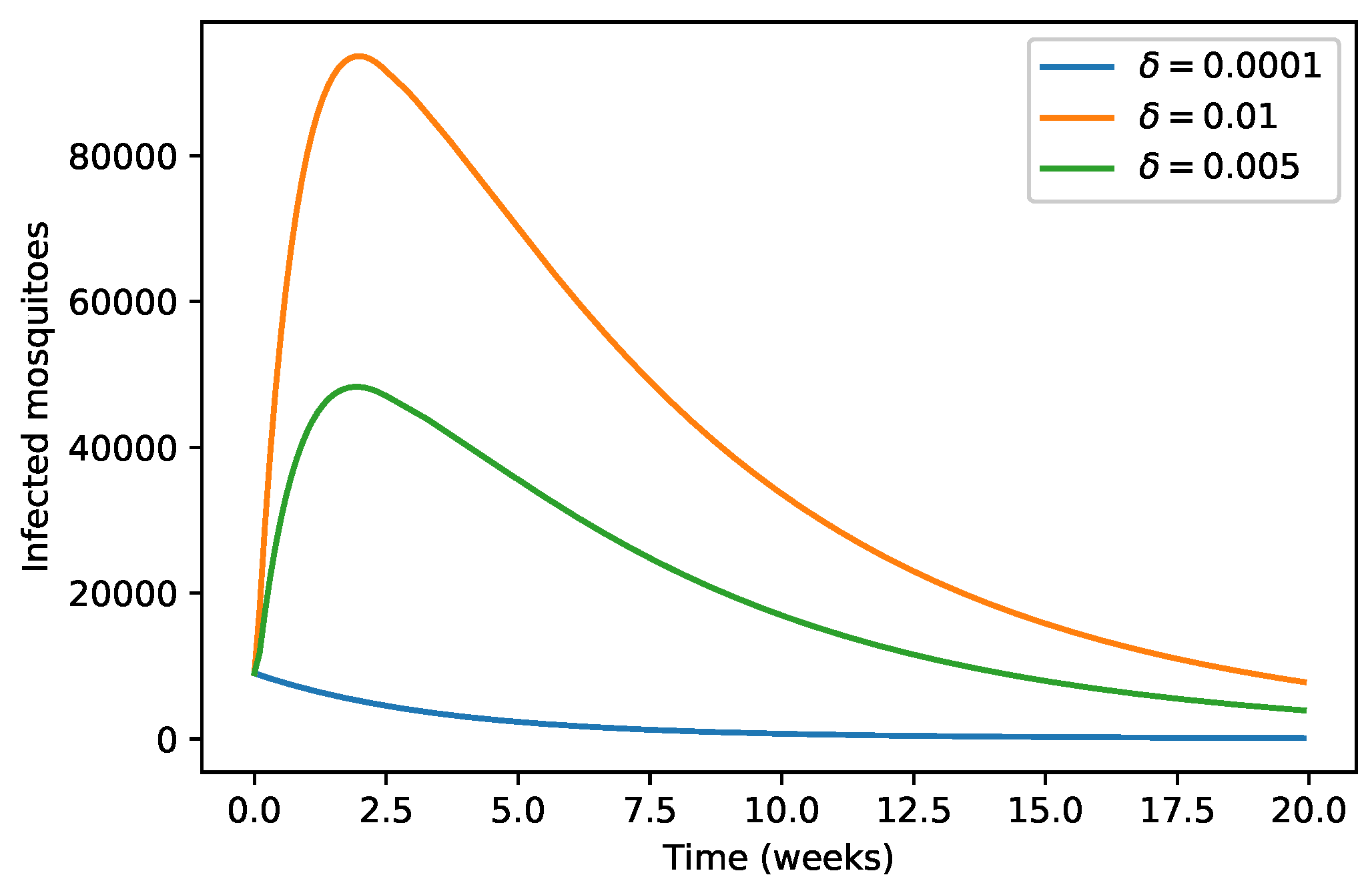
| 40,000 | 500 | 12,000 | 10,000 | 9000 | 7000 | 1000 | |
| 45,000 | 700 | 15,000 | 12,000 | 11,000 | 10,000 | 12,000 | |
| 35,000 | 300 | 10,000 | 7000 | 6000 | 5000 | 800 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ndaïrou, F.; Area, I.; Torres, D.F.M. Mathematical Modeling of Japanese Encephalitis under Aquatic Environmental Effects. Mathematics 2020, 8, 1880. https://doi.org/10.3390/math8111880
Ndaïrou F, Area I, Torres DFM. Mathematical Modeling of Japanese Encephalitis under Aquatic Environmental Effects. Mathematics. 2020; 8(11):1880. https://doi.org/10.3390/math8111880
Chicago/Turabian StyleNdaïrou, Faïçal, Iván Area, and Delfim F. M. Torres. 2020. "Mathematical Modeling of Japanese Encephalitis under Aquatic Environmental Effects" Mathematics 8, no. 11: 1880. https://doi.org/10.3390/math8111880
APA StyleNdaïrou, F., Area, I., & Torres, D. F. M. (2020). Mathematical Modeling of Japanese Encephalitis under Aquatic Environmental Effects. Mathematics, 8(11), 1880. https://doi.org/10.3390/math8111880







